Abstract
The Drosophila wing imaginal disc is subdivided along the proximodistal axis into the distal pouch, the hinge, the surrounding pleura, and the notum. While the genetic pathways that specify the identity of each of these domains have been well studied, the mechanisms that coordinate the relative expansion of these domains are not well understood. Here we investigated the role of the stat92E signal transducer and activator of transcription in wing proximodistal development. We find that Stat92E is active ubiquitously in early wing imaginal discs, where it acts to inhibit the induction of ectopic wing fields. Subsequently, Stat92E activity is down regulated in the notum and distal pouch. These dynamics coincide with and contribute to the proportional subdivision and expansion of these primordia. As development proceeds, Stat92E activity becomes restricted to the hinge, where it promotes normal expansion of the hinge, and restricts expansion of the notum. We also find that stat92E is required autonomously to specify dorsal pleura identity and inhibit notum identity to properly subdivide the body wall. Our data suggest that Stat92E activity is regulated along the proximodistal axis to pattern this axis and control the relative expansion of the pouch, hinge, and notum.
Keywords: Notum, pleura, hinge, pouch, Upd, Bowl, Eyg, Mirr, Zfh2
Introduction
The subdivision of the proximodistal (PD) axis of developing appendages is critical for their physiological roles in locomotion, sensing, feeding and reproduction. The elaboration of appendage structure depends on signals that emanate from localized sources termed organizers (Lawrence et al., 1996; Mann and Morata, 2000). These signals subdivide nascent appendages into progressively smaller domains along their anteroposterior (AP), dorsoventral (DV) and PD axes. The PD axis of the wing imaginal disc is subdivided into the distal pouch that forms the wing blade, the hinge that connects the blade to the body wall, the pleura that surrounds the hinge and forms the lateral plate of the body wall, and the notum that forms part of the dorsal body wall (Fig. 1A-C). While the genetic pathways that specify the identity of these domains have been well studied, the mechanisms that control the expansion of each of these domains remain unclear.
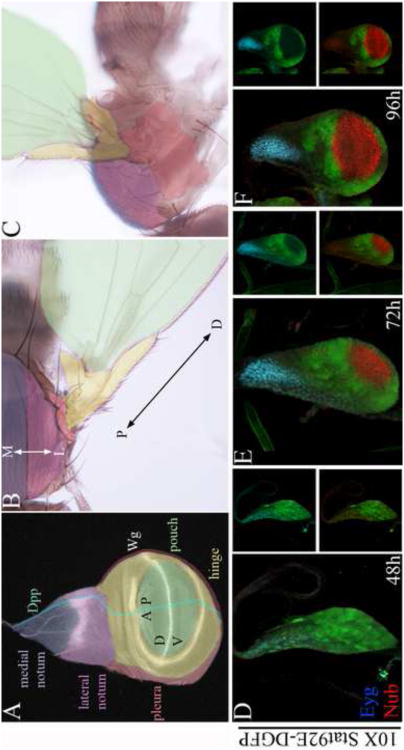
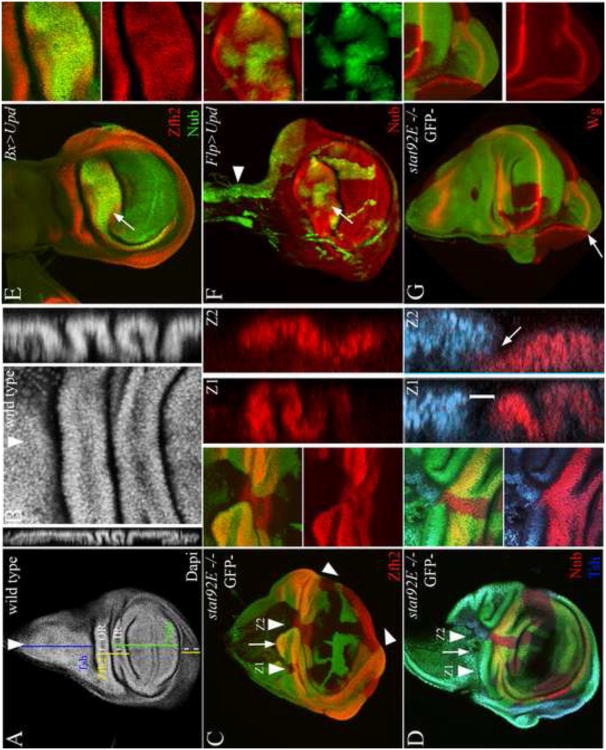
Figure 1. Downregulation of Stat92E activity coincides with the expansion of the pouch and notum.
(A-C) Schematics of third in star wing disc (A) and its derivatives in adult flies shown in dorsal (B) and lateral (C) orientations. The third in star wing disc (A) is subdivided along the PD axis into the distal pouch (green), the intermediate hinge (yellow),the pleura (red), and the proximal notum (lateral notum in mauve, and medial notum in purple). wg expression (white) defines the border between the medial and lateral notum, the hinge, and the DV compartment boundary in the wing field. dpp (cyan) is expressed in the anterior compartment along the AP compartment boundary. (B-C) The notum and presumptive pleura give rise to dorsal and lateral parts of the adult mesothorax, respectively, while the wing field gives rise to the wing hinge and blade. The notum mediolateral (ML) and wing proximodistal (PD) axes are indicated by double arrows in B. (D-F) 10× Stat92E-DGFP (green), Nub (red), Eyg (cyan). Downregulation of Stat92E activity in the presumptive notum and pouch coincided with the expansion of these primordial marked with Eyg and Nub, respectively.
The PD axis of the wing imaginal disc is initially patterned by two opposing signals, Wingless (Wg) and epidermal growth factor (EGF), which specify the wing field and body wall, respectively. wg induces the expression of the zinc finger genes elbow (el) and no ocelli (noc) (el/noc) in the early wing primordium. In turn, el/noc represses the expression of the zinc finger gene teashirt (tsh) in distal cells (Weihe et al., 2004; Wu and Cohen, 2002) to specify wing identity and to inhibit body wall identity (Azpiazu and Morata, 2000; Casares and Mann, 2000). In the notum, the neuregulin-like signal Vein (Vn) activates epidermal growth factor (EGF) receptor signaling to induce expression of the Iroquois Complex (iro-C) homeobox genes to specify notum fate and inhibit wing fate (Simcox et al., 1996; Wang et al., 2000; Zecca and Struhl, 2002a; Zecca and Struhl, 2002b). Each of these domains is then progressively subdivided into smaller PD sub domains by the activities of secreted signals and transcription factors.
The elaboration of the wing PD axis depends on signaling centers that are established along both the DV and AP compartment boundaries. Activation of Notch (N) signaling along the DV compartment boundary induces the expression of the wing selector gene vestigial (vg) in a narrow domain that centers on this boundary (Couso et al., 1995; Kim et al., 1996; Klein and Arias, 1998; Williams et al., 1994). Activation of vg by both the Wg signal and the Bone Morphogenetic Protein (BMP)-like signal Decapentaplegic (Dpp) further expands the range of vg function (Zecca and Struhl, 2007a; Zecca and Struhl, 2007b). Within the wing field, vg activates a set of genes required for the elaboration of the wing PD axis in nested circular domains (Kolzer et al., 2003; Ng et al., 1995; St Pierre et al., 2002; Terriente et al., 2007; Terriente et al., 2008). Different combinations of the wing PD genes progressively subdivide the wing field, from distal to proximal, into the pouch, the distal hinge and the proximal hinge (Cho and Irvine, 2004; Jakobi et al., 1993; Kolzer et al., 2003; Terriente et al., 2008; Dichtel-Danjoy et al., 2009; Perea et al., 2009; Rodriguez del Alamo et al., 2002; Terriente et al., 2007).
The notum is also subdivided into lateral and medial domains, which can be viewed as the most proximal subdivisions of the wing PD axis (Fig. 1A-C). We refer to this axis as the notum mediolateral (ML) axis, and refer to the entire axis spanning both the wing and notum as the wing PD/ML axis. Signaling centers that are established along the notum margins elaborate both the notum ML and AP axes. The Dpp signal is distributed in a medial to lateral gradient at early stages and organizes the notum ML axis. dpp promotes expression of the GATA and FoG genes pannier (pnr) and u-shaped (ush) in the medial notum, and limits the expression of the iro-C genes to the lateral notum (Fromental-Ramain et al., 2008; García-García et al., 1999; Letizia et al., 2007). Wg is induced in the lateral notum along the interface with the medial notum (Sato and Saigo, 2000; Tomoyasu et al., 2000) and is required to control cell fate in this region (García-García et al., 1999). As the pathways that specify and subdivide the wing and notum have been well characterized, we explored the mechanisms that control the relative expansion of these primordia.
The JAK/STAT pathway controls numerous developmental processes including the patterning, growth and morphogenesis of epithelial sheets during embryonic, larval and adult life (Arbouzova and Zeidler, 2006; Hombria and Brown, 2002; Hombria and Sotillos, 2008). Canonical JAK/STAT signaling is initiated by the binding of the extracellular ligand Unpaired (Upd) genes (Upd1-3) to the transmembrane Domeless (Dome) receptor. This binding activates the receptor associated Janus kinase (JAK) family member hopscotch (hop). Activated JAKs phosphorylate themselves and associated receptors to generate docking sites for the signal transducer Stat92E. Following recruitment and phosphorylation by the receptor-JAK complex, Stat92E dimers assemble and translocate to the nucleus. Nuclear Stat92E dimers bind conserved sequences in downstream target genes to regulate gene expression (Levy and Darnell, 2002). Unphosphorylated Stat92E can translocate to the nucleus and associate with chromatin remodeling factors to stabilize transcriptionally repressed heterochromatin independently of canonical JAK/STAT signaling. Receptor activation of the JAK/STAT pathway reduces the association of Stat92E with chromatin causing global chromatin instability and changes in gene expression (Li, 2008; Shi et al., 2006; Shi et al., 2008).
To test the role of stat92E in the elaboration of the wing PD/ML axis, we examined the dynamics of Stat92E activity and the requirements for stat92E in wing development from early stages of development using genetic loss- and gain-of-function analyses. We find that Stat92E is active ubiquitously at early stages and is then downregulated in a medial to lateral direction in the notum, and in a distal to proximal direction in the pouch. We provide evidence that the dynamics of stat92E downregulation controls the relative expansion of gene expression domains along the wing PD/ML axis. We also show that the early ubiquitous activity of Stat92E is required to inhibit ectopic wing induction in ectopic locations, while the later restriction of Stat92E activity to the hinge and pleura is required to promote the expansion of the hinge and the specification of the dorsal pleura. Together, these results suggest novel roles for stat92E in patterning and coordinating the expansion of the various subdivisions of the wing PD/ML axis.
Results
Downregulation of Stat92E activity coincides with the expansion of the presumptive wing pouch and notum
To understand the role of the JAK/STAT pathway in wing PD axis development, we compared the evolution of Stat92E activity to the gene expression patterns of the PD genes Nubbin (Nub) and Eyegone (Eyg). Nub marks the wing pouch and dorsal hinge (Ng et al., 1995), while Eyg marks a broad anterior part of the notum (Aldaz et al., 2003). To monitor canonical JAK/STAT signaling activity, we utilized a previously characterized reporter composed of 10 tandem Stat92E DNA binding sites inserted upstream of a minimal promoter and a GFP reporter (10× Stat92E-GFP) (Bach et al., 2007). We found that at second in star, Stat92E was active throughout the disc proper of the wing disc (Fig. 1D). By early third in star, Stat92E activity was repressed in the distal pouch and proximal notum, coincident with the induction of Nub and Eyg in these primordia, respectively (Fig. 1E-F). Analysis of Stat92E protein distribution at third in star revealed nuclear accumulation in the wing hinge, pleura and the anterior border of the notum, and cytoplasmic accumulation in the wing and notum (Sup. Fig. 1). Though slightly more restricted, we detected a similar pattern of expression of the Stat92E activating ligand upd, using Upd-GAL4 driving GFP (Sup. Fig. 2). To confirm these dynamics, we performed a lineage analysis of upd-expressing cells, using the G-TRACE system (Evans et al., 2009). We found that most wing disc cells originated from upd-GAL4 expressing cells except for a small cell population near the disc stalk (Sup. Fig. 3A). This cell population expanded at later stages to occupy most of the medial notum and part of the lateral notum (Sup. Fig. 3B). These dynamics suggested roles for stat92E in the elaboration of the wing PD axis.
stat92E suppresses the induction and elaboration of the wing PD axis during early stages of wing development
To test whether Stat92E activity plays a functional role in wing PD development, we analyzed the clonal phenotypes of the strong stat92E85C9 mutant allele. Because the stat92E mutant clones grow poorly in a wild type background (Mukherjee et al., 2005), we generated the clones in a Minute background to provide them with a growth advantage (Garcia-Bellido et al., 1973; Morata and Ripoll, 1975). We found that a subset of the clones induced at 48-72h AEL in a Minute background (equivalent to 34-58h AEL in a wild type background) led to the formation of ectopic wings from the posterior part of the notum in adult flies (Sup. Fig. 6B). Analysis of third in star imaginal discs with molecular markers revealed induction of ectopic wing fields from the posterior region of the notum (Fig. 2). Ectopic wing fields were positively marked by the expression of Nub, Wg and the T-box protein Dorsocross2 (Doc2), (Fig. 2B, D and F, respectively). Clones associated with ectopic wing fields were often small and localized to the disc proper near the disc stalk (Fig. 2B and 2F) and/or the posterior disc margins (Fig. 2D). Wg and Doc2 were expressed in both the pouch and hinge of ectopic wings, indicating that the PD axis was elaborated within ectopic wing fields. Interestingly, the ectopic wings did not substitute for the native notum, as revealed by the expression of both Wg and the Iro-C protein Mirror (Mirr) and Doc2 in the notum (asterisks in Fig. 2D and 2F, respectively). The induction of the notum and wing is dependent on inductive interactions across the AP compartment boundary, which in the notum localizes to the posterior margin (Klein and Arias, 1998; Ng et al., 1996; Williams et al., 1993). Our data suggest that stat92E inhibits the competence of this region of the notum to respond to this wing field-inducing signal from the posterior compartment. Ectopic wings were composed of mostly wild type cells suggesting that following wing induction non-wing cells were recruited to the wing field using a non-autonomous mechanism.
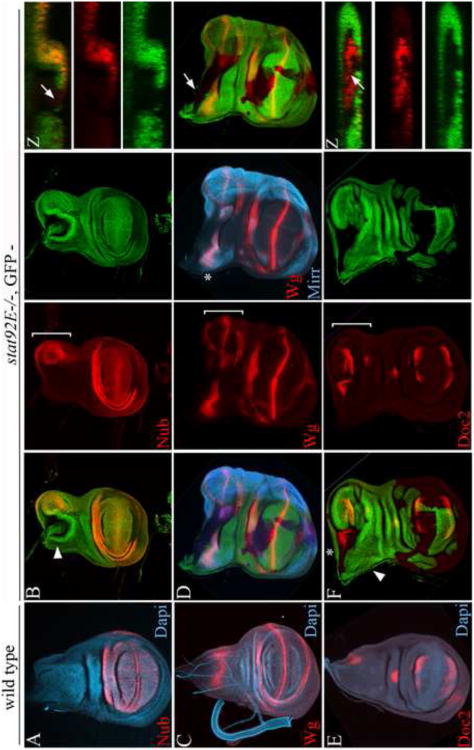
Figure 2. stat92E inhibits the induction of ectopic wing fields.
(A, C, E) Wild type wing discs, and (B, D, F) wing discs bearing negatively marked stat92E mutant clones generated at first to second in star, and stained for Nub (A-B),Wg (C), both Wg and Mirr (D), and Doc2 (E-F). Arrowheads in B and F point to planes of Z sections shown in corresponding insets. (A) Nub is expressed in the pouch and distal hinge. (C) Wg is expressed along the DV compartment boundary in the pouch, in two concentric rings in the hinge and at an intermediate position in the notum. Lateral notum is marked with Mirr in D. (E) Doc2 is expressed in dorsal and ventral rims in the pouch, in a spot in the central region of the dorsal hinge, and in a narrow domain along the posterior notum margin. (B, D, F) Early stat92E mutant clones that were generated in the disc proper near the disc stalk led to the induction of ectopic wing fields in the posterior part of the notum (demarcated by brackets in red channel). Ectopic wings were marked by the stereotypic expression of Nub, Wg and Doc2. Note that the notum specific expression of Mirr and Wg (asterisk in D), as well as Doc2 (asterisk in F) persisted in discs baring ectopic wings fields.
To confirm these findings, we analyzed the stat92Ej6C8 and stat92E397 alleles by clonal analysis. We found that these alleles produce partially overlapping phenotypes with the stronger phenotypes produced by the stat92E85C9 and stat92E397 alleles and the weaker phenotypes by the stat92Ej6C8 allele (Sup. Fig. 4, Sup. Tables 1-2 and Materials and Methods). Additionally, we were able to rescue the stat92E85C9 mutant clone phenotypes using a genomic DNA that spans the stat92E locus indicating that the clonal phenotypes resulted from the loss of stat92E and not from an associated mutation (Sup. Fig. 5 and Materials and Methods). For the remainder of analyses, we continued to focus on the stat92E85C9 allele.
Upd is sufficient to inhibit the expansion of the wing and notum
The formation and expansion of the pouch and notum coincided with the downregulation of Stat92E activity in these regions (Fig. 1), and the early loss of stat92E function resulted in induction of ectopic wing fields (Fig. 2). These observations suggested that stat92E suppresses that formation of the wing and notum at early stages. To determine whether Stat92E activity was sufficient to inhibit the specification of the wing and/or notum, we broadly expressed the JAK/STAT activating ligand upd from early stages with the broadly expressed Esg-GAL4 driver. We found that in severe cases, the Esg>Upd discs formed tiny rudiments smaller in size than the leg discs (Fig. 3B-C, E, H). In less severe cases the wing remained larger than the leg, but the morphologies of the wing and notum were impaired (Fig. 3F, I). We examined the expression of Nub and Zfh2 as well as Mirr and Eyg to determine if the wing field and the notum were properly patterned in these discs. Zfh2 is expressed broadly in the early wing field. At later stages Zfh2 becomes restricted to the hinge (Fig. 3A) as expression of pouch-specific genes, such as Nub, are induced in distal cells (Terriente et al., 2008; Wu and Cohen, 2002). We found that Nub was expressed in distal cells in these rudiments as in wild type discs (data not shown). However, Zfh2 was expressed more broadly across the wing field (Fig. 3B-C compared to wild type in 3A) and failed to be properly restricted to the hinge. Thus, constitutive upd expression in the pouch disrupted the elaboration of the wing PD axis.
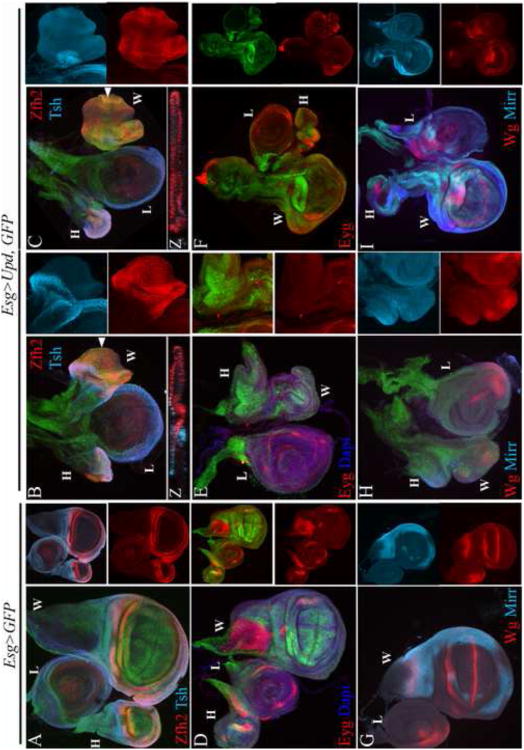
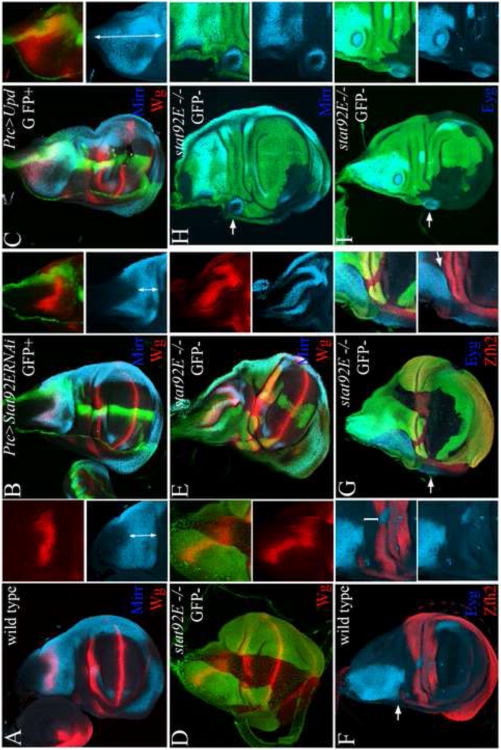
Figure 3. The Stat92E activating signal upd is sufficient to suppress the elaboration of the wing PD axis and notum ML axis.
(A, D, G) Wild type; (A) Zfh2 highlights the hinge and Tsh the surrounding body wall; (D) Eyg highlights a broad part of the anterior notum; (G) Mirr highlights the lateral notum and Wg an intermediate position along the notum ML axis, as well as the hinge and DV compartment boundary in the wing field. (B-C, E-F, H-I) Esg-GAL4/UAS-Upd; UAS-GFP (Esg>Upd, GFP). Broad and constitutive expression of upd in the wing field led to severe (middle column: B, E, H, right column: C) to moderate (right column: F, I) reduction in wing size and defects in wing and notum patterning. Broad upd expression prevented the proper restriction of the Zfh2 domain to the hinge (B-C, and corresponding Z sections). In addition, broad upd expression led to loss or a severe reduction of Eyg (E-F) as well as (H-I) Mirr and Wg expression in the presumptive notum. H-haltere, L-leg, W-Wing.
We detected a similar disruption of ML axis elaboration in the notum. The Pax protein Eyg marks a broad anterior part of the notum, while Mirr marks the lateral part (Fig. 3D and G, respectively) (Aldaz et al., 2003; Kehl et al., 1998). We found that Eyg expression was lost in Esg>Upd rudiments in both severely and moderately affected wing discs (Fig. 3E-F compared to wild type in Fig. 3D). Esg>GFP expression is excluded from the Eyg domain of late third instar wing discs in the notum (Fig. 3D). However, it was not excluded from the Esg>Upd rudiments, and Eyg was not expressed in these rudiments (Fig. 3E-F). Likewise, neither Wg nor Mirr were expressed in these discs suggesting that upd expression prevented the elaboration of the intermediate and lateral subdivisions of the notum (Fig. 3H-I compare to wild type in Fig. 3G). Thus, in both the pouch and the notum, suppression of Stat92E activity is required for the elaboration of the PD and ML axes, respectively.
stat92E restricts the scope of the anterior border of the notum
The dynamics of Stat92E activity and the upd gain-of-function phenotypes suggested additional roles for stat92E in the patterning and expansion of the wing and notum. To test this, we generated stat92E mutant clones at later stages of development and examined the clonal phenotypes in adult flies and developing imaginal discs. We found that stat92E mutant clones generated at 72-96 hours AEL in a Minute background (equivalent to 51-74 hours AEL in a wild type background) caused severe defects in dorsal closure and cuticle differentiation along the dorsal midline (Sup. Fig. 6C). The anterior and posterior margins of the notum contribute to the closure of the two hemi-not a along the dorsal midline, suggesting that stat92E affects the specification, subdivision or expansion of these regions of the wing disc. The Odd-skipped protein Bowl is expressed along the anterior border of the notum, adjacent to cells expressing the Notch signal Delta (Dl) (Fig 4A), where it contributes to dorsal closure (DelSignore et al., 2012). We therefore asked whether stat92E mutant clones affect the expression of the Odd-skipped protein Bowl in the notum.
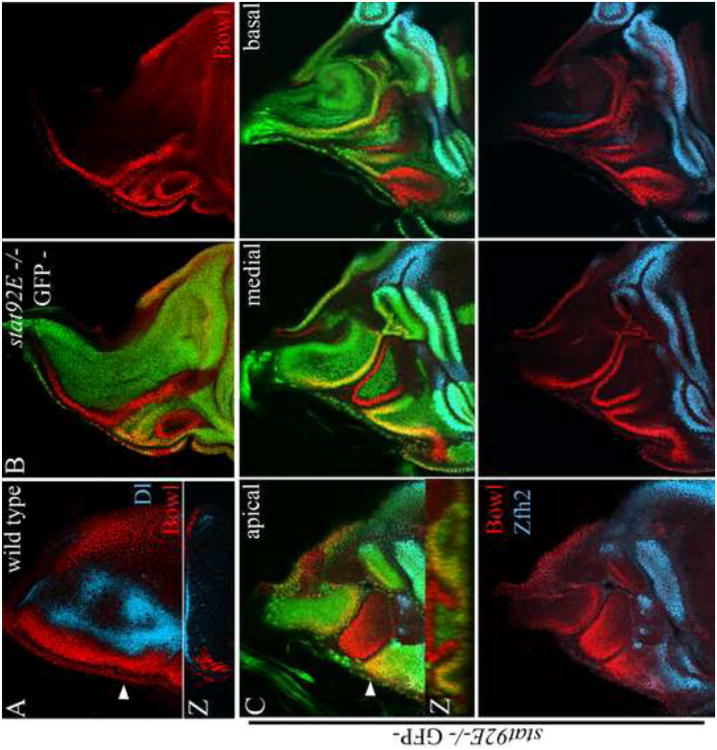
Figure 4. stat92E inhibits the expansion of the anterior border of the notum.
(A) Wild type; Bowl (red) accumulates along the anterior border of the notum adjacent to Dl expressing cells (cyan). (B-C) Negatively marked stat92E mutant clones; Bowl (red), Zfh2 (cyan). In stat92E mutant clones that spanned the anterior border of the notum, the Bowl domain expanded posteriorly toward the central part of the notum and laterally toward the hinge. Bowl was also expressed in wild type cells along clone borders (yellow). Images in C are optical sections at the apical, medial and basal planes of a single wing disc. Arrowheads in A, C point to plane of Z-sections shown in corresponding insets.
Mutant clones generated at 72-96h caused invaginations between the mutant clones and adjacent wild type cells in developing wing imaginal discs (Figs. 4-6). Clones that were generated in the Bowl domain protruded into the central part of the notum and expanded laterally toward the hinge (Fig. 4B-C). Bowl accumulated in these clones and in wild type cells along clone borders. We speculate that the non-autonomous induction of Bowl was mediated by the induction of the related family member drumstick (drm) in the clones, which has been shown to promote Bowl accumulation both autonomously and non-autonomously in the notum (DelSignore et al., 2012). Clones that spanned the posterior notum margin protruded only weakly into the central part of the notum. These results indicate that stat92E inhibits the expansion of the anterior border of the notum.
Figure 6. stat92E promotes the expansion and fine subdivision of the hinge.
(A-B) Wild type wing disc stained with Dapi to highlight the folded appearance of the mature dorsal hinge. Arrowheads indicate planes of Z sections shown in corresponding insets. The Nub, Zfh2, Wg and Tsh expression domains are indicated schematically in A. (C) The scope of the dorsal hinge was reduced in stat92E mutant clones. Corresponding Z-sections reveal a reduced folding of the Zfh2 domain in mutant clones (Z2) compared to wild type tissue (Z1). Also note the thinning of the ventral hinge in a stat92E mutant clone (demarcated with opposing arrowheads). (D) An intervening domain intercalates between the Nub and Tsh domains in wild type tissue (demarcated with a bracket in Z1). This intervening domain was missing in stat92E mutant clones (arrow in Z2 shows absence of the intervening domain in stat92E mutant clone). (E) Expression of upd with Bx-GAL4 caused expansion of the distal part of the dorsal hinge (arrow). (F) Likewise, ectopic expression of upd in FLP-out clones caused expansion of the dorsal hinge (arrow). However, upd clones that were generated in the notum severely perturbed notum size and morphology (arrowhead). (G) Wg expression in the hinge was maintained in the stat92E mutant clones. The contour of Wg expression in patches of wild type cells was displaced proximally (arrow) relative to surrounding patches of stat92E mutant cells.
stat92E promotes expansion of the lateral notum and inhibits expansion of the medial notum
The downregulation of Stat92E activity in the notum coincided with the expansion and patterning of the notum mediolateral axis (Fig. 1D-I). We therefore asked whether stat92E affects the organization of this axis. Dpp induces the expression of ush and pnr at high and low thresholds in the medial notum, and represses the expression of mirr in this region. wg is induced at an intermediate position along the border with the medial notum (Fig. 5A). Depletion of stat92E in the Ptc domain led to the displacement of the Mirr and Wg domains toward the hinge (Fig. 5B). Likewise, the Mirr and Wg domains were displaced laterally toward the hinge in stat92E mutant clones (Fig. 5D-E). The lateral displacement of the Wg domain coincided with a reduction in the scope of the lateral notum. Reciprocally, ectopic expression of upd in the Ptc domain led to the displacement of the Mirr and Wg domains in an opposite direction toward the disc stalk, with a coincidental loss of the medial notum (Fig. 5C). The displacement of the Mirr and Wg domains was also observed at a distance from the Ptc domain. It is possible that this apparent non-autonomy reflects the broader expression of Ptc-GAL4 at earlier stages of development. Consistent with this idea, lineage tracing revealed that most of the notum originates from the Ptc-GAL4 expressing cells (Evans et al., 2009). We conclude that stat92E restricts the expansion of the medial notum and promotes the expansion of the lateral notum, consistent with the medial to lateral repression of Stat92E activity during notum development (Fig. 1).
Figure 5. stat92E restricts the expansion of the medial notum and specifies the dorsal pleura cell-autonomously.
(A) Mirr (cyan) is expressed in the lateral notum and Wg (red) in the lateral notum along the border with the medial notum. (B) Depletion of Stat92E activity in the Ptc domain (green) displaced the Mirr and Wg domains toward the hinge. (C) Expression of upd in the Ptc domain resulted in the opposite displacement of the Mirr and Wg domains toward the disc stalk. Double arrows demarcate the scope of the lateral notum marked with Mirr in A-C. To quantify the effects, we measured the scope of the lateral notum relative to scope of the entire notum ML axis and found that a reduction in Ptc>StatRNAi (0.275 +/− 0.05 SE, N=7) and expansion in Ptc>Upd (0.86 +/− 0.1 SE, N=6) compared to wild type (0.395 +/− 0.5 SE, N=8). (D-E) The Wg and Mirr expression domains were also displaced laterally in stat92E mutant clones. (F) Eyg (cyan) is expressed in the notum in a broad anterior domain, and Zfh2 (red) is expressed in the hinge. An intervening domain separates the Eyg and Zfh2 domains (bracket in inset in F). Only along the anterior margin the Eyg and Zfh2 domains are adjacent (arrow in F). (G-I) In large stat92E clones that spanned the Eyg and Zfh2 domains, the intervening domain between the Eyg and Zfh2 domains was missing (arrow in inset in G). In addition, the Eyg domain was displaced laterally into the pleura (arrow in G). (H) Mirr was expressed ectopically in stat92E mutant clones laterally to the Mirr domain along the wing margin (arrow). (I) Eyg was expressed ectopically in stat92E mutant clones in the dorsal pleura (arrows).
As Stat92E activity remains high in the hinge throughout development (Fig.1), we asked whether stat92E was required to limit the scope of the lateral notum along the notum/hinge interface. In the wild type disc, the lateral notum and dorsal pleura marked by Eyg and Zfh2 are adjacent along the anterior disc margin (arrow in Fig. 5F). Further posterior, a small region separates the Eyg and Zfh2 domains (bracket in Fig. 5F inset). In large stat92E mutant clones that spanned the Eyg and Zfh2 domains this intervening domain between the Eyg and Zfh2 domains was missing. As a consequence, the Eyg and Zfh2 domains were adjacent in the clones (arrow in Fig. 5G inset). In addition, the Eyg domain extended along the anterior margin from the notum into the dorsal pleura (arrow in Fig. 5G). Similarly, the broad expression of UAS-Stat92ERNAi with C311-GAL4 resulted in the expansion of the Eyg domain toward the dorsal hinge and pleura and coincided with the downregulation of Zfh2 expression in this region (Sup.Fig. 7). Mirr and Eyg were also expressed cell-autonomously in smaller stat92E mutant clones that were generated in the dorsalpleura (arrows in Fig. 5H and I, respectively). These phenotypes coincided with the patterning defects in the pleura surrounding the hinge in adult flies baring stat92E mutant clones (Sup. Fig. 6E-F). Thus, stat92E is required to limit the lateral expansion of the Eyg domain and to promote the expansion of the intervening domain between the Eyg domain and the dorsal hinge. stat92E also acts cell-autonomously to promote dorsal pleura identity and inhibit notum identity in this region.
stat92E promotes the expansion of the hinge
While Stat92E activity is downregulated in the pouch and notum, high levels of Stat92E activity persist in the hinge during all stages of development, suggesting additional roles for stat92E in hinge development. The mature hinge is subdivided morphologically along its PD axis by a series of epithelial folds (Fig. 6A, zoom of the dorsal hinge in Fig. 6B), and is subdivided molecularly by the restricted expression of different combinations of gene products (Fig. 6A). Zfh2 is expressed broadly in the hinge (Terriente et al., 2008; Whitworth and Russell, 2003), while Nub is restricted to the distal part of the dorsal hinge (Ng et al., 1995), and Wg to two concentric rings within the distal and the proximal parts of the hinge (Baker, 1988; Couso et al., 1993). Tsh is expressed proximal to the Zfh2 domain in the presumptive body wall (Azpiazu and Morata, 2000; Casares and Mann, 2000). Early in development Nub and Tsh are expressed in nearly adjacent domains. As development proceeds, the intervening domain (bracketed in Z1 Fig. 6D) expands and displaces the Nub and Tsh domains from one another (Zirin and Mann, 2007). stat92E mutant clones that were generated in the dorsal hinge caused a reduction in the scope of the Zfh2 domain (Fig. 6C), as well as a reduction or loss of the intervening domain between the Nub and Tsh domains (Fig. 6D arrow in Z2). Likewise, clones that were generated in the ventral hinge caused a reduction in the scope of the Zfh2 domain (opposing arrows, Fig. 6C).
To determine if Stat92E activation is sufficient to promote hinge expansion, we broadly expressed upd with Bx-GAL4 in a region that spans the dorsal hinge. Ectopic expression of upd in this region promoted the expansion of the distal subdivision of the dorsal hinge (arrow, Fig. 6E). Likewise, expression of upd in FLP-out clones, using a combination of the FLP-FRT and UAS-GAL4 systems, led to the expansion of both the dorsal and ventral hinge (Fig. 6F).
Wg is expressed in two concentric rings within the distal and proximal hinge (Baker, 1988; Couso et al., 1993; Couso et al., 1994; Dichtel-Danjoy et al., 2009; Perea et al., 2009; Rodriguez del Alamo et al, 2002; Terriente et al., 2008) where it promotes hinge cell proliferation (Neumann and Cohen, 1996). To determine if stat92E affects hinge growth by promoting the expression of the hinge mitogenic signal Wg (Neumann and Cohen, 1996), we examined Wg protein expression in stat92E mutant clones. We found that Wg expression was generally maintained in the stat92E mutant clones ruling out the possibility that stat92E promotes hinge growth by regulating Wg production (Fig. 6G also see Fig. 5D). We note that the contour of Wg expression was displaced proximally in patches of wild type cells relative to the contour of Wg expression in surrounding mutant cells (arrow in Fig. 6G), further supporting a role for stat92E in hinge expansion.
Requirements for canonical JAK/STAT signaling in wing PD axis development
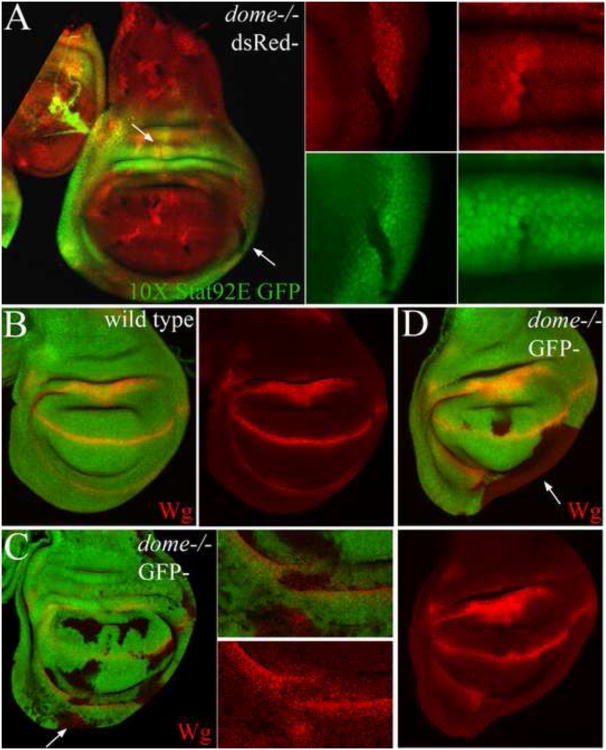
Stat92E has been shown to translocate to the nucleus independent of receptor activation via a non-canonical mechanism (Li, 2008; Shi et al., 2006; Shi et al., 2008). To determine the role of canonical JAK/STAT signaling in wing development we examined the role of upd and the upd receptor dome in wing PD patterning. Broad depletion of upd in the wing by RNAi resulted in a mild outstretched wing phenotype (data not shown), suggesting a mild requirement for upd in hinge development. Similarly, expressing a dominant negative dome receptor (UAS-domeΔCYT) (Brown et al., 2001) with Tsh-GAL4 resulted in a mild to moderate reduction of notum size (Sup. Fig. 8G-H). By contrast, depleting stat92E function by RNAi caused a range of phenotypes which were generally more severe than the Tsh>domeΔCYT phenotypes (Sup. Fig. 8D-F). In severe cases, the entire wing disc formed a tiny rudiment and the pouch failed to expand (Sup. Fig. 8F). Further analysis of dome function by clonal analysis revealed only mild requirements for dome in wing PD patterning (Fig. 7). Mutant clones for the strong dome468 allele generated in a wild type background were recovered readily in the pouch and notum. However, only small clones were recovered in the hinge, while mostly wild type twin spot clones were recovered in the dorsal hinge (Fig. 7A). dome mutant clones that were generated in a Minute background caused only mild patterning defects. Clones that spanned the hinge disrupted the normal pattern of Wg expression (arrow, Fig. 7C), while a subset of large clones caused a reduction in the growth of the ventral hinge (arrow, Fig. 7D). Furthermore, dome mutant clones in both the dorsal and ventral hinge lost expression of 10× Stat92E-GFP reporter activity (Fig. 7A). However, in general we did not recover phenotypes as severe as those generated by the stat92E mutant clones such as wing duplication or severe reduction in the growth of the dorsal hinge.
Figure 7. The role of canonical JAK/STAT Signaling in the elaboration of the wing PD/ML axis.
(A) Negatively marked dome468 mutant cell clones generated in a wild-type background survived and grew poorly in the dorsal hinge. Expression of the 10× Stat92E-GFP reporter was missing in dome clones in either the dorsal or ventral hinge (arrows). (B) Wg expression in a wild type wing field. (C-D) Negatively marked dome mutant clones generated in a Minute background caused mild defects in Wg expression in clones that spanned the hinge region (C). Similar defects in Wg expression were observed in the ventral hinge in stat92E mutant clones (see examples in Fig. 2D and Fig. 5D). (D) In rare cases, the hinge was more severely affected in the clones (in 2 large clones in 45 discs examined).
Our data is largely consistent with a canonical role for stat92E in wing development with the exception of the dome468 loss-of-function phenotypes. However, there are several possible explanations for the difference between the dome468 and stat92E clonal phenotypes that are consistent with a canonical role for stat92E in wing development. It is conceivable that the dome468 clones produce a more severe loss of growth and proliferation due to stronger loss of stat92E function that precludes formation of more dramatic clonal phenotypes. In addition, the lack of the receptor could cause an increase of the pool of the non-phosphorylated cytoplasmic Stat92E leading to an increase in the level of unphosphorylated heterochromatin bound protein. This effect could mask the requirements of stat92E in the canonical pathway. Additional experiments will be necessary to define the role of stat92E in the canonical and non-canonical pathways during wing development.
Discussion
stat92E promotes cell proliferation in the second instar larval wing disc and inhibits cell proliferation in the pouch at later stages (Mukherjee et al., 2005). stat92E activity levels also regulate cell competition, such that stat92E mutant cells are eliminated by cell competition, while cells with hyper-activated Stat92E eliminate surrounding wild type cells (Rodrigues et al, 2012). While the above studies assigned general roles for stat92E in control of cell proliferation and cell competition, the current study relates the temporal pattern of Stat92E activity to the patterning and expansion of the wing PD axis. We show that Stat92E is active ubiquitously at early stages and acts to inhibit the induction of ectopic wing fields. Subsequently, Stat92E activity is repressed in the notum and pouch to allow for the proper expansion of these primordia. At later stages, Stat92E is active in and surrounding the hinge and is required for the expansion of the hinge and the specification of the dorsal pleura. Thus, Stat92E is regulated along the wing PD/ML axis to coordinate the expansion of this axis and control its proper subdivision.
The integration of Stat92E activity with other patterning signals
Secreted signals that emanate from localized organizers operate in concert to pattern fields of cells (Lawrence et al., 1996; Mann and Morata, 2000). In some contexts, organizer signals emanate from field boundaries and act reciprocally and antagonistically to organize pattern across the intervening field. In relatively simple systems, such as the patterning of the fly embryonic epidermis and vertebrate neural tubes, two organizers are established at field boundaries to pattern the intervening field of cells (Hatini & DiNardo, 2001; Wilson and Maden, 2005; Gomez-Skarmeta et al., 2003). In more complex systems, such as the patterning of the vertebrate limb bud, the coordinated activities of more than two organizers are required to elaborate the pattern (Duboc and Logan, 2009). Our findings suggest that Dpp, the Odd-skipped proteins and Stat92E interact functionally to organize both the AP and ML axes of the notum (Fig. 8B). Dpp and the Odd-skipped proteins act antagonistically and reciprocally to organize the notum AP axis (DelSignore et al., 2012). We now find that stat92E restricts the scope of the Odd-skipped domain and thereby the organization of the notum AP axis (Fig. 4). In addition, we find that stat92E acts antagonistically to Dpp to pattern the notum ML axis (Fig. 5).
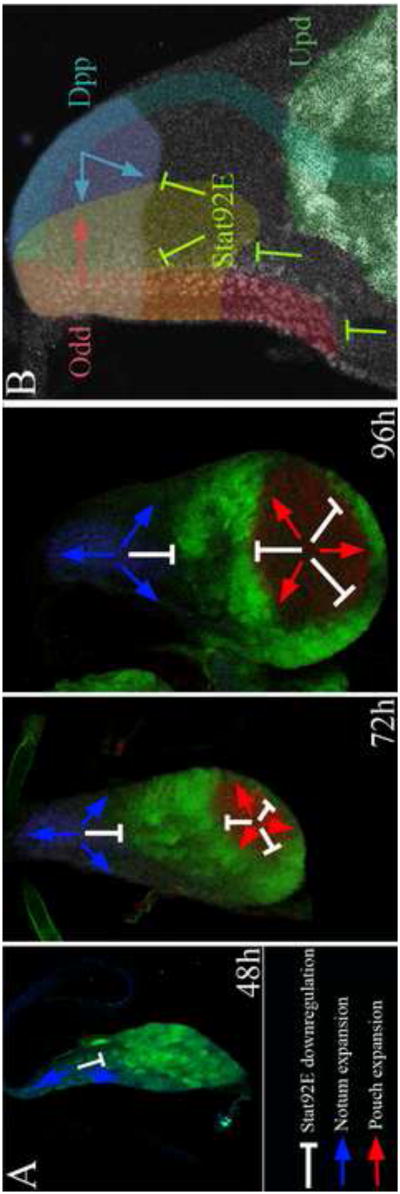
Figure 8. Model illustrating stat92E regulation and role in elaboration of the wing PD/ML axis.
(A) A cartoon depicting the relationship between the progressive downregulation of Stat92E activity (white inhibitory arrows) and the expansion of the notum (blue arrows) and pouch (red arrows). The dynamics of Stat92E donwregulation coincided and contributed to the expansion of the pouch and notum, while the persistent activity of Stat92E in the hinge promoted the expansion of this domain. (B) A cartoon depicting the activity of Stat92E in notum patterning in relationship to the role of dpp and the odd-skipped genes. dpp plays two distinct roles in notum patterning. dpp promotes medial notum identity and represses lateral identity. In addition, dpp promotes anterior notum identity and represses posterior identity. The odd-skipped genes have been proposed to regulate the production of an unknown signal at the anterior border of the notum, which acts reciprocally and antagonistically to dpp to promote anterior identity and repress posterior identity. We provide evidence that stat92E restricts the lateral expansion of medial notum and thus acts antagonistically to dpp to pattern the notum ML axis. stat92E also restricts the scope of the Bowl domain to the anterior border of the notum and thereby the patterning of the notum AP axis.
Unpaired signals, which activate the JAK/STAT pathway, are produced in several localized sources surrounding the pouch and the lateral border of the notum and Stat92E is activated in and surrounding these sources (Fig. 1 and Sup. Fig. 2). Moreover, the dynamics of canonical Stat92E activation coincides with the requirement of stat92E in wing development. Thus, our data is largely consistent with a canonical role for stat92E in wing development. According to this model the coordinated activities of the three pattern organizing signals that emanate from the three borders of the notum are coordinated to control notum growth and patterning (Fig. 8B). stat92E regulates wg expression to pattern the eye, and wg and dpp expression to pattern the leg and antenna (Ayala-Camargo et al., 2007; Ekas et al., 2006). Thus, stat92E appears to regulate the expression or modulate the activity of other organizer signals to pattern developing imaginal discs. Although unlikely, it remains possible that stat92E acts primarily through a non-canonical mechanism, a model that rules out the idea that a third organizer expressing upd signals is required for the elaboration of the wing PD/ML axis. Instead, this model predicts that a balance of non-canonical Stat92E activity and a temporally regulated pattern of Stat92E repression are superimposed on existing patterning systems to elaborate this axis (Fig. 8).
stat92E controls the relative expansion of gene expression domains along the notum ML axis
The pouch is organized by several signals that are produced along the AP and DV compartment boundaries (Lawrence et al., 1996; Mann and Morata, 2000), while the notum is organized by signals that emanate from the anterior and posterior borders of the notum (DelSignore et al., 2012; Fromental-Ramain et al., 2008; García-García et al., 1999; Letizia et al., 2007). However, it remains unclear how the programs of gene expression controlled by these organizers are coordinated to control relative expansion of these primordia. We provide evidence that Stat92E activity is regulated across the wing disc PD/ML axis to coordinate the relative expansion of the pouch, hinge, and notum. Our data reveals that Stat92E activity is downregulated in the presumptive notum and pouch with dynamics that coincide with the expansion of these primordia (Fig. 8A). The broad constitutive activation of canonical Stat92E signaling blocked the induction of Eyg, Mirr and Wg expression in the notum as well as the general expansion of the notum (Fig. 3E-F and H-I, respectively). In the wing field, constitutive upd expression impaired the expansion of the wing field and the restriction of the Zfh2 domain to an intermediate PD position (Fig. 3B-C). These results suggest that the downregulation of Stat92E activity in the notum and pouch are required for the proper expansion and patterning of these domains from early stages.
Analysis of stat92E mutant clones that were generated at later stages revealed that stat92E affects the relative scope of various domains along the notum ML axis and wing PD axis. stat92E restricted the scope of the medial notum and promoted the expansion of the lateral notum. In addition, stat92E was required to promote the expansion of the lateral part of the notum between the Eyg domain and the dorsal hinge (Fig. 5F-G) indicating a more general role for stat92E in controlling the scope of various gene expression domains along the entire notum ML axis (Fig. 8). How might stat92E restrict the expansion of medial notum identities while promoting expansion of lateral identities? It is conceivable that stat92E inhibits cell proliferation in early wing discs and downregulation of Stat92E activity stimulates cell proliferation by relief-of-repression at later stages. According to this model, the subdivision and expansion of the notum are coupled with the downregulation of Stat92E activity in a medial to lateral direction. This model predicts that the untimely removal of Stat92E activity in early wing disc would lead to the untimely proliferation and expansion of medial notum identities at the expanse of lateral identities as a result of precocious relief-of-repression in lateral cells.
While stat92E restricted the expansion of the notum, it was required to promote the expansion of the hinge. In particular, stat92E was required for the generation of the intervening domain between the Tsh and Nub domains in the dorsal hinge as well as the expansion of the ventral hinge. The failure to generate the intervening domain could result from the lateral expansion of the Tsh domain at the expense of the intervening domain. Alternatively, it could result from a role for stat92E in controlling cell proliferation in this region of the wing hinge. Consistent with the latter, both dome and stat92E mutant clones survived and grew poorly in this region of the wing disc (Fig. 7A) (Mukherjee et al., 2005). This observation suggests differential roles for stat92E along the wing PD/ML axis including a negative role in notum and pouch expansion and a positive role in hinge expansion.
stat92E suppresses ectopic wing PD axis specification
In addition to specific roles in the elaboration of the notum and wing ML/PD axis, we also find that stat92E represses inappropriate induction of ectopic wing fields (Fig. 2). Stat92E is active ubiquitously in early wing discs and inhibits the induction of ectopic wing fields in part of the wing disc that normally adopts body wall identity (Figs. 1-2, Sup. Fig. 6B). Conversely, ectopic activation of Stat92E activity suppressed the elaboration of the wing PD axis (Fig. 3). Vn-dependent EGF receptor signaling promotes notum identity and antagonizes wing identity (Wang et al., 2000; Zecca and Struhl, 2002a, 2002b). stat92E mutant clones in the presumptive notum caused induction of ectopic wing fields despite the specification of the notum in these wing discs (Fig. 2). This observation indicates that the Vn-dependent mechanisms that specify the notum are insufficient to inhibit wing induction, and implies that additional mechanisms are required to inhibit wing induction in this region. The induction of the wing field depends on signals that are delivered by the posterior compartment to the adjacent anterior compartment in early wing discs (Klein and Arias, 1998; Ng et al., 1996; Williams et al., 1993). Our data indicate that wing-inducing signals are also delivered in the notum, but the competence of the notum to respond to these signals is blocked by stat92E. It has been previously reported that wing inducing signals are also delivered in the peripodial epithelium (PE) of the wing imaginal disc, but the competence of this tissue to respond to these signals is blocked by the odd-skipped genes (Nusinow et al., 2008). We therefore propose that the broad competence of the early wing disc to respond to wing inducing signals is antagonized in the presumptive notum by stat92E and in the PE by the odd-skipped genes. This may be significant at early stages of development when the wing disc is relatively small and the organizing-centers that specify the notum and wing reside in close proximity to one another. During this stage stat92E may act as a buffer to prevent inappropriate pathway activation in ectopic locations.
Material and Methods
Generation and analysis of stat92E mutant alleles by mosaic analysis
The Rps3Plac190 Minute mutation prolongs larval development by 51 hours (Saeboe-Larssen et al., 1998). Mitotic stat92E clones were induced using the Minute FLP/FRT technique at 48-72, 72-96 hours AEL, which correspond to approximately first and second larval in star in the Rps3Plac190 Minute background. Flies of the genotype y w hs-FLP; FRT82B Ubi-GFP Rps3Plac190 /TM6C, Sb were used to induce marked Minute FLP/FRT clones.
We examined the clonal phenotypes of the stat92E85C9, stat92E397, stat92Ej6C8, and stat92E06346 mutant alleles to rule out the possibility that phenotypes arose from independent genetically linked mutations. We found that stat92E85C9, stat92E397 and stat92Ej6C8 mutant clones generated at second and early third in star caused morphological defects in the appendages and body wall of adult flies (Sup. Fig. 6, Sup. Tables 1-2). Analysis of mutant clones in developing imaginal discs revealed strong morphological defects and changes in expression of patterning markers in stat92E85C9 and stat92E397 mutant clones and only weaker morphological anomalies in stat92Ej6C8 clones (Sup. Fig. 4, Sup. Tables 1-2). In comparison, neither stat92E06346 mutant clones nor control clones produced any obvious defects (data not shown). To rule out the possibility that the stat92E clonal phenotypes resulted from an associated mutation we rescued the strongest stat92E85C9 clonal phenotypes with genomic DNA that spans the stat92E locus (P[acman] Bac clone CH321-73F24). A super folder EGFP (sf-EGFP) was fused in frame with stat92E coding region in this genomic DNA by recombineering (gift from R. Spokony), and the modified Bac was inserted by site-specific recombination at 22A3. The distribution of the Stat92E:sf-EGFP protein coded by the Bac coincided with the distribution of the endogenous protein and the expression of the 10× Stat92E-GPF reporter in developing wing imaginal discs (Sup. Fig. 5, and data not shown). Expression of the Stat92E:sf-EGFP protein in the stat92E85C9 mutant clones rescued to wild type the patterning defects and the abnormal morphology of the clones (Sup. Fig. 5; 27 wing discs bearing clones stained with Dapi and Wg were analyzed). We thus conclude that the stat92E85C9 clonal phenotypes resulted from the loss of stat92E function and not from an associated mutation. We note that the rescue does not exclude the possibility that the stat92E85C9 chromosome carry a second mutation close to the stat92E locus though this possibility is unlikely.
The nature of the stat92E alleles is consistent with why some produce strong phenotypes and others do not. Both stat92E06346 and stat92Ej6C8 have P-element insertions 5′ to the start site of the stat92E gene, which presumably lower stat92E gene transcription (Hou et al., 1996; Spradling et al., 1999). The 761 amino acid Stat92E protein is composed on an N-terminal region, a coiled-coil domain, a DNA-binding domain, a linker region, an SH2 domain, and a C-terminal transactivation domain. The stat9285C9 allele is caused by a substitution of an Arg at position 442 to a Pro in the DNA binding domain and this substitution abolishes the transcriptional activity of the protein in vitro (Ekas et al., 2010). The stat92E397 allele is caused by substitution of a Trp to a stop codon at position 594 3′ of the DNA binding domain resulting in truncation of part of the SH2 domain and the C-terminal transactivation domain (Silver and Montell, 2001). The in vitro transcriptional activity of this variant has not been examined. Analysis of the zygotic lethal phase of these stat92E alleles placed over a chromosomal deficiency that removes the stat92E locus suggested the following stat92E allelic series:85C9=397>j6C8>06346. In support of this conclusion it has been reported that the stat92E85C9 and stat92E397 clones generated stronger phenotypes than stat92E06346 or stat92Ej6C8 clones in the eye-antennal, wing or leg disc (Ayala-Camargo et al., 2007; Ekas et al., 2006). Our analysis in the wing imaginal disc is largely consistent with these reports (Sup. Tables 1-2).
Analysis of dome function by clonal analysis
Flies of the genotype Ubi-mRFPhsFLP FRT19A were used to generate dome468 mutant clones in a wild type background. Females of the genotype Ubi-GFP M(1)ospFRT19A/FM7; hsFLP/TM6b (G. Struhl) were crossed to dome468FRT19A/FM7 mutant males that were rescued to viability with a genomic fragment spanning the dome locus inserted at 65B2 (Dp DC365) in trans to a TM6b balancer chromosome. Marked Minute FLP/FRT clones were induced in female larvae of the genotype Ubi-GFP M(1)ospFRT19A/dome468FRT19A; hsFLP/+.45 wing imaginal discs stained with Wg and Dapi were examined for phenotypes.
UAS transgenes
Flies of the genotype UAS-RedStinger, UAS-FLP, Ubi>STOP>Stinger were used to trace the lineage of Upd-GAL4 expressing cells (green) and simultaneously detect the source of upd expression (red) (Evans et al., 2009). UAS-updRNAi (VDRC ID-3282), UAS-Stat92ERNAi (VDRC ID-43866, ID-43867, strong and weak insertion, respectively), UAS-domeΔCYT3.2 (J. Castelli-Gair) where used to inhibit canonical JAK/STAT signaling. UAS-upd was used to activate canonical signaling (Harrison et al., 1998). Esg>Upd and Esg>domeΔCYT3.2 larvae were raised for 5-6 days at 30°C prior to dissection.
GAL4 lines
The following drivers were used: UAS-GFP; Tsh-GAL4md621, Esg-GAL4NP5130, UAS-GFP, C311-GAL4 (G. Schubiger); UAS-GFP, Ptc-GAL4 (Speicher et al., 1994), UAS-GFP; Ubx-GAL4 (Pallavi and Shashidhara, 2003) and Bx-GAL4ms1096. Tsh-GAL4md621 is broadly expressed and excluded from the pouch and distal hinge, Esg-GAL4NP5130 is broadly expressed and excluded from a broad anterior domain in the notum, C311-GAL4 is broadly expressed and is upregulated along the anterior notum margin, Ptc-GAL4 is expressed in a narrow domain along the AP compartment boundary, Ubx-GAL4 is expressed in the peripodial epithelium and Bx-GAL4ms1096 is expressed at high levels in the dorsal hinge and pouch and at lower levels in the ventral wing field and notum. upd-GAL4 (E132) was used to highlight the sources of upd expression.
Immunofluorescence, reporter lines and microscopy
10× Stat92E reporter driving expression of GFP (10× Stat92E-GFP) or destabilized GFP (10× Stat92E-DCFP) were used to monitor the pattern of Stat92E activity (Bach et al., 2007). Both reporters were expressed in a similar pattern. Antibody staining protocols have been described elsewhere (Hatini et al., 2005). Primary antibodies used were: mouse anti-Nub (Ng et al., 1995), rabbit anti-Tsh (Wu and Cohen, 2002), rat anti-Zfh2 (M. Lundell), mouse anti-Wg (4D4, DSHB), rabbit anti-Bowl (DelSignore et al., 2012), guinea pig anti-Eyg (Aldaz et al., 2003), rabbit anti-Doc2 (M. Frasch), mouse anti-Ubx (R. White), mouse anti-Tup (DSHB), rat anti-Mirr (H. McNeill) and rabbit anti-Stat92E (Cell Signaling).
Typically 50-100 wing imaginal discs were stained, mounted and inspected per experiment. Of those, 5-10 representative imaginal disc were imaged by confocal microscopy for detailed analysis. Imaginal discs were scanned using a Zeiss LSM510 confocal microscope in multitracking mode and images were assembled and adjusted using Adobe Photoshop CS3.
Supplementary Material
At third in star, the Stat92E protein was enriched in nuclei in the hinge and in a narrow domain along the anterior border of the notum. However, in the pouch and most of the notum Stat92E localized to the cytoplasm. Arrowheads point to planes of Z-sections shown in corresponding insets. Asterisks in insets point to regions with enriched nuclear accumulation of Stat92E.
(A-D) Upd-GAL4 UAS GFP (Upd>GFP; green); Bowl (red). Optical sections at the plane of the disc proper (DP) and overlying peripodial epithelium (PE) in A-B. (A) At 48h, Upd>GFP was broadly expressed in the presumptive pouch, hinge, and the lateral part of the notum. Upd>GFP expression was excluded from the medial notum and the PE. (B) At 60h, Upd>GFP expression was downregulated in the distal pouch and notum. (C-D) At 96-120h, Upd>GFP expression was restricted to the lateral border of the notum and several sources surrounding the pouch. Bowl and Upd>GFP were expressed in adjacent domains along the anterior margin in an interlocking pattern (arrows in C and D).
Upd>RFP (red); Upd>GFP lineage-trace (green).(A) Early and (B) late third in star. Lineage tracing shows that most cells in the wing disc originate from upd-expressing cells except for a small cell population near the disc stalk (arrow in A), which expanded preferentially to form a large part of the medial and lateral notum at later stages (arrow in B).
We examined the clonal phenotypes of several stat92E mutant alleles to rule out the possibility that phenotypes arose from independent genetically linked mutations (also see Material and Methods). (A) stat92E397 mutant clones generated strong defects in epithelial morphology and changes in expression of patterning markers in L3 wing discs that recapitulated the stat92E85C9 clonal phenotypes. (B) stat92Ej6C8 mutant clones caused milder defects (see Sup. Table 1-2). Note the lateral expansion of the Bowl domain (red) in stat92E397 mutant clones compared to fairly normal accumulation of Bowl in stat92Ej6C8 mutant clones.
(A-B) Two examples of wing imaginal disc containing negatively marked stat92E85C9 clones expressing stat92E from a genomic DNA transgene that spans that stat92E locus. The stat92E reading frame in this genomic DNA was fused to super-folder EGFP (sf-EGFP). Note low level of Stat92E:sf-EGFP protein accumulation in the stat92E85C9 mutant clones spanning the hinge and the lateral part of the notum. The expression of the Stat92E:sf-EGFP rescued the abnormal morphology of the clones and the expression of molecular markers to wild type. We conclude that the stat92E85C9 clonal phenotype resulted from the loss of stat92E itself and not from an associated mutation.
(A-F) Adult flies; (A, D) wild type and (B-C, D-F) flies bearing stat92E mutant clones; (A-C) dorsal views; (D-F) lateral views. stat92E mutant clones led to several classes of phenotype including: (B) formation of ectopic wings from the posterior part of the notum (arrow) corresponding to disc phenotypes shown in Fig 2, (C) dorsal closure defect and defect in cuticle differentiation in the notum (arrow point to dorsal midline), possibly corresponding to disc phenotypes shown in Fig. 5B-C, and (E-F) differentiation of cuticular protrusions decorated with bristles characteristic of the notum in place of naked cuticle characteristic of the pleura surrounding the hinge (arrows) corresponding to disc phenotypes shown in Fig. 5G-I.
(A) C311-GAL4, UAS-GFP (C311>GFP); Optical section at the plane of the disc proper (DP) and the peripodial epithelium (PE). C311>GFP expression is mildly upregulated along the anterior margin lateral to the notum (arrows). (B-C) C311-GAL4 UAS-Stat92ERNAi (C311>Stat92ERNAi); Depletion of stat92E function with C311-GAL4 led to the lateral expansion of the Eyg domain toward the pleura (arrows in B) and the thinning of the Zfh2 domain in this region (arrows in C).
(A-H) Late third in star wing discs. (A) 10× Stat92E-GFP. Stat92E was active along the lateral border of the notum, the pleura and hinge in wild type imaginal discs. Low levels of Stat92E activity persisted into late third in star along the anterior border of the notum (demarcated with arrowheads in A, C). (B) Ptc-GAL4; UAS-GFP (Ptc>GAL4). (C) Ptc>Stat92E RNAi; 10× Stat92E-GFP. Expression of Stat92E RNAi in the Ptc domain strongly reduced 10× Stat92E-GFP reporter activity in this domain (arrow). (D-F) Tsh-GAL4/UAS-Stat92E RNAi; UAS-GFP. Depleting stat92E activity in the Tsh domain led to a mild (D), moderate (E) or severe (F) reduction of wing size. (G-H) Tsh-GAL4/UAS-DomeΔCYT; UAS-GFP. Inhibition of JAK/STAT signaling with a dominant negative Dome receptor led to a mild to moderate reduction of the notum. Scale bars = 50μm.
Comparing the recovery of adult flies baring stat92E85C9, stat92E397 and stat92j6C8 mutant clones induced at second (L2) and early third in star (EL3) larval stages in a Minute background. 82B refers to the FRT82B chromosome and U>G to the Ubi-GFP cell marker. The recovery of adults bearing stat92E85C9 mutant clones was lower compared to adult bearing stat92E397 and stat92Ej6C8 mutant clones. The incidence of dead pharates bearing stat92E85C9 and stat92E397 was higher compared to dead pharates bearing stat92Ej6C8 mutant clones.
Clones were induced at second (L2) and early third in star (EL3) larval stages in a Minute (M) background and scored for phenotypes at early adult stages. 82B refers to the FRT82B chromosome and U>G to the Ubi-GFP cell marker. Adult bearing stat92E mutant clones displayed stereotypic defects in leg, eye and wing derivatives. Eye phenotypes included rough eyes, small eyes and replacement of retinal tissue with head cuticle. Leg defects included shortening and bending of leg segments. Pleura defects included replacement of pleura tissue surrounding the hinge with notum tissue, while notum defects included notum closure defects. N refers to the number of flies bearing clones. Note that the stat92Ej6C8 mutant clones caused mostly leg and eye defects and outstretched (Os) wings, while stat92E85C9 and stat92E397 mutant clones caused in addition wing duplication and replacement of pleura with notum tissue. We interpret these data to suggest that the different stat92E alleles do not produce different phenotypes but rather partially overlapping phenotypes with the stronger phenotypes produced by the stat92E85C9 and stat92E397 alleles and the weaker phenotypes by the stat92Ej6C8 allele.
Genetically marked mutant clones were induced at second (L2) and early third larval in star (EL3) and scored for the location of the clones, the number of clones scored per location, and the number and percentage of clones producing each phenotype. Wing duplications were detected with Nub, Wg, and Doc2. Hinge morphology was analyzed in imaginal discs stained with Wg and Zfh2. The scope of the dorsal hinge was examined in Z-sections in mutant clones compared to adjacent wild type cells. Notum subdivision was examined using Eyg and Wg expression and the scope of the anterior notum margin using Bowl expression.
Highlights.
► stat92E suppresses wing induction in early wing discs. ► Stat92E repression coincides with the expansion of the pouch and notum. ► stat92E controls the relative expansion of the lateral and medial notum. ► stat92E promotes hinge expansion.
Acknowledgments
We thank S. DiNardo, G. Struhl, J. Hombria, E. Matunis, E. Bach, the Bloomington Stock Center, and the Vienna Drosophila Research Center (VDRC) for fly stocks, E. Bach, G. Campbell, S. Cohen, M. Lundell, N. Azpiazu, M. Frasch, H. McNeill and R. White and the Developmental Studies Hybridoma Bank (DSHB) for antibodies, and members of the lab for comments on the manuscript. This work was supported by a grant from the NIH to V.H. (R01GM06806).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldaz S, Morata G, Azpiazu N. The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development. 2003;130:4473–82. doi: 10.1242/dev.00643. [DOI] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–16. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Ayala-Camargo A, Ekas LA, Flaherty MS, Baeg GH, Bach EA. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236:2721–30. doi: 10.1002/dvdy.21230. [DOI] [PubMed] [Google Scholar]
- Azpiazu N, Morata G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development. 2000;127:2685–93. doi: 10.1242/dev.127.12.2685. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–31. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Baker NE. Localization of transcripts from the wingless gene in whole Drosophila embryos. Development. 1988;103:289–98. doi: 10.1242/dev.103.2.289. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–5. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development. 2000;127:1499–508. doi: 10.1242/dev.127.7.1499. [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bate M, Martinez Arias A. A wingless-dependent polar coordinate system in Drosophila imaginal discs. Science. 1993;259:484–489. doi: 10.1126/science.8424170. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bishop SA, Martinez Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- Couso JP, Knust E, Martinez Arias A. Serrate and wingless cooperate to induce vestigial gene expression and wing formation in Drosophila. Curr Biol. 1995;5:1437–48. doi: 10.1016/s0960-9822(95)00281-8. [DOI] [PubMed] [Google Scholar]
- DelSignore S, Hayashi T, Hatini V. odd-skipped genes and lines organize the notum anterior-posterior axis using autonomous and non-autonomous mechanisms. Mechanisms of Development. 2012 doi: 10.1016/j.mod.2012.05.001. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtel-Danjoy ML, Caldeira J, Casares F. SoxF is part of a novel negative-feedback loop in the wingless pathway that controls proliferation in the Drosophila wing disc. Development. 2009;136:761–9. doi: 10.1242/dev.032854. [DOI] [PubMed] [Google Scholar]
- Duboc V, Logan MP. Building limb morphology through integration of signaling modules. Curr Opin Genet Dev. 2009;19:497–503. doi: 10.1016/j.gde.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–9. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Ekas LA, Cardozo TJ, Flaherty MS, McMillan EA, Gonsalves FC, Bach EA. Characterization of a dominant-active STAT that promotes tumorigenesis in Drosophila. Developmental Biology. 2010;344:621–636. doi: 10.1016/j.ydbio.2010.05.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, Yackle K, Cespedes A, Hartenstein V, Call GB, Banerjee U. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–5. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromental-Ramain C, Vanolst L, Delaporte C, Ramain P. pannier encodes two structurally related isoforms that are differentially expressed during Drosophila development and display distinct functions during thorax patterning. Mech Dev. 2008;125:43–57. doi: 10.1016/j.mod.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Ripoll P, Morata AG. Developmental compartmentalization of the wing disc of Drosophila. Nature New Biol. 1973;245:251–253. doi: 10.1038/newbio245251a0. [DOI] [PubMed] [Google Scholar]
- García-García MJ, Ramain P, Simpson P, Modolell J. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development. 1999;126:3523–3532. doi: 10.1242/dev.126.16.3523. [DOI] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–63. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–18. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–75. doi: 10.1016/s0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- Hombria JC, Sotillos S. Disclosing JAK/STAT links to cell adhesion and cell polarity. Semin Cell Dev Biol. 2008;19:370–8. doi: 10.1016/j.semcdb.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Jakobi R, Fuss K, Guckenbiehl R, Hermann V, Keller S, Malisch S, Patinius W, Winkler M. Training in electronic data processing for nurse administrators. Krankenpflege (Frankf) 1993;47:363–5. [PubMed] [Google Scholar]
- Kehl BT, Cho KO, Choi KW. mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development. 1998;125:1217–27. doi: 10.1242/dev.125.7.1217. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–8. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Klein T, Arias AM. Different spatial and temporal interactions between Notch, wingless, and vestigial specify proximal and distal pattern elements of the wing in Drosophila. Dev Biol. 1998;194:196–212. doi: 10.1006/dbio.1997.8829. [DOI] [PubMed] [Google Scholar]
- Kolzer S, Fuss B, Hoch M, Klein T. Defective proventriculus is required for pattern formation along the proximodistal axis, cell proliferation and formation of veins in the Drosophila wing. Development. 2003;130:4135–47. doi: 10.1242/dev.00608. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Sanson B, Vincent JP. Compartments, wingless and engrailed: patterning the ventral epidermis of Drosophila embryos. Development. 1996;122:4095–103. doi: 10.1242/dev.122.12.4095. [DOI] [PubMed] [Google Scholar]
- Letizia A, Barrio R, campuzano S. Antagonistic and cooperative actions of the EGFR and Dpp pathways on the iroquois genes regulate Drosophila mesothorax specification and patterning. Development. 2007;134:1337–1346. doi: 10.1242/dev.02823. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–51. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–71. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- Mukherjee T, Hombria JC, Zeidler MP. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24:2503–11. doi: 10.1038/sj.onc.1208487. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development. 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea FJ, Cohen SM. Nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development. 1995;121:589–99. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea FJ, Vincent JP, Wu J, Cohen SM. Specification of the wing by localized expression of wingless protein. Nature. 1996;381:316–8. doi: 10.1038/381316a0. [DOI] [PubMed] [Google Scholar]
- Nusinow D, Greenberg L, Hatini V. Reciprocal roles for bowl and lines in specifying the peripodial epithelium and the disc proper of the Drosophila wing primordium. Development. 2008;135:3031–41. doi: 10.1242/dev.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallavi SK, Shashidhara LS. Egfr/Ras pathway mediates interactions between peripodial and disc proper cells in Drosophila wing discs. Development. 2003;130:4931–41. doi: 10.1242/dev.00719. [DOI] [PubMed] [Google Scholar]
- Perea D, Terriente J, Diaz-Benjumea FJ. Temporal and spatial windows delimit activation of the outer ring of wingless in the Drosophila wing. Dev Biol. 2009;328:445–55. doi: 10.1016/j.ydbio.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Rodrigues AB, Zoranovic T, Ayala-Camargo A, Grewal S, Reyes-Robles T, Krasny M, Wu DC, Johnston LA, Bach EA. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139:4051–61. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez del Alamo A, Terriente J, Galindo MI, Couso JP, Diaz-Benjumea FJ. Different mechanisms initiate and maintain wingless expression in the Drosophila wing hinge. Development. 2002;129:3995–4004. doi: 10.1242/dev.129.17.3995. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics. 1998;148:1215–24. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38:1071–6. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10:489–96. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Simcox AA, Grumbling G, Schnepp B, Bennington-Mathias C, Hersperger E, Shearn A. Molecular, phenotypic, and expression analysis of vein, a gene required for growth of the Drosophila wing disc. Dev Biol. 1996;177:475–89. doi: 10.1006/dbio.1996.0179. [DOI] [PubMed] [Google Scholar]
- Speicher SA, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 1994;120:535–44. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Galindo MI, Couso JP, Thor S. Control of Drosophila imaginal disc development by rotund and roughened eye: differentially expressed transcripts of the same gene encoding functionally distinct zinc finger proteins. Development. 2002;129:1273–81. doi: 10.1242/dev.129.5.1273. [DOI] [PubMed] [Google Scholar]
- Terriente J, Magarinos M, Diaz-Benjumea FJ. Nab controls the activity of the zinc-finger transcription factors Squeeze and Rotund in Drosophila development. Development. 2007;134:1845–52. doi: 10.1242/dev.003830. [DOI] [PubMed] [Google Scholar]
- Terriente J, Perea D, Suzanne M, Diaz-Benjumea FJ. The Drosophila gene zfh2 is required to establish proximal-distal domains in the wing disc. Dev Biol. 2008;320:102–12. doi: 10.1016/j.ydbio.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Wang SH, Simcox A, Campbell G. Dual role for Drosophila epidermal growth factor receptor signaling in early wing disc development. Genes Dev. 2000;14:2271–6. doi: 10.1101/gad.827000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe U, Dorfman R, Wernet MF, Cohen SM, Milan M. Proximodistal subdivision of Drosophila legs and wings: the elbow-no ocelli gene complex. Development. 2004;131:767–74. doi: 10.1242/dev.00979. [DOI] [PubMed] [Google Scholar]
- Whitworth AJ, Russell S. Temporally dynamic response to Wingless directs the sequential elaboration of the proximodistal axis of the Drosophila wing. Dev Biol. 2003;254:277–88. doi: 10.1016/s0012-1606(02)00036-2. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock S, Carroll SB. Pattern formation in a secondary field: a hierarchy of regulatory genes subdivides the developing Drosphila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Vorwerk K, Carroll SB. Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Repression of Teashirt marks the initiation of wing development. Development. 2002;129:2411–8. doi: 10.1242/dev.129.10.2411. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development. 2002a;129:1369–76. doi: 10.1242/dev.129.6.1369. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Subdivision of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development. 2002b;129:1357–68. doi: 10.1242/dev.129.6.1357. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Control of Drosophila wing growth by the vestigial quadrant enhancer. Development. 2007a;134:3011–20. doi: 10.1242/dev.006445. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feed-forward circuit of vestigial autoregulation. Development. 2007b;134:3001–10. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- Zirin JD, Mann RS. Nubbin and Teashirt mark barriers to clonal growth along the proximal-distal axis of the Drosophila wing. Dev Biol. 2007;304:745–58. doi: 10.1016/j.ydbio.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
At third in star, the Stat92E protein was enriched in nuclei in the hinge and in a narrow domain along the anterior border of the notum. However, in the pouch and most of the notum Stat92E localized to the cytoplasm. Arrowheads point to planes of Z-sections shown in corresponding insets. Asterisks in insets point to regions with enriched nuclear accumulation of Stat92E.
(A-D) Upd-GAL4 UAS GFP (Upd>GFP; green); Bowl (red). Optical sections at the plane of the disc proper (DP) and overlying peripodial epithelium (PE) in A-B. (A) At 48h, Upd>GFP was broadly expressed in the presumptive pouch, hinge, and the lateral part of the notum. Upd>GFP expression was excluded from the medial notum and the PE. (B) At 60h, Upd>GFP expression was downregulated in the distal pouch and notum. (C-D) At 96-120h, Upd>GFP expression was restricted to the lateral border of the notum and several sources surrounding the pouch. Bowl and Upd>GFP were expressed in adjacent domains along the anterior margin in an interlocking pattern (arrows in C and D).
Upd>RFP (red); Upd>GFP lineage-trace (green).(A) Early and (B) late third in star. Lineage tracing shows that most cells in the wing disc originate from upd-expressing cells except for a small cell population near the disc stalk (arrow in A), which expanded preferentially to form a large part of the medial and lateral notum at later stages (arrow in B).
We examined the clonal phenotypes of several stat92E mutant alleles to rule out the possibility that phenotypes arose from independent genetically linked mutations (also see Material and Methods). (A) stat92E397 mutant clones generated strong defects in epithelial morphology and changes in expression of patterning markers in L3 wing discs that recapitulated the stat92E85C9 clonal phenotypes. (B) stat92Ej6C8 mutant clones caused milder defects (see Sup. Table 1-2). Note the lateral expansion of the Bowl domain (red) in stat92E397 mutant clones compared to fairly normal accumulation of Bowl in stat92Ej6C8 mutant clones.
(A-B) Two examples of wing imaginal disc containing negatively marked stat92E85C9 clones expressing stat92E from a genomic DNA transgene that spans that stat92E locus. The stat92E reading frame in this genomic DNA was fused to super-folder EGFP (sf-EGFP). Note low level of Stat92E:sf-EGFP protein accumulation in the stat92E85C9 mutant clones spanning the hinge and the lateral part of the notum. The expression of the Stat92E:sf-EGFP rescued the abnormal morphology of the clones and the expression of molecular markers to wild type. We conclude that the stat92E85C9 clonal phenotype resulted from the loss of stat92E itself and not from an associated mutation.
(A-F) Adult flies; (A, D) wild type and (B-C, D-F) flies bearing stat92E mutant clones; (A-C) dorsal views; (D-F) lateral views. stat92E mutant clones led to several classes of phenotype including: (B) formation of ectopic wings from the posterior part of the notum (arrow) corresponding to disc phenotypes shown in Fig 2, (C) dorsal closure defect and defect in cuticle differentiation in the notum (arrow point to dorsal midline), possibly corresponding to disc phenotypes shown in Fig. 5B-C, and (E-F) differentiation of cuticular protrusions decorated with bristles characteristic of the notum in place of naked cuticle characteristic of the pleura surrounding the hinge (arrows) corresponding to disc phenotypes shown in Fig. 5G-I.
(A) C311-GAL4, UAS-GFP (C311>GFP); Optical section at the plane of the disc proper (DP) and the peripodial epithelium (PE). C311>GFP expression is mildly upregulated along the anterior margin lateral to the notum (arrows). (B-C) C311-GAL4 UAS-Stat92ERNAi (C311>Stat92ERNAi); Depletion of stat92E function with C311-GAL4 led to the lateral expansion of the Eyg domain toward the pleura (arrows in B) and the thinning of the Zfh2 domain in this region (arrows in C).
(A-H) Late third in star wing discs. (A) 10× Stat92E-GFP. Stat92E was active along the lateral border of the notum, the pleura and hinge in wild type imaginal discs. Low levels of Stat92E activity persisted into late third in star along the anterior border of the notum (demarcated with arrowheads in A, C). (B) Ptc-GAL4; UAS-GFP (Ptc>GAL4). (C) Ptc>Stat92E RNAi; 10× Stat92E-GFP. Expression of Stat92E RNAi in the Ptc domain strongly reduced 10× Stat92E-GFP reporter activity in this domain (arrow). (D-F) Tsh-GAL4/UAS-Stat92E RNAi; UAS-GFP. Depleting stat92E activity in the Tsh domain led to a mild (D), moderate (E) or severe (F) reduction of wing size. (G-H) Tsh-GAL4/UAS-DomeΔCYT; UAS-GFP. Inhibition of JAK/STAT signaling with a dominant negative Dome receptor led to a mild to moderate reduction of the notum. Scale bars = 50μm.
Comparing the recovery of adult flies baring stat92E85C9, stat92E397 and stat92j6C8 mutant clones induced at second (L2) and early third in star (EL3) larval stages in a Minute background. 82B refers to the FRT82B chromosome and U>G to the Ubi-GFP cell marker. The recovery of adults bearing stat92E85C9 mutant clones was lower compared to adult bearing stat92E397 and stat92Ej6C8 mutant clones. The incidence of dead pharates bearing stat92E85C9 and stat92E397 was higher compared to dead pharates bearing stat92Ej6C8 mutant clones.
Clones were induced at second (L2) and early third in star (EL3) larval stages in a Minute (M) background and scored for phenotypes at early adult stages. 82B refers to the FRT82B chromosome and U>G to the Ubi-GFP cell marker. Adult bearing stat92E mutant clones displayed stereotypic defects in leg, eye and wing derivatives. Eye phenotypes included rough eyes, small eyes and replacement of retinal tissue with head cuticle. Leg defects included shortening and bending of leg segments. Pleura defects included replacement of pleura tissue surrounding the hinge with notum tissue, while notum defects included notum closure defects. N refers to the number of flies bearing clones. Note that the stat92Ej6C8 mutant clones caused mostly leg and eye defects and outstretched (Os) wings, while stat92E85C9 and stat92E397 mutant clones caused in addition wing duplication and replacement of pleura with notum tissue. We interpret these data to suggest that the different stat92E alleles do not produce different phenotypes but rather partially overlapping phenotypes with the stronger phenotypes produced by the stat92E85C9 and stat92E397 alleles and the weaker phenotypes by the stat92Ej6C8 allele.
Genetically marked mutant clones were induced at second (L2) and early third larval in star (EL3) and scored for the location of the clones, the number of clones scored per location, and the number and percentage of clones producing each phenotype. Wing duplications were detected with Nub, Wg, and Doc2. Hinge morphology was analyzed in imaginal discs stained with Wg and Zfh2. The scope of the dorsal hinge was examined in Z-sections in mutant clones compared to adjacent wild type cells. Notum subdivision was examined using Eyg and Wg expression and the scope of the anterior notum margin using Bowl expression.