Abstract
Lymphedema is caused by dysfunction of lymphatic vessels, leading to disabling swelling that occurs mostly on the extremities. Lymphedema can be either primary (congenital) or secondary (acquired). Familial primary lymphedema commonly segregates in an autosomal dominant or recessive manner. It can also occur in combination with other clinical features. Nine mutated genes have been identified in different isolated or syndromic forms of lymphedema. However, the prevalence of primary lymphedema that can be explained by these genetic alterations is unknown. In this study, we investigated 7 of these putative genes. We screened 78 index patients from families with inherited lymphedema for mutations in FLT4, GJC2, FOXC2, SOX18, GATA2, CCBE1, and PTPN14. Altogether, we discovered 28 mutations explaining 36% of the cases. Additionally, 149 patients with sporadic primary lymphedema were screened for FLT4, FOXC2, SOX18, CCBE1, and PTPN14. Twelve mutations were found that explain 8% of the cases. Still unidentified is the genetic cause of primary lymphedema in 64% of patients with a family history and 92% of sporadic cases. Identification of those genes is important for understanding of etiopathogenesis, stratification of treatments and generation of disease models. Interestingly, most of the proteins that are encoded by the genes mutated in primary lymphedema seem to act in a single functional pathway involving VEGFR3 signaling. This underscores the important role this pathway plays in lymphatic development and function and suggests that the unknown genes also have a role.
Key Words: Functional pathway, Genetic, Mutation, Phenotype
The lymphatic system is a blind-ended endothelium-lined network [Butler et al., 2009] that collects and transports extravasated fluid, plasma proteins and immune cells back to the blood circulation [Karkkainen et al., 2001]. Developmental defects or dysfunction of the lymphatic system cause lymphedema [Brouillard and Vikkula, 2007; Boon and Vikkula, 2012], a disorder characterized by abnormal swelling of one or more extremities due to impaired transport of the lymph [Irrthum et al., 2000]. Lymphedema can be primary (congenital) or secondary (acquired). Hereditary lymphedema is more easily recognized when it occurs as an autosomal dominant trait, even if there is reduced penetrance and variable expressivity [Irrthum et al., 2000]. Parental consanguinity suggests recessive inheritance [Ghalamkarpour et al., 2009b]. Lymphedema can occur in association with other clinical features or as part of a syndrome [Irrthum et al., 2003]. Currently, 9 genes have been linked to different types of primary lymphedema: FLT4 (VEGFR3), GJC2, FOXC2, SOX18, GATA2, CCBE1, PTPN14, KIF11, and the lastly identified VEGFC.
FLT4 and GJC2 (fms-related tyrosine kinase 4, encoding VEGFR3, and gap junction protein gamma-2, encoding connexin-47, CX47, respectively) code for proteins localized on the cellular membrane. FLT4 is mutated in Nonne-Milroy disease (OMIM 153100), characterized by congenital bilateral lower limb lymphedema. All reported mutations are localized within the 2 intracellular tyrosine kinase domains of the encoded VEGFR3 receptor [Butler et al., 2007, Butler et al., 2009]. Cells expressing mutant VEGFR3 show inhibited autophosphorylation of the receptor, indicating downregulation of VEGFR3 signaling [Irrthum et al., 2000; Karkkainen and Petrova, 2000; Ghalamkarpour et al., 2009b]. Nonsynonymous mutations in GJC2 were discovered in a few families with late-onset autosomal dominant lymphedema (OMIM 613480) affecting all 4 extremities (‘four-limb lymphedema’) and sometimes associated with saphenous vein insufficiency, blepharoptosis, and involvement of the face or genitalia. Some families showed reduced penetrance [Ferrell et al., 2010; Ostergaard et al., 2011a]. The GJC2 amino acid substitutions likely cause gain-of-function, as loss-of-function mutations in GJC2 are found in patients with inherited autosomal recessive Pelizaeus-Merzbacher-like disease (PMLD, OMIM 608804), a hypomyelinating disorder of the central nervous system [Uhlenberg et al., 2004].
The second group of lymphedema-causing genes encodes 3 transcription factors (FOXC2, SOX18 and GATA2). FOXC2 (forkhead box C2) is mutated in puberty or late-onset primary lymphedema associated with distichiasis (LDS, OMIM 153400). FOXC2 regulates the expression of genes involved in cell growth, proliferation, differentiation, and longevity [Fang et al., 2000]. The majority of the FOXC2 mutations are insertions, deletions or nonsense mutations, leading to mRNA decay or truncated loss-of-function proteins [Dagenais et al., 2004; Ghalamkarpour et al., 2009a; van Steensel et al., 2009]. FOXC2 suppresses PDGFB production [Shimoda et al., 2011], and loss of its activity leads to accumulation of vascular smooth muscle cells in collecting lymphatics of knock-out mice and also in patients [Petrova et al., 2004; Norrmen et al., 2009].
Recessive and dominant mutations in SOX18 (SRY-box 18) cause the hypotrichosis-lymphedema-telangiectasia syndrome (HLTS, OMIM 607823), characterized by congenital lymphedema, reduced body hair including the absence of eyelashes and eyebrows, and localized cutaneous telangiectasias. This transcription factor plays an important role in early blood vessel modeling [Downes et al., 2009] as well as in the differentiation of lymphatic endothelial progenitor cells from venous precursors [Francois et al., 2008]. The 3 published SOX18 mutations are localized in the DNA-binding domain (recessive) or in the transactivation domain (truncating/dominant) [Irrthum et al., 2003]. The Sox18-/- mice exhibit only a minor coat defect [Pennisi et al., 2000a]. In contrast, the spontaneous ragged mutant of Sox18 has defective lymphatic and cardiovascular tissues and hair follicle defects [Pennisi et al., 2000b].
Mutations in GATA2 (GATA-binding protein 2) have been linked to a predisposition to myelodysplastic syndrome (MDS, OMIM 614286) and to acute myeloid leukemia (AML, OMIM 601626). Subsequently, mutations were identified in patients with primary lymphedema with myelodysplasia (also known as Emberger syndrome, OMIM 614038) [Hahn et al., 2011; Ostergaard et al., 2011b] and the monocytopenia with mycobacterial infection syndrome (MonoMAC, OMIM 614172), associated with dendritic cell, monocyte, B lymphocyte and natural killer lymphocyte deficiency (DCML) [Kazenwadel et al., 2012]. The phenotypes are not exclusive, underscored by a Japanese patient with a GATA2 mutation with MonoMAC and Emberger syndromes [Ishida et al., 2012]. There are no obvious genotype-to-phenotype correlations [Hyde and Liu, 2011; Holme et al., 2012].
The gene coding for the extracellular protein CCBE1 (collagen and calcium-binding EGF domain-1) that enhances the lymphangiogenic effects of VEGFC in vivo [Bos et al., 2011] is essential for fetal liver erythropoiesis [Zou et al., 2013]. CCBE1 was identified to be important for lymphatic development by genetic knock-down screening in zebrafish [Hogan et al., 2009]. In human patients, CCBE1 homozygous and compound heterozygous mutations cause the Hennekam lymphangiectasia-lymphedema syndrome (OMIM 235510), characterized by severe peripheral lymphedema associated with intestinal lymphangiectasias, characteristic facial features, growth and mental retardation, and hydrops fetalis [Hennekam et al., 1989; Alders et al., 2009, Alders et al., 2013; Connell et al., 2010].
An intracellular phosphatase (PTPN14) was shown to interact with the VEGFC-receptor VEGFR3 by co-immunoprecipitation upon activation by VEGFC. An intragenic deletion encompassing both sides of exon 7 of PTPN14 (protein tyrosine phosphatase, non-receptor type 14) was identified in a consanguineous family with autosomal recessive choanal atresia and lymphedema (OMIM 613611) [Au et al., 2010]. This mutation causes a frameshift p.Ser194Argfs*19.
Whole exome sequencing successfully revealed 2 additional genes, mutations of which cause primary lymphedema. Firstly, KIF11 mutations (kinesin family member 11, a DNA-interacting protein) were discovered to cause MLCRD (microcephaly, lymphedema, chorioretinal dysplasia) and CDMMR (chorioretinal dysplasia, microcephaly and mental retardation), 2 allelic syndromes that have now been regrouped as MCLMR (microcephaly with or without chorioretinopathy, lymphedema, or mental retardation, OMIM 152950) [Ostergaard et al., 2012]. MCLMR can be sporadic or inherited as an autosomal dominant trait, and the mutations are predicted to result in loss-of-function of EG5, the encoded homotetrameric kinesin motor protein playing a role in mitotic spindle assembly and function [Ostergaard et al., 2012]. The second gene, VEGFC (vascular endothelial growth factor C), is the major ligand of VEGFR3. A frameshift mutation was identified in 1 family, resulting in loss-of-function of the mutant allele [Gordon et al., 2013].
All the gene identifications reported to date have been performed using specific and limited numbers of families; thus, it is impossible to determine the proportion of all inherited primary lymphedema that can be explained by mutations in these genes. Therefore, we screened a total of 78 familial index patients for 7 of the genes that were known at the beginning of this study. We screened another 149 samples with presumably sporadic primary lymphedema or lymphedema of unknown mode of inheritance for 5 of the genes.
Materials and Methods
Sample Collection
DNA or whole blood samples were collected from patients seen in collaborating clinics. Informed consent was obtained from each participant, as approved by the ethical committee of the Medical Faculty at the Université catholique de Louvain (Brussels, Belgium) and the respective local committees. DNA was extracted manually from whole blood using a DNA purification kit (Gentra) or Wizard genomic DNA purification kit (Promega).
Screening Method
A series of 78 index patients from families with primary hereditary lymphedema was screened for mutations in FLT4, GJC2, FOXC2, SOX18, GATA2, CCBE1, and PTPN14. The screenings were performed sequentially. The new samples were included in screens of the remaining genes (table 1). Once a mutation was found, the sample was not tested for the remaining genes. In some cases, patients exhibiting distinct signs/symptoms associated with a disease were prioritized for specific genes (e.g. hypotrichosis or alopecia for SOX18) (online suppl. table 1, www.karger.com/doi/10.1159/000354097). In addition to the familial samples, we screened 5 genes (FLT4, FOXC2, SOX18, CCBE1, and PTPN14) in a cohort of 149 patients in which the primary lymphedema was presumably sporadic or the mode of inheritance was unknown (table 1). The recently discovered KIF11 and VEGFC genes were not included in the study.
Table 1.
227 samples were screened for 7 lymphedema genes and 40 (17%) mutations were found
| Gene | Families |
Sporadic and unknown inheritance |
||
|---|---|---|---|---|
| samples tested | mutations | samples tested | mutations | |
| FLT4 | 71 | 16 | 101 | 7 |
| GJC2 | 55 | 1 | – | – |
| FOXC2 | 64 | 5 | 135 | 4 |
| SOX18 | 22 | 3 | 63 | 1 |
| GATA2 | 45 | 1 | – | – |
| CCBE1 | 55 | 2 | 86 | 0 |
| PTPN14 | 37 | 0 | 118 | 0 |
| Total | 78 | 28 (35.9%) | 149 | 12 (8%) |
The coding regions of the genes (exons and splice-sites) were screened either by denaturing high performance liquid chromatography (DHPLC, Wave System 3500A, Transgenomic), high resolution melting (HRM) with a LightCycler® 480 (Roche Applied) or direct sequencing (CEQ2000 capillary sequencer (Beckman Coulter) or a 3130xl Genetic Analyzer (Life Technologies)). For the FLT4 receptor, we screened only the kinase domains (encoded by exons 17-25), for which all the known mutations have been reported. Amplicons with an abnormal DHPLC or HRM profile were re-amplified and sequenced. Sequences were analyzed with Sequencher® 4.5 (Gene Codes Inc.) or with CLC Main Workbench 6© (CLC Bio) software.
Variant Analysis
All variants were searched against dbSNP135 (http://www.ncbi.nlm.nih.gov/projects/SNP/) [Sherry et al., 2001]. The effects of amino acid changes on proteins were predicted using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) [Sunyaev et al., 1999; Ramensky et al., 2002; Adzhubei et al., 2010]. MutationTaster (http://www.mutationtaster.org/) [Schwarz et al., 2010] was also used, in particular to predict the effects of splice-site changes. The nomenclature of the mutations follows the guidelines of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/) and was verified with Mutalyzer (https://mutalyzer.nl/) [Wildeman et al., 2008]. When DNAs of other family members were available, cosegregation or de novo nature was checked. For the sporadic cases, paternity tests were performed using the PowerPlex16 HS system (Promega) on a 3130xl Genetic Analyzer (Life Technologies). All mutations have been submitted to the Vascular Anomalies Mutation Database (www.icp.ucl.ac.be/vikkula/VAdb/).
Results
We found an FLT4 (VEGFR3) mutation in 16 families (table 1; 6 mutations published previously) [Irrthum et al., 2000, Irrthum et al., 2003; Ghalamkarpour et al., 2006, Ghalamkarpour et al., 2009a, Ghalamkarpour et al., 2009b]. These mutations included 12 distinct nonsynonymous changes (two of which were found in 2 families), 1 in-frame deletion (p.Phe1108del) and 1 intronic change in a consensus splice site (c.2647+2T>C) (table 2). Of the 12 substitutions, 11 were predicted to alter the function of the receptor, whereas the last one (p.Glu1106Lys) is ‘possibly damaging’. According to MutationTaster, the intronic splice-site change is predicted to be ‘disease-causing’. Some of these substitutions are listed in dbSNP135 as ‘probably pathogenic’ alleles (table 2).
Table 2.
Mutations found in familial cases of lymphedema ordered according to amino acid position
| Gene Sample ID | Nucleotide change | Amino acid change | Protein effect | dbSNP135 | Cosegregationa | Mutation reported by |
|---|---|---|---|---|---|---|
| FLT4 | ||||||
| Family-1 | c.2554G>T | p.Gly852Cys | probably damaging | not reported | NA | – |
| Family-2 | c.2554G>T | p.Gly852Cys | probably damaging | not reported | NA | – |
| Family-3b | c.2563G>Ac | p.Ala855Thrc | probably damaging | rs121909657d | ♀♂ | Ghalamkarpour et al., 2009be |
| Family-4 | c.2569G>C | p.Gly857Arg | probably damaging | not reported | NA | Karkkainen et al., 2000 |
| Family-5 | c.2585C>T | p.Ala862Val | probably damaging | not reported | ♂ | – |
| Family-6 | c.2626G>T | p.Val876Leu | probably damaging | not reported | ♀ | – |
| Family-7 | c.2632G>A | p.Val878Met | probably damaging | rs121909654d | ♂ | Ghalamkarpour et al., 2006e |
| Family-8 | c.2647 + 2T>C | – | splice-site | not reported | NA | – |
| Family-9 | c.2797G>C | p.Gly933Arg | probably damaging | not reported | ♀ | Evans et al., 2003 |
| Family-10 | c.3104A>G | p.His1035Arg | probably damaging | rs121909653d | ♂ | Irrthum et al., 2000e |
| Family-11 | c.3121C>T | p.Arg1041Trp | probably damaging | rs121909650d | ♂ | Evans et al., 2003 |
| Family-12 | c.3121C>T | p.Arg1041Trp | probably damaging | rs121909650d | NA | Evans et al., 2003 |
| Family-13 | c.3121C>G | p.Arg1041Gly | probably damaging | not reported | NA | – |
| Family-14 | c.3257T>C | p.Ile1086Thr | probably damaging | rs121909655d | ♂ | Ghalamkarpour et al., 2006e |
| Family-15b | c.3316G>A | p.Glu1106Lys | possibly damaging | rs121909656d | ♀ | Spiegel et al., 2006e |
| Family-16 | c.3323_3325del | p.Phe1108del | in-frame deletion | not reported | ♂ | Evans et al., 2003; |
| Ghalamkarpour et al., 2006e | ||||||
| GJC2 | ||||||
| Family-17 | c.143C>T | p.Ser48Leu | probably damaging | not reported | NA | Ferrel et al., 2010 |
| FOXC2 | ||||||
| Family-18b | c.595dupc | p.His199Profs*264c | frameshift | not reported | ♀ | Finegold et al., 2001 |
| Family-19 | c.902_923dup | p.Pro309Glyfs*161 | frameshift | not reported | NA | – |
| Family-20 | c.982_983del | p.Gly328Leufs*134 | frameshift | not reported | ♂ | – |
| Family-21 | c.983dup | p.Tyr329Leufs*134 | frameshift | not reported | ♀ | – |
| Family-22 | c.1315_1316dup | p.Phe440Argfs*33 | frameshift | not reported | NA | – |
| SOX18 | ||||||
| Family-23b | c.283T>Ac | p.Trp95Argc | probably damaging | rs28936693d | ♀♂ | Irrthum et al., 2003e |
| Family-24b | c.310G>Cc | p.Ala104Proc | probably damaging | rs28936692d | ♀♂ | Irrthum et al., 2003e |
| Family-25 | c.720C>A | p.Cys240* | premature stop codon | rs74315430d | present in sibling | Irrthum et al., 2003e |
| GATA2 | ||||||
| Family-26 | c.130G>T | p.Glu44* | premature stop codon | not reported | present in sibling | – |
| CCBE1 | ||||||
| Family-27b | c.223T>Ac | p.Cys75Serc | probably damaging | rs121908250d | ♀♂ | Alders et al., 2009e |
| Family-28b | c.520T>Cc | p.Cys174Argc | probably damaging | rs121908254d | ♀♂ | Alders et al., 2009 |
| PTPN14 | ||||||
| No familial mutation found. | ||||||
NA = Family members not available; ♀ = mutation maternally inherited and present in other family members; ♂ = mutation paternally inherited and present in other family members; ♀♂ = consanguineous inheritance.
Consanguineous family.
Homozygous mutation.
Reported by dbSNP135 as probably pathogenic allele.
Patient published by our group.
Only 1 mutation was found in GJC2 (p.Ser48Leu) (table 2). The same change has been reported in 2 other families [Ferrell et al., 2010; Ostergaard et al., 2011a]. It is predicted to be ‘probably damaging’ for the protein function.
In FOXC2, we found 4 nucleotide insertions and 1 deletion, all leading to reading frameshifts, expected to produce a truncated protein (table 2). Among the samples screened for SOX18 (table 1), we identified 3 families with a mutation: 2 with amino acid substitutions (p.Trp95Arg and p.Ala104Pro) and 1 with a nonsense change (p.Cys240*) (table 2). The substitutions were homozygous mutations in consanguineous families, whereas the nonsense mutation was dominant [Irrthum et al., 2003]. All are reported as ‘pathogenic’ in dbSNP135. In the third transcription factor gene, GATA2, we identified only 1 mutation, which leads to a premature termination codon (p.Glu44*).
Within the 55 families tested for CCBE1 (table 1), we found 2 homozygous substitutions: p.Cys75Ser and p.Cys174Arg. Both were predicted to be ‘probably damaging’ and referred to as a ‘probably pathogenic’ allele in dbSNP135 (table 2). No pathogenic change was identified in PTPN14 (table 2).
We also screened a series of 149 samples of index patients presumed to have sporadic lymphedema or with an unknown family history (table 1). We identified 7 patients with nonsynonymous VEGFR3 mutations, predicted to be ‘probably damaging’ (table 3). Among these, only p.Pro1114Leu was reported in dbSNP135 as a ‘probably pathogenic’ allele. We also found 4 patients with a FOXC2 mutation (table 3). These included 1 deletion (p.Tyr305Cysfs*155), 1 duplication (p.Tyr313Argfs*152), 1 in-frame deletion (p.Arg165_Arg167del) and 1 premature stop codon (p.Gln315*). In SOX18, we identified 1 premature stop codon (p.Cys240*), which had previously been found in a familial case. No mutation was detected in CCBE1 or PTPN14 (tables 1, 3) in this non-familial screen. In all, 7 mutations appeared de novo, while 1 was inherited from an unaffected parent, therefore showing incomplete penetrance (Patient-8). The inheritance of 3 others could not be assessed (table 3).
Table 3.
Mutations found in sporadic cases and samples with unknown mode of inheritance ordered according to amino acid position
| Gene Sample ID | Nucleotide change | Amino acid change | Protein effect | dbSNP135 | Cosegregationa | Paternity attested | Mutation reported by |
|---|---|---|---|---|---|---|---|
| FLT4 | |||||||
| Patient-1 | c.2587T>C | p.Ser863Pro | probably damaging | not reported | de novo | yes | – |
| Patient-2 | c.2677C>G | p.Leu893Val | probably damaging | not reported | de novo | yes | Ghalamkarpour et al., 2009ad |
| Patient-3b | c.2819G>C | p.Arg940Pro | probably damaging | not reported | NA | – | – |
| Patient-4 | c.3105C>G | p.His1035Gln | probably damaging | not reported | de novo | yes | Ghalamkarpour et al., 2006d |
| Patient-5 | c.3341C>T | p.Pro1114Leu | probably damaging | rs121909652c | de novo | yes | Karkkainen et al., 2000 |
| Patient-6 | c.3391G>A | p.Gly1131Ser | probably damaging | not reported | de novo | yes | – |
| Patient-7 | c.3410C>T | p.Pro1137Leu | probably damaging | not reported | NA | – | Evans et al., 2003; Ghalamkarpour et al., 2009ad |
| FOXC2 | |||||||
| Patient-8 | c.493_501del | p.Arg165_Arg167del | in-frame deletion | not reported | incomplete | yes | – |
| penetrance | |||||||
| Patient-9 | c.914_921del | p.Tyr305Cysfs*155 | frameshift | not reported | NA | – | Bell et al., 2001 |
| Patient-10 | c.930_936dup | p.Tyr313Argfs*152 | frameshift | not reported | de novo | yes | Finegold et al., 2001 |
| Patient-11 | c.943C>T | p.Gln315* | premature stop codon | not reported | de novo | yes | Ghalamkarpour et al., 2009ad |
| SOX18 | |||||||
| Patient-12 | p.Cys240* | premature stop codon | rs74315430c | de novo | yes | Irrthum et al., 2003d | |
| CCBE1 | |||||||
| No mutation found. | |||||||
| PTPN14 | |||||||
| No mutation found. | |||||||
NA = Family members not available.
Unknown mode of inheritance.
Reported by dbSNP135 as probably pathogenic allele.
Patient published by our group.
Discussion
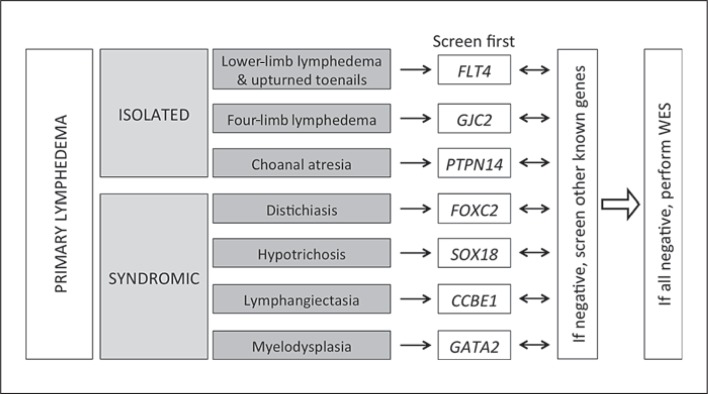
We screened 78 families with primary hereditary lymphedema for 7genes reported to be mutated in some families. We discovered mutations in FLT4 (16 patients mutated/71 samples tested), GJC2 (1/55), FOXC2 (5/64), SOX18 (3/22), GATA2 (1/45), and CCBE1 (2/55). Overall, a mutation was found in one-third (n = 28) of the familial cases (table 1). These numbers are lower when the mode of inheritance is sporadic or unknown. Yet, in 149 such samples tested, a mutation was found in 12 (8%), and all but 3 were conclusively de novo. These included 7 mutations in FLT4 (6.9%, 7/101), 4 in FOXC2 (2.9%, 4/135) and 1 in SOX18 (1.6%, 1/63) (table 1). In aggregate, only about 17% of the primary lymphedema samples can be explained by a change in the 7 genes tested. Thus, when evaluating a patient with primary lymphedema, family history is an important guide to genetic screening. Associated symptoms direct to the most likely gene(s) (fig. 1). For a large proportion of the families, the mutated gene remains unknown.
Fig. 1.
Scheme for the genetic analysis of lymphedema patients. WES = Whole exome sequencing.
We also evaluated possible genotype-phenotype correlations and protein function. The majority (56%) of the lymphedema-causing mutations were discovered in the transmembrane receptor VEGFR3, encoded by FLT4. Most of the mutations are amino acid substitutions that alter conserved residues located in the 2 intracellular tyrosine kinase domains. They can occur either in familial or sporadic cases. The only exceptions were a recessive mutation in the ATP-binding domain (p.Ala855Tyr) for which a homozygous state is required to decrease receptor activity, as well as 2 less common types of FLT4 changes: an in-frame deletion (p.Phe1108del) found in 3 distinct families [Evans et al., 2003; Ghalamkarpour et al., 2006; Connell et al., 2009], and an intronic splice-site change (c.2647+2T>C). By cloning and RT-PCR, it has been demonstrated that a deletion in a similar position of another splice-site (c.2542+2delT, formerly IVS17+2delT) generates an abnormal splicing product: intronic insertion and exon 17 skipping [Futatani et al., 2008]. The phenotype caused by the identified VEGFR3 mutations is as described for Nonne-Milroy disease in earlier reports [Brice et al., 2005; Ghalamkarpour et al., 2006]. Most patients had bilateral congenital lower-limb lymphedema, predominantly on the dorsal feet. In some older patients it progressed higher on the leg. Familial history is not obligatory [Ghalamkarpour et al., 2006], as de novo mutations were identified in 7 patients. Several patients had associated features: hydrocele (n = 5), upturned toenails (n = 4) and saphenous insufficiency (n = 1) (table 4).
Table 4.
Phenotypes associated with mutations in the known lymphedema genes
| Gene | Disease name (OMIM No.) | Type of lymphedema | Associated symptoms |
|---|---|---|---|
| FLT4 | Nonne-Milroy disease (153100) | congenital or early-onset lower limb lymphedema, predominantly at the dorsum of the feet | hydrocele, prominent veins, infection, upturned toenails and hydrops fetalis |
| GJC2 | hereditary lymphedema (613480) | postpubertal lower or upper limb lymphedema | vein insufficiency, blepharoptosis, involvement of face and genitalia |
| FOXC2 | lymphedema-distichiasis syndrome (153400) | pubertal or late-onset lower limb lymphedema | double row of eyelashes, cardiovascular anomalies, cleft palate, blepharoptosis, hydrops fetalis, cystic hygroma and bone defects |
| SOX18 | hypotrichosis-lymphedema-telangiectasia syndrome (607823) | lower limb lymphedema | hypotrichosis, telangiectasia and blepharoptosis |
| GATA2 | primary lymphedema with myelodysplasia (Emberger syndrome) (614038) | lower limb primary lymphedema | hydrocele, predisposition to myelodysplastic syndrome and acute myeloid leukemia, deafness and warts |
| CCBE1 | Hennekam syndrome (235510) | upper and lower limb lymphedema | mental retardation and lymphangiectasia |
| PTPN14 | choanal atresia and lymphedema (613611) | lower limb lymphedema | choanal atresia |
CCBE1 mutations were identified in 2 families, accounting for 4.8% of the mutated patients. In zebrafish, the expression of ccbe1 overlaps with that of vegfc (the VEGFR3 ligand) in the somatic mesoderm [Hogan et al., 2009], and in mice, CCBE1 enhances the lymphatic responses to VEGFC [Bos et al., 2011]. All CCBE1-mutated patients clearly had Hennekam syndrome [Alders et al., 2009], and no change in this gene was found among the sporadic primary lymphedema patients. Thus, future screens of CCBE1 could be restricted to patients with generalized lymphatic dysplasia, with or without facial anomalies, with a recessive mode of inheritance [Alders et al., 2009; Connell et al., 2010] (table 4).
Mutations in the 3 transcription factor genes (FOXC2, SOX18 and GATA2) account for 35% of the mutations identified. The majority result in premature truncation of the protein (tables 2, 3) and likely loss-of-function or dominant-negative effect, assuming that the shortened protein is expressed and stable, and can bind to its target sequences or dimerize with the wild-type protein. Mutations in transcription factors commonly have pleiotropic effects. Accordingly, all the patients exhibited syndromic phenotypes (table 4). FOXC2 patients had lymphedema and distichiasis [Fang et al., 2000; Bell et al., 2001], and patients with mutations in SOX18 had hypotrichosis with lymphedema and telangiectasia [Irrthum et al., 2003]. One patient with GATA2 mutation did not have Emberger syndrome [Hahn et al., 2011; Ostergaard et al., 2011b] (table 4). Patients with a mutation in one of these 3 genes had characteristic features of their respective syndrome; however, the contrary was not always the case. Some individuals with clear signs of these phenotypes were not found to have a change in one of these genes, suggesting technical limitations in detecting all mutations, or more likely, locus heterogeneity.
GJC2 (encoding connexin-47) is mutated in patients with various phenotypes. It causes autosomal dominant, often late-onset lymphedema of lower and upper extremities. Associated symptoms include blepharoptosis, recurrent cellulitis, venous insufficiency, and facial and genital involvement (table 4) [Ferrell et al., 2010; Ostergaard et al., 2011a]. In our sequential series, a GJC2 mutation accounted for only 2.4% of mutated patients (a single family with a mutation). The index case had a late-onset (age 30) bilateral lymphedema, localized to the ankles.
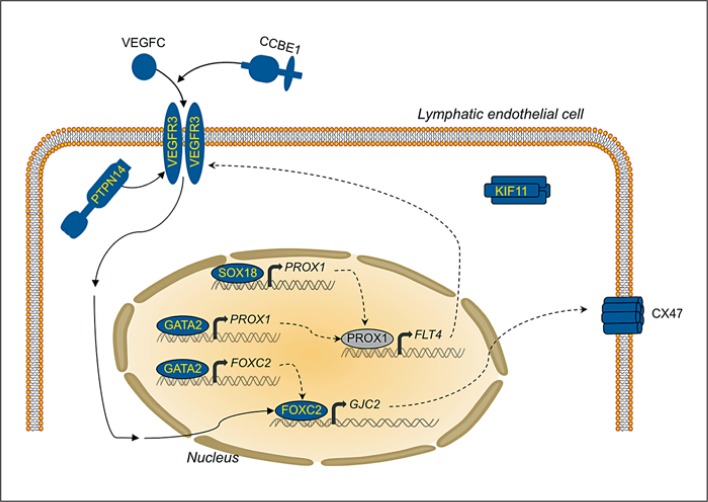
No PTPN14 mutation was found in our series. PTPN14 has a dynamic subcellular localization in vitro. It is nuclear in proliferating cells, but concentrated at intercellular junctions in confluent cells [Wadham et al., 2000; Benzinou et al., 2012]. In vivo, there is an overlap in expression of PTPN14 and VEGFC, and it has been shown that VEGFC stimulation enhances recruitment of PTPN14 to a complex including VEGFR3 (fig. 2). The only mutation reported so far in this gene is a homozygous deletion of exon 7, resulting in loss-of-function in a family with juvenile-onset lymphedema, choanal atresia, developmental delay, and pericardial effusion in some affected individuals [Au et al.,2010]. It may be that this is a very rare private mutation; PTPN14 mutations probably explain only a very small proportion of primary hereditary lymphedema.
Fig. 2.
The functions of the proteins encoded by the genes mutated in primary lymphedema cluster around a central molecular pathway: the VEGFR3 signaling.
VEGFR3 signaling is a theme common to the functions of the proteins encoded by the 9 genes known to be mutated in patients with primary lymphedema (fig. 2). The functional link varies. It can be at the level of the ligand/receptor (VEGFR3, CCBE1), modification of receptor activity (PTPN14), production of the receptor (SOX18 and GATA2 via PROX1), (in)direct control of regulation of transcription (FOXC2, also via GATA2), or a downstream target gene (GJC2, and FLT4 itself). Genetic interactions between the different molecules have been demonstrated in combined heterozygous mice or double knock-down zebrafish [Bos et al., 2011; Cermenati et al., 2013; Hagerling et al., 2013]. The link is less obvious between KIF11, which participates in mitotic spindle assembly, and mutations of which cause MCLMR [Ostergaard et al., 2012]. A comprehensive view suggests that regulation of the VEGFR3 signaling pathway is central to the molecular mechanism of primary lymphedema. This is underscored by the loss-of-function mutation discovered in VEGFC in a family with autosomal dominant Milroy-like primary lymphedema [Gordon et al., 2013].
Conclusion
This study shows that mutations in 7 lymphedema-causing genes explain about one-third of all familial primary lymphedemas. Although this proportion is increased due to the newly discovered genes KIF11 and VEGFC, more than half of hereditary primary lymphedemas remain unexplained. Based on our analyses, the proteins encoded by mutated genes cluster around a central molecular pathway: VEGFR3 signaling (fig. 2). The detailed functional roles among all the players are as yet unclear. It is likely that the genes to be identified will also encode proteins that interact with this central pathway. This concept should help guide discovery of more genes, and their identification will divulge the precise regulation and function of VEGFR3 signaling.
It is clear that primary (hereditary) lymphedema exhibits high locus heterogeneity due to numerous private mutations. The emerging notion that some patients with secondary lymphedema also harbor germline changes in genes that are mutated in primary lymphedema extends this pathogenetic view to explain the more common forms of lymphedema [Finegold et al., 2012]. Whole exome sequencing should help to overcome this heterogeneity and aid in the hunt for the missing genes responsible for primary lymphedema. Someday, pinpointed screening of a complete lymphedema gene panel may become a useful strategy to prevent primary, and secondary, lymphedema.
Acknowledgements
We are grateful to all family members who participated in this study. We thank N. Yilmaz and T. Duchateau for their work during their internship, and the clinicians who sent blood or DNA samples: S. Townshend, A. Quichet, A. Maat-Kievit, P. Jensen, C. Coubes, C. Mignot, F. Inzana, G. Repetto, J.E. Garcia Ortiz, L. Lehtonen, W. Wuyts, G.E. Tiller, L.E. Wehner, M.S. El-Khateeb, F. Blei, J. Perdu, P. Sarda, L. Olivier-Faivre, and K. Karaer. The authors thank Anne Van Egeren, Dominique Cottem, Audrey Debue, and Stephane Messe for their expert technical assistance, Raphaël Helaers for the IT support and Liliana Niculescu for her excellent secretarial help. M.J.S. is a doctoral student supported by the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (FRIA). The authors' studies were partially funded by the Interuniversity Attraction Poles initiated by the Belgian Federal Science Policy, network P7/43; the F.R.S.-FNRS (Fonds de la Recherche Scientifique), all to M.V.; and la Communauté française de Wallonie-Bruxelles and de la Lotterie nationale, Belgium.
References
- 1.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alders M, Hogan BM, Gjini E, Salehi F, Al-Gazali L, et al. Mutations in CCBE1 cause generalized lymph vessel dysplasia in humans. Nat Genet. 2009;41:1272–1274. doi: 10.1038/ng.484. [DOI] [PubMed] [Google Scholar]
- 3.Alders M, Mendola A, Ades L, Al Gazali L, Bellini C, et al. Evaluation of clinical manifestations in patients with severe lymphedema with and without CCBE1 mutations. Mol Syndromol. 2013;4:107–113. doi: 10.1159/000342486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Au AC, Hernandez PA, Lieber E, Nadroo AM, Shen YM, et al. Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans. Am J Hum Genet. 2010;87:436–444. doi: 10.1016/j.ajhg.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell R, Brice G, Child AH, Murday VA, Mansour S, et al. Analysis of lymphoedema-distichiasis families for FOXC2 mutations reveals small insertions and deletions throughout the gene. Hum Genet. 2001;108:546–551. doi: 10.1007/s004390100528. [DOI] [PubMed] [Google Scholar]
- 6.Benzinou M, Clermont FF, Letteboer TG, Kim JH, Espejel S, et al. Mouse and human strategies identify PTPN14 as a modifier of angiogenesis and hereditary haemorrhagic telangiectasia. Nat Commun. 2012;3:616. doi: 10.1038/ncomms1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon LM, Vikkula M. Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, Wolff K. Fitzpatrick's Dermatology in General Medicine. ed 8. New York: McGraw-Hill Professional Publishing; 2012. Vascular Anomalies. [Google Scholar]
- 8.Bos FL, Caunt M, Peterson-Maduro J, Planas-Paz L, Kowalski J, et al. CCBE1 is essential for mammalian lymphatic vascular development and enhances the lymphangiogenic effect of vascular endothelial growth factor-C in vivo. Circ Res. 2011;109:486–491. doi: 10.1161/CIRCRESAHA.111.250738. [DOI] [PubMed] [Google Scholar]
- 9.Brice G, Child AH, Evans A, Bell R, Mansour S, et al. Milroy disease and the VEGFR-3 mutation phenotype. J Med Genet. 2005;42:98–102. doi: 10.1136/jmg.2004.024802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouillard P, Vikkula M. Genetic causes of vascular malformations. Hum Mol Genet. 2007;16:R140–R149. doi: 10.1093/hmg/ddm211. [DOI] [PubMed] [Google Scholar]
- 11.Butler MG, Dagenais SL, Rockson SG, Glover TW. A novel VEGFR3 mutation causes Milroy disease. Am J Med Genet A. 2007;143A:1212–1217. doi: 10.1002/ajmg.a.31703. [DOI] [PubMed] [Google Scholar]
- 12.Butler MG, Isogai S, Weinstein BM. Lymphatic development. Birth Defects Res C Embryo Today. 2009;87:222–231. doi: 10.1002/bdrc.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cermenati S, Moleri S, Neyt C, Bresciani E, Carra S, et al. Sox18 genetically interacts with VegfC to regulate lymphangiogenesis in zebrafish. Arterioscler Thromb Vasc Biol. 2013;33:1238–1247. doi: 10.1161/ATVBAHA.112.300254. [DOI] [PubMed] [Google Scholar]
- 14.Connell FC, Ostergaard P, Carver C, Brice G, Williams N, et al. Analysis of the coding regions of VEGFR3 and VEGFC in Milroy disease and other primary lymphoedemas. Hum Genet. 2009;124:625–631. doi: 10.1007/s00439-008-0586-5. [DOI] [PubMed] [Google Scholar]
- 15.Connell F, Kalidas K, Ostergaard P, Brice G, Homfray T, et al. Linkage and sequence analysis indicate that CCBE1 is mutated in recessively inherited generalised lymphatic dysplasia. Hum Genet. 2010;127:231–241. doi: 10.1007/s00439-009-0766-y. [DOI] [PubMed] [Google Scholar]
- 16.Dagenais SL, Hartsough RL, Erickson RP, Witte MH, Butler MG, Glover TW. Foxc2 is expressed in developing lymphatic vessels and other tissues associated with lymphedema-distichiasis syndrome. Gene Expr Patterns. 2004;4:611–619. doi: 10.1016/j.modgep.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Downes M, Francois M, Ferguson C, Parton RG, Koopman P. Vascular defects in a mouse model of hypotrichosis-lymphedema-telangiectasia syndrome indicate a role for SOX18 in blood vessel maturation. Hum Mol Genet. 2009;18:2839–2850. doi: 10.1093/hmg/ddp219. [DOI] [PubMed] [Google Scholar]
- 18.Evans AL, Bell R, Brice G, Comeglio P, Lipede C, et al. Identification of eight novel VEGFR-3 mutations in families with primary congenital lymphoedema. J Med Genet. 2003;40:697–703. doi: 10.1136/jmg.40.9.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, et al. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet. 2000;67:1382–1388. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrell RE, Baty CJ, Kimak MA, Karlsson JM, Lawrence EC, et al. GJC2 missense mutations cause human lymphedema. Am J Hum Genet. 2010;86:943–948. doi: 10.1016/j.ajhg.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finegold D, Baty C, Knickelbein K, Perschke S, Noon S, et al. Connexin 47 mutations increase risk for secondary lymphedema following breast cancer treatment. Clin Cancer Res. 2012;18:2382–2890. doi: 10.1158/1078-0432.CCR-11-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francois M, Caprini A, Hosking B, Orsenigo F, Wilhelm D, et al. Sox18 induces development of the lymphatic vasculature in mice. Nature. 2008;456:643–647. doi: 10.1038/nature07391. [DOI] [PubMed] [Google Scholar]
- 23.Futatani T, Nii E, Obata M, Ichida F, Okabe Y, et al. Molecular characterization of two novel VEGFR3 mutations in Japanese families with Milroy's disease. Pediatr Int. 2008;50:116–118. doi: 10.1111/j.1442-200X.2007.02505.x. [DOI] [PubMed] [Google Scholar]
- 24.Ghalamkarpour A, Morlot S, Raas-Rothschild A, Utkus A, Mulliken JB, et al. Hereditary lymphedema type I associated with VEGFR3 mutation: the first de novo case and atypical presentations. Clin Genet. 2006;70:330–335. doi: 10.1111/j.1399-0004.2006.00687.x. [DOI] [PubMed] [Google Scholar]
- 25.Ghalamkarpour A, Debauche C, Haan E, Van Regemorter N, Sznajer Y, et al. Sporadic in utero generalized edema caused by mutations in the lymphangiogenic genes VEGFR3 and FOXC2. J Pediatr. 2009a;155:90–93. doi: 10.1016/j.jpeds.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Ghalamkarpour A, Holnthoner W, Saharinen P, Boon LM, Mulliken JB, et al. Recessive primary congenital lymphoedema caused by a VEGFR3 mutation. J Med Genet. 2009b;46:399–404. doi: 10.1136/jmg.2008.064469. [DOI] [PubMed] [Google Scholar]
- 27.Gordon K, Schulte D, Brice G, Simpson MA, Roukens MG, et al. Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant Milroy-like primary lymphedema. Circ Res. 2013;112:956–960. doi: 10.1161/CIRCRESAHA.113.300350. [DOI] [PubMed] [Google Scholar]
- 28.Hagerling R, Pollmann C, Andreas M, Schmidt C, Nurmi H, et al. A novel multistep mechanism for initial lymphangiogenesis in mouse embryos based on ultramicroscopy. EMBO J. 2013;32:629–644. doi: 10.1038/emboj.2012.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn CN, Chong CE, Carmichael CL, Wilkins EJ, Brautigan PJ, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hennekam RC, Geerdink RA, Hamel BC, Hennekam FA, Kraus P, et al. Autosomal recessive intestinal lymphangiectasia and lymphedema, with facial anomalies and mental retardation. Am J Med Genet. 1989;34:593–600. doi: 10.1002/ajmg.1320340429. [DOI] [PubMed] [Google Scholar]
- 31.Hogan BM, Bos FL, Bussmann J, Witte M, Chi NC, et al. Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat Genet. 2009;41:396–398. doi: 10.1038/ng.321. [DOI] [PubMed] [Google Scholar]
- 32.Holme H, Hossain U, Kirwan M, Walne A, Vulliamy T, Dokal I. Marked genetic heterogeneity in familial myelodysplasia/acute myeloid leukaemia. Br J Haematol. 2012;158:242–248. doi: 10.1111/j.1365-2141.2012.09136.x. [DOI] [PubMed] [Google Scholar]
- 33.Hyde RK, Liu PP. GATA2 mutations lead to MDS and AML. Nat Genet. 2011;43:926–927. doi: 10.1038/ng.949. [DOI] [PubMed] [Google Scholar]
- 34.Irrthum A, Karkkainen MJ, Devriendt K, Alitalo K, Vikkula M. Congenital hereditary lymphedema caused by a mutation that inactivates VEGFR3 tyrosine kinase. Am J Hum Genet. 2000;67:295–301. doi: 10.1086/303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irrthum A, Devriendt K, Chitayat D, Matthijs G, Glade C, et al. Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet. 2003;72:1470–1478. doi: 10.1086/375614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishida H, Imai K, Honma K, Tamura SI, Imamura T, et al. GATA-2 anomaly and clinical phenotype of a sporadic case of lymphedema, dendritic cell, monocyte, B- and NK-cell (DCML) deficiency, and myelodysplasia. Eur J Pediatr. 2012;171:1273–1276. doi: 10.1007/s00431-012-1715-7. [DOI] [PubMed] [Google Scholar]
- 37.Karkkainen MJ, Petrova TV. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene. 2000;19:5598–5605. doi: 10.1038/sj.onc.1203855. [DOI] [PubMed] [Google Scholar]
- 38.Karkkainen MJ, Jussila L, Ferrell RE, Finegold DN, Alitalo K. Molecular regulation of lymphangiogenesis and targets for tissue oedema. Trends Mol Med. 2001;7:18–22. doi: 10.1016/s1471-4914(00)01864-5. [DOI] [PubMed] [Google Scholar]
- 39.Kazenwadel J, Secker GA, Liu YJ, Rosenfeld JA, Wildin RS, et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood. 2012;119:1283–1291. doi: 10.1182/blood-2011-08-374363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norrmen C, Ivanov KI, Cheng J, Zangger N, Delorenzi M, et al. FOXC2 controls formation and maturation of lymphatic collecting vessels through cooperation with NFATc1. J Cell Biol. 2009;185:439–457. doi: 10.1083/jcb.200901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostergaard P, Simpson MA, Brice G, Mansour S, Connell FC, et al. Rapid identification of mutations in GJC2 in primary lymphoedema using whole exome sequencing combined with linkage analysis with delineation of the phenotype. J Med Genet. 2011a;48:251–255. doi: 10.1136/jmg.2010.085563. [DOI] [PubMed] [Google Scholar]
- 42.Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011b;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 43.Ostergaard P, Simpson MA, Mendola A, Vasudevan P, Connell FC, et al. Mutations in KIF11 cause autosomal-dominant microcephaly variably associated with congenital lymphedema and chorioretinopathy. Am J Hum Genet. 2012;90:356–362. doi: 10.1016/j.ajhg.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pennisi D, Bowles J, Nagy A, Muscat G, Koopman P. Mice null for Sox18 are viable and display a mild coat defect. Mol Cell Biol. 2000a;20:9331–9336. doi: 10.1128/mcb.20.24.9331-9336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, et al. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 2000b;24:434–437. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- 46.Petrova TV, Karpanen T, Norrmen C, Mellor R, Tamakoshi T, et al. Defective valves and abnormal mural cell recruitment underlie lymphatic vascular failure in lymphedema distichiasis. Nat Med. 2004;10:974–981. doi: 10.1038/nm1094. [DOI] [PubMed] [Google Scholar]
- 47.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 49.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimoda H, Bernas MJ, Witte MH. Dysmorphogenesis of lymph nodes in Foxc2 haploinsufficient mice. Histochem Cell Biol. 2011;135:603–613. doi: 10.1007/s00418-011-0819-x. [DOI] [PubMed] [Google Scholar]
- 51.Sunyaev SR, Eisenhaber F, Rodchenkov IV, Eisenhaber B, Tumanyan VG, Kuznetsov EN. PSIC: profile extraction from sequence alignments with position-specific counts of independent observations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
- 52.Uhlenberg B, Schuelke M, Ruschendorf F, Ruf N, Kaindl AM, et al. Mutations in the gene encoding gap junction protein alpha 12 (connexin 46.6) cause Pelizaeus-Merzbacher-like disease. Am J Hum Genet. 2004;75:251–260. doi: 10.1086/422763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Steensel MA, Damstra RJ, Heitink MV, Bladergroen RS, Veraart J, et al. Novel missense mutations in the FOXC2 gene alter transcriptional activity. Hum Mutat. 2009;30:E1002–E1009. doi: 10.1002/humu.21127. [DOI] [PubMed] [Google Scholar]
- 54.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. Translocation of protein tyrosine phosphatase Pez/PTPD2/PTP36 to the nucleus is associated with induction of cell proliferation. J Cell Sci. 2000;113:3117–3123. doi: 10.1242/jcs.113.17.3117. [DOI] [PubMed] [Google Scholar]
- 55.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 56.Zou Z, Enis DR, Bui H, Khandros E, Kumar V, et al. The secreted lymphangiogenic factor CCBE1 is essential for fetal liver erythropoiesis. Blood. 2013;121:3228–3236. doi: 10.1182/blood-2012-10-462689. [DOI] [PMC free article] [PubMed] [Google Scholar]