Abstract
Aim
Identification of genes that contribute to secondary palate development provide a better understanding of the etiology of palatal clefts. Gene-expression profiling of the murine palate from gestational days 12–14 (GD12–14), a critical period in palate development, identified Sox4 as a differentially expressed gene. In this study, we have examined if the differential expression of Sox4 in the palate is due to changes in DNA methylation.
Materials & methods
In situ hybridization analysis was used to localize the expression of Sox4 in the developing murine secondary palate. CpG methylation profiling of a 1.8-kb upstream region of Sox4 in the secondary palate from GD12–14 and transfection analysis in murine embryonic maxillary mesenchymal cells using Sox4 deletion, mutant and in vitro methylated plasmid constructs were used to identify critical CpG residues regulating Sox4 expression in the palate.
Results
Spatiotemporal analysis revealed that Sox4 is expressed in the medial edge epithelium and presumptive rugae-forming regions of the palate from GD12 to GD13. Following palatal shelf fusion on GD14, Sox4 was expressed exclusively in the epithelia of the palatal rugae, structures that serve as signaling centers for the anteroposterior extension of the palate, and that are thought to serve as neural stem cell niches. Methylation of a 1.8-kb region upstream of Sox4, containing the putative promoter, completely eliminated promoter activity. CpG methylation profiling of the 1.8-kb region identified a CpG-poor region (DMR4) that exhibited significant differential methylation during palate development, consistent with changes in Sox4 mRNA expression. Changes in the methylation of DMR4 were attributed primarily to CpGs 83 and 85.
Conclusion
Our studies indicate that Sox4 is an epigenetically regulated gene that likely integrates multiple signaling systems for mediating palatal fusion, palatal extension and/or the maintenance of the neural stem cell niche in the rugae.
Keywords: cleft palate, CpG methylation, epigenetics, medial edge epithelium, orofacial, rugae, secondary palate, Sox4
Orofacial clefts, such as cleft lip with or without cleft palate (CP) and isolated CP, are amongst the most prevalent of birth defects [1]. Affected individuals encounter a myriad of complications including feeding, dental and speech difficulties; recurrent ear infections or hearing loss; as well as bearing the stigma of their facial appearance in society. Isolated CP primarily arises due to aberrant development of the secondary palate. The key phases in secondary palate development include growth of the palatal processes from the oral aspect of the maxillary process of the first branchial arch, morphogenesis and remodeling of palatal processes resulting in their elevation above the tongue, and subsequent adhesion and fusion of the processes – aspects that are highly conserved in most vertebrates. In mice, these morphogenetic processes occur during gestational days (GDs) 12–14.
Genes essential for secondary palate development encode transcription factors, growth and signaling molecules and their receptors, and extracellular matrix components [2–7], and include members of key signaling systems, such as the Wnt-, TGF-β-, PDGF-, FGF- and Shh-signaling pathways [2,6,8,9]. Cross-talk among these signaling systems in the developing orofacial region dictates the spatiotemporal expression of a diverse array of genes that eventually contribute to the normal development of a secondary palate. Orofacial clefts can be categorized as nonsyndromic or as part of several (as many as 700) syndromes [10]. The genetic contribution to cleft lip with or without CP and CP has been estimated to be between 20 and 50% with environmental mechanisms likely accounting for the remainder [7,11]. Maternal exposure to alcohol, tobacco smoke, drugs, infection and chemicals (e.g., pesticides), and inadequate consumption of nutrients (e.g., trace elements and folate) during pregnancy can be deleterious to the developing embryo or fetus [3,4,7,8,12,13]. Epigenetic mechanisms such as DNA methylation, chromatin modification and/or microRNA expression can influence gene expression by responding to environmental conditions. Thus, it is reasonable to speculate that, for example, altered DNA methylation in susceptible genes during the key phases of palatal development, as a result of environmental toxicity, could contribute to the etiology of CP.
To begin to address the role of epigenetics in regulating ontogenesis of the secondary palate, we conducted a study that identified differentially expressed mRNAs in the murine developing palate (GD12–GD14) [14]. Using high-density microarray-based mRNA profiling, a number of differentially expressed genes, including Sox4, were identified. Sox4 mRNA levels were found to decrease approximately twofold from GD12 to GD13, in the secondary palate, after which their levels remained constant from GD13 to GD14 (Table 1). Very little information exists regarding the expression and/or function of Sox4 in the developing palate. However, as a member of the SOX (SRY-related high mobility group box) family of transcription factors, Sox4 is known to be vital for embryogenesis and cell fate decisions [15,16]. Aberrant expression of Sox4 has been noted in a variety of cancers [16–25], and evidence exists that Sox4 may be involved in cell differentiation, proliferation and survival [15], processes that also underlie tissue and organ development, such as that of the secondary palate. Furthermore, Sox4 is regulated by TGF-β [17,19], a key signaling molecule that plays a critical role in palate development, and defects in which often result in CP [14,26–28].
Table 1.
Sox4 mRNA levels during development of the murine secondary palate as determined by mRNA profiling and confirmed by quantitative RT-PCR analysis.
Sox4 transcript levels on GD13 relative to those on GD12 (set as onefold).
Sox4 transcript levels on GD14 relative to those on GD13 (set as onefold).
Fold change of ≥1.5 and a p-value of ≤0.05 were considered statistically significant (bold).
qRT-PCR: Quantitative real-time PCR.
Data derived from [14].
In the current study, we examined expression of Sox4 within the developing murine secondary palate and investigated whether DNA methylation could account for the differential Sox4 expression observed in this tissue. We show that Sox4 is transiently expressed in the medial edge epithelium (MEE) of the palatal shelf during outgrowth and remodeling, and in the palatal rugae after palatal shelf fusion. We further demonstrate that Sox4 expression during development of the secondary palate is regulated by differential methylation of critical CpG residues localized distal to the CpG island or the putative promoter. This is the first report to demonstrate differential methylation of a gene during development of the secondary palate, and provide support for the hypothesis that normal as well as abnormal palate development may be regulated at an epigenetic level.
Materials & methods
Materials
Sodium bisulfite (catalog number S9000) and Hydroquinone (Cat. No. H9003) were purchased from Sigma-Aldrich Corp.; PCR primers were synthesized by Integrated DNA Technologies, Inc. Unless otherwise specified, all chemicals and reagents were of analytical grade and procured from established manufacturers. The mouse Shh cDNA, cloned in pBSKII and used for the preparation of riboprobes, was a gift from ZJ Lan (University of Louisville, KY, USA). The pCpGL-basic vector was a gift from Michael Rehli (Universitatsklinikum Regensburg, Regensburg, Germany).
Animals
Mature male and female ICR (CD-1) strain mice (Harlan) were maintained in rooms with a 12-h light/dark cycle (20°C, 50% humidity) with ad libitum access to food and water in an Association for Assessment and Accredition of Laboratory Animal Care (AAALAC)-accredited vivarium, and were utilized as per Institutional Animal Care and Use Committee (IACUC)-approved Animal Use Protocols. Timed pregnancies were established by overnight mating of a single mature male with two nulliparous females. The presence of a vaginal plug the following morning was considered evidence of mating and the time considered GD 0 (GD0). At GD12, GD13 and GD14, dams were euthanized by CO2 asphyxiation and embryos were collected in ice-cold calcium/magnesium-free phosphate buffered saline (PBS) and processed immediately.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as described [29]. Murine secondary palates were isolated from fetuses on GD12, GD13 and GD14, transferred to, and rinsed with PBS buffer in 24-well plates, and then immersed in 4% paraformaldehyde overnight at 4°C. Tissues were dehydrated in increasing concentrations of methanol and digested with 10 μg/ml proteinase K for 15 min (GD12), 20 min (GD13) or 25 min (GD14). For detection of Sox4 mRNA expression, tissues were rehydrated and hybridized overnight with either sense or antisense Sox4 riboprobes. Sox4 riboprobes were generated from a plasmid, mSox4/pGEM-T, constructed by cloning an approximately 800-bp cDNA fragment, generated from mouse liver total RNA using Sox4-specific primers, into the pGEM-T Easy vector (Promega). The sense probe was generated after digestion of the plasmid with SalI and transcription with T7 RNA polymerase, whereas the antisense probe was generated using SP6 RNA polymerase after digestion of the plasmid with NcoI. Digoxigenin (DIG)-labeled riboprobes were generated using the DIG RNA labeling kit (Cat. No. 11175025910; Roche). Following extensive washes, using standard protocols, tissues were incubated overnight with anti-DIG AP antibody (Cat. No.11093274910; Roche) and color developed with nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolyl phosphate (Cat. No. 11681451001; Roche). Tissues, in 80% glycerol/0.1% sodium azide in PBS, were visualized and photographed with a Nikon SMZ1500 stereomicroscope equipped with a Nikon DXM1200F digital camera controlled by ACT-1v 2.62 software (Nikon, Inc.).
Sectional in situ hybridization
Sectional in situ hybridization was performed with Shh and Sox4 riboprobes on sagittal sections of fetal murine heads (GD12–GD14) as previously described [29]. The Shh riboprobes were generated from a plasmid harboring the mouse Shh cDNA cloned in pBSKII (see ‘Materials’ section).
Isolation of secondary palates & genomic DNA
Murine secondary palates were isolated, dissected and suspended in ice-cold PBS, as previously described [30]. The developmental stage of the secondary palates was determined to ensure sample consistency among the biological replicates (GD12 – palatal processes vertically oriented on either side of the tongue; GD13 – palatal processes larger and in the process of reorientation, or horizontally oriented above the tongue but unfused; GD14 – palatal processes fused to each other). Genomic DNA was isolated from 20–40 mg of pooled secondary palatal tissue for each GD using the QIAamp DNA Mini kit (Cat. No. 51304; Qiagen Inc.).
Bisulfite conversion, DNA sequencing & determination of Sox4 CpG methylation profiles
A total of 10–15 μg genomic DNA was digested with EcoRI (that does not digest the genomic region under investigation) and subjected to the ‘bisulfite’ reaction in the presence of 3.1 M sodium bisulfite, pH 5.0 and 0.5 mM hydroquinone at 55°C for 16 h under mineral oil [31]. DNA was purified using the QIAquick PCR purification kit (Cat. No. 28104; Qiagen), denatured in 0.3 M NaOH, ethanol precipitated, dissolved in Tris-EDTA buffer, pH 8 and used for PCR amplification using primers specific for the bisulfite-modified DNA. Primers (Table 2) were designed using the BiSearch program [32]. Each amplicon was ligated into pGEM-T Easy vector (Cat. No. A1360; Promega) and transformed into Escherichia coli DH5α competent cells. Approximately ten random colonies were selected for plasmid DNA preparation using the Spin Miniprep kit (Cat. No. 27106; Qiagen), sequenced at the University of Louisville Sequencing Core facility, and methylated CpG residues identified. CpG methylation level was defined as the percentage of methylation of a CpG residue in all clones analyzed. Methylation level of an amplicon was defined as the percentage of methylated CpG residues present at all CpG sites in all clones analyzed.
Table 2.
Primers used to determine the CpG methylation profile of the murine Sox4 upstream region.
| Primer | Sequence | Amplicon number (size) | Total number of CpG sites |
|---|---|---|---|
| Sox4-9 | 5′-TAATCTATTATACCATAACCCCR | 1 (317 bp) | 39 |
| Sox4-325 | 5′-GGTGGGGGAAAGATTTGTTT | ||
|
| |||
| Sox4-591 | 5′-GGTAGAGTTTTTTTATTGTAG | 2 (324 bp) | 13 |
| Sox4-914 | 5′-CAACTATACTTCTAACCCTTTTA | ||
|
| |||
| Sox4-899 | 5′-GTTAGAAGTATAGTTGAGAT | 3 (153 bp) | 16 |
| Sox4-1051 | 5′-ATACATTAACATCCTCTTCA | ||
|
| |||
| Sox4-1049 | 5′-TATGAAGTTATTAAGGGGAGA | 4 (397 bp) | 3 |
| Sox4-1445 | 5′-AAAAAAAAACCCTACCTTTC | ||
|
| |||
| Sox4-1786 | 5′-TAAAAGATTAAGAAGAGAAAGGG | 5 (339 bp) | 9 |
| Sox4-2124 | 5′-CAAACRAAAACCACCAAAAAAAC | ||
|
| |||
| Sox4-2098 | 5′-GAAAGTTTTTTTGGTGGTTTT | 6 (400 bp) | 8 |
| Sox4-2497 | 5′-AATCCCAATACTCAAATCTT | ||
Primers are listed pairwise. Both primers when used together give rise to the indicated amplicon product.
Construction of Sox4 expression vectors & determination of promoter activity in murine embryonic maxillary mesenchymal cells
Since the murine Sox4 promoter was not characterized, an approximately 1.8-kb DNA sequence (accession number: NC_000079.6, NCBI) upstream of the translational start site (ATG) was analyzed. A fragment harboring the 1.8-kb sequence was amplified by PCR using mouse genomic DNA and primers tagged with NheI and BglII restriction sites. After digestion with NheI and BglII, the resultant fragment was cloned into NheI and BglII digested pGL3-Basic luciferase vector (Promega). After transformation into E. coli DH5α cells, colonies harboring the plasmid were identified, the sequence verified by DNA sequencing and the plasmid (termed Sox4/pGL3) was prepared using Qiagen's Plasmid Maxi kit (Cat. No. 12163). A total of 2 μg/ml of the plasmid containing the Sox4 promoter (Sox4/pGL3) and 50 ng/ml of a control vector (pRL-CMV; Cat. No. E2261; Promega) expressing Renilla luciferase were transfected into freshly isolated murine embryonic maxillary mesenchymal (MEMM) cells, prepared essentially as described [33]. Transfection was performed in 2-ml media containing 3 × 105 cells using FuGENE6 Reagent (Cat. No. 11815091001; Roche) in six-well plates. Cells were harvested after 48 h, and firefly and Renilla luciferase activities were determined using the Dual Luciferase Reporter Assay system (Cat. No. E1910; Promega) in a Turner 20/20 luminometer (Promega). Normalized luciferase activities (ratio of firefly to Renilla luciferase activities) were determined. The experiment was repeated three-times with duplicates and expressed as mean ± SD. The reporter activity of Sox4/pGL3 was expressed relative to a similarly transfected control vector, pGL3-Promoter vector (Cat. No. E1761; Promega), whose luciferase activity is under the control of the SV40 promoter. The vector without both Sox4 differentially methylated regions (DMRs; Sox4Δ/pGL3) was constructed by removal of an 854-bp fragment from the 5′ end of Sox4/pGL3 and then religating the vector fragment. This was accomplished by digesting Sox4/pGL3 with NheI, blunting the ends with Klenow polymerase, digesting with PvuII, and religating the resultant vector fragment. Additionally, site-specific mutations were introduced into CpGs 83 and 85 in Sox4/pGL3 using the QuikChange® site-directed mutagenesis kit (Cat. No. 200519–5; Stratagene) to generate Sox4Mut83/pGL3 and Sox4Mut85/pGL3, respectively. CpG residues were converted to CpT and the mutated sequence was confirmed by DNA sequencing. Transfection was performed four times, in duplicate. Statistical significance was analyzed by unpaired, two-tailed Student's t-test using GraphPad software (GraphPad Software, Inc., CA, USA). Values are expressed as mean ± SD.
Methylation analysis of the Sox4 DMR
Sox4/pGL3 was digested with NheI, blunt-ended with Klenow polymerase and then digested with BglII to release the approximately 1.8-kb Sox4 promoter-containing fragment. The fragment was cloned into pCpGL-Basic vector [34], which was first digested with BamHI, then blunt-ended and digested with BglII to generate compatible cloning ends. The resultant plasmid, referred to as Sox4/pCpGL, was transformed into E. coli PIR cells (Cat. No. C1010-10; Invitrogen) and colonies harboring the plasmid were identified. Plasmid DNA was prepared using Qiagen's Maxiprep kit after growth in PIR cells. The advantage of using the pCpGL vector is that CpG residues to be methylated are contributed only by the insert (i.e., the 1.8-kb Sox4 upstream region), as the vector harbors no CpG residues [34]. Thus, any effect on promoter activity can be directly attributed to CpG residues present in the insert and not to the vector. This also facilitates methylation of the entire plasmid, obviating the need for first isolating and methylating the insert, then religating the methylated insert back into the vector, followed by isolation and purification of the ligated plasmid for transfection analysis. Sox4/pCpGL was methylated using SssI methylase (Cat. No. M02263; New England BioLabs), according to the protocol provided by the manufacturer. The efficacy of DNA methylation was assessed by digestion of the unmethylated and methylated plasmids with AclI (Cat. No. R0598S; New England BioLabs) and BstUI (Cat. No. R0518S; New England BioLabs). The plasmids were purified using Qiagen's PCR kit and the DNA used for transfection in MEMM cells, as described above. Transfection was carried out four-times using duplicate wells. Statistical significance was analyzed by unpaired, two-tailed Student's t-test using GraphPad software. Values are expressed as mean ± SD.
Results
Spatiotemporal expression of Sox4 in the developing murine secondary palate
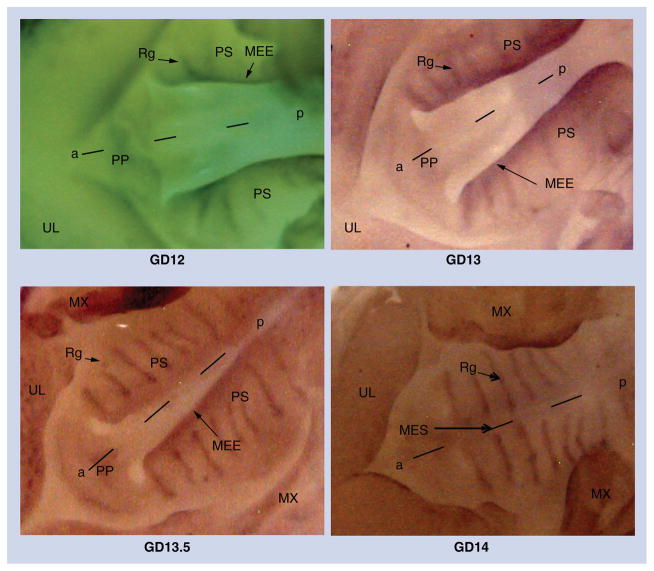
The expression of Sox4 in the developing secondary palate (GD12–GD14) was determined using Sox4 riboprobes. On GD12 (Figure 1), Sox4 expression was visible in the MEE of the palatal processes, with streams of Sox4-expressing cells appearing to ‘migrate’ from the MEE toward regions of the prospective rugae (Figure 1, compare GD12 and GD13). From GD13 to 13.5, Sox4 expression was predominantly observed in the rugae (8–9 in number), and its expression in the MEE began to dissipate. On GD14, upon palatal fusion, Sox4 expression was localized only in the rugae with no traces of expression in the medial edge seam (MES).
Figure 1. Whole-mount in situ hybridization with Sox4 riboprobes on GD12–GD14 palatal processes.
Sox4 expression in the developing murine secondary palate was determined from GD12 to GD14 using antisense riboprobes. On GD12 and GD13, Sox4 expression is visible in the MEE and the developing rugae. At GD13.5, Sox4 expression is more distinct in the rugae and expression persists at the medial edge epithelium. After palatal fusion on GD14, expression is confined only to the rugae with no evidence of expression at the MES where the palatal shelves have fused. Representative photomicrographs depict ventral views of the oral region of GD12–GD14 mouse embryos subsequent to removal of the mandible and tongue.
a–p: Anteroposterior axis; MEE: Medial edge epithelium; MES: Medial edge seam; MX: Maxilla; PP: Primary palate; PS: Palatal shelf; Rg: Ruga; UL: Upper lip.
Sectional in situ hybridization of Sox4 expression in the developing murine secondary palate
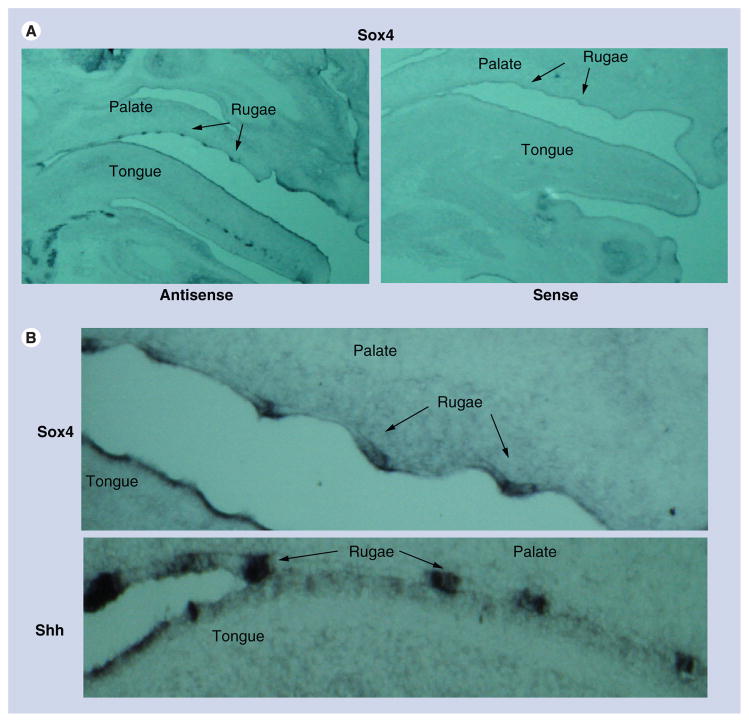
Coronal sections of secondary palates indicated intense Sox4 expression in the inferior margins of vertically directed palatal processes (arrows; (presumptive MEE) on both GD12 and GD13 (Figure 2A & B). Sagittal sections of GD14 secondary palates indicated that Sox4 expression in the secondary palate was confined to the palatal rugae (Figure 3A). Comparison of Sox4 expression in the rugae to that of Shh, which encodes a secreted signaling protein expressed primarily in the rugae epithelium, indicated that Sox4 expression was localized to the epithelial thickenings of the palatal rugae (Figure 3B).
Figure 2. Sectional in situ hybridization with Sox4 riboprobes on coronal sections of GD12 and GD13 palatal processes.
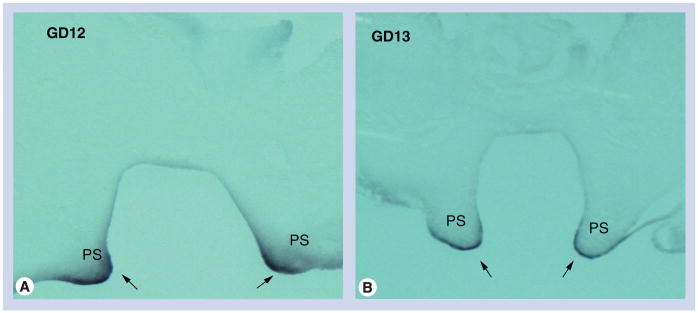
Coronal sections of secondary palates from (A) GD12 and (B) GD13 mice were hybridized with Sox4 antisense riboprobes. Representative photomicrographs indicate intense Sox4 expression in the inferior margins of vertically directed palatal processes (presumptive medial edge epithelium) on both GD12 and GD13 PS (arrows).
PS: Palatal shelf.
Figure 3. Sectional in situ hybridization with Shh and Sox4 riboprobes on sagittal sections of GD14 oral/facial region and palatal processes.
(A) Sagittal sections of oral/facial region from GD14 mice probed with Sox4 sense and antisense riboprobes. Representative photomicrographs indicate that Sox4 expression, in the secondary palate, is specifically localized to the rugae of the palate on GD14. (B) Sagittal sections of secondary palates from GD14 mice probed with Sox4 and Shh antisense riboprobes. Similar to Shh (which is expressed in the rugae epithelium to direct anteroposterior extension of the palate), Sox4 is also found to be expressed in the epithelial thickenings of the palatal rugae.
Sox4 mRNA levels during development of the murine secondary palate
Using mRNA gene-expression profiling strategies, Sox4 was found to be differentially expressed in developing palatal tissue, and was thus considered as a candidate gene contributing to genesis of the mammalian palate [14]. Sox4 mRNA levels decreased twofold from GD12 to GD13 in the secondary palate, and subsequent expression remained unchanged in the tissue from GD13 to GD14 (Table 1). This expression pattern could be hypothesized as being attributed to increased methylation of the 5′ flanking/promoter regions of Sox4 in the secondary palate from GD12 to GD13, followed by methylation levels remaining relatively stable between GD13 and GD14.
CpG methylation profiling of the Sox4 5′ flanking region
To assess if methylation of the 5′ flanking/promoter regions of Sox4 could account for the observed mRNA changes during palatogenesis, genomic DNA was isolated from GD12, GD13 and GD14 murine secondary palates and subjected to bisulfite modification. Approximately 1.8 kb of the 5′ flanking region was analyzed by PCR amplification of bisulfite-modified DNA. Six different amplicons (amplicons 1–6) were generated spanning the 5′ flanking region, with amplicon 1 being the most proximal. The methylation level of each amplicon in GD12, GD13 and GD14 palatal tissue was compared (Figure 4). Amplicon 1 partially localized within the CpG island (determined by CpG plot [101]) was unmethylated in the tissue on all 3 days of gestation, with methylation levels ranging between 0.6 and 1.5%. Likewise, amplicon 2 was unmethylated in the tissue on all 3 GDs (0–0.8%). By contrast, the distal amplicons (amplicons 5 and 6) were almost completely methylated, exhibiting 86–90% and 96% methylation levels, respectively. No significant differences in methylation levels were observed for amplicons 1, 2, 5 and 6 from GD12 to GD14 (significant changes in methylation levels were based on an arbitrary cutoff of ≥25%). Amplicons 3 and 4, however, localized between the highly methylated and the poorly methylated regions of the 5′ flanking/promoter regions of Sox4, exhibited prominent differences in methylation levels between GD12 and GD13, but not between GD13 and GD14. Methylation levels of these two amplicons, however, diverged in opposite directions – that of amplicon 3 decreased from 33 to 14% (19% change), whereas that of amplicon 4 increased from 70 to 100% (30% change) between GD12 and GD13 (Figure 4). Notably, none of the six amplicons exhibited significant differences in methylation levels between GD13 and GD14, suggesting that methylation levels had stabilized by GD13. It is interesting to note that the observed methylation changes are not randomly distributed, but rather localized to regions of close proximity (i.e., in adjacent amplicons 3 and 4) on GD12 and GD13. Amplicons 3 and 4 were, hereafter, termed DMR3 and DMR4, respectively. DMR3 and DMR4 contained 16 and 3 CpG residues, respectively (Box 1 & Table 3). Since methylation of CpG residues in upstream/promoter regions is typically associated with gene silencing, we hypothesized that the increase in methylation of CpG residues in DMR4 from GD12 to GD13 (Table 3) could account for the decrease in Sox4 mRNA levels observed during this period (Table 1).
Figure 4. CpG methylation profiling of the 5′ upstream region of Sox4 in the developing murine secondary palate.
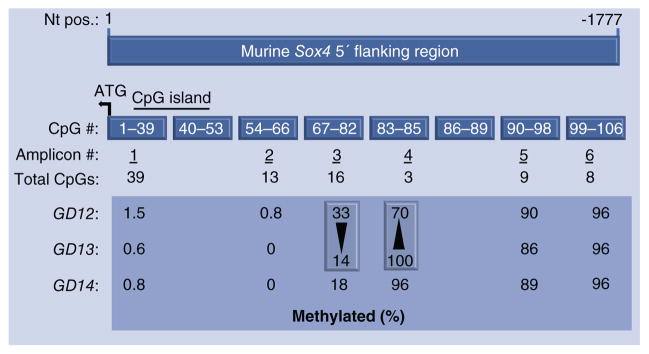
A 1777 nt region upstream of the translational start site (+1) of murine Sox4 (accession number: NC_000079.6, NCBI) was used for CpG methylation profiling. After ‘bisulfite’ treatment of genomic DNA, six PCR amplicons were generated, spanning the 5′ upstream region. The 1777 nt region contained a total of 106 CpG residues, numbered sequentially from nt +1. Amplicon 1 represents the most proximal region and includes CpGs 1–39. A CpG island (horizontal bar) spans CpG residues 13–51. Regions containing CpGs 40–53 and 86–89 were not amplified (see text). The large box compares CpG methylation levels (%) in genomic DNA derived from secondary palates on GD12, GD13 and GD14. The two vertical boxes indicate the two amplicons exhibiting the most pronounced changes in methylation levels between GD12 and GD13 (note that methylation levels between GD13 and GD14 for these two amplicons are almost identical). Notably, amplicon 3 exhibited a decrease in methylation (33–14%) and amplicon 4 exhibited an increase (70–100%). The increase in methylation in amplicon 4 was consistent with decreasing mRNA expression observed between GD12 and GD13.
Table 3.
Methylation levels of individual CpG residues in amplicons 3 and 4 of Sox4 5′ upstream region.
| CpG residues | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||||
| Amplicon 3 | Amplicon 4 | ||||||||||||||||||
| Number | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 |
|
| |||||||||||||||||||
| GD12 | 0 | 0 | 0 | 0 | 0 | 70† | 90† | 90† | 60† | 60† | 50† | 70† | 20 | 10 | 0 | 0 | 60† | 80† | 70† |
|
| |||||||||||||||||||
| GD13 | 0 | 0 | 0 | 0 | 0 | 20† | 40† | 40† | 30† | 30† | 20† | 30† | 20 | 0 | 0 | 0 | 100† | 100† | 100† |
|
| |||||||||||||||||||
| GD14 | 0 | 0 | 10 | 0 | 0 | 30 | 30 | 40 | 40 | 40 | 40 | 70† | 11 | 0 | 0 | 0 | 89 | 100 | 100 |
CpG residues that exhibit >25% change in methylation levels between adjacent gestational days.
Box 1. Localization of amplicons 3 and 4 and key CpG sites in the 5′ upstream region of Sox4.
| 5′:agcacagacacacgggagacaatgggtaagaaagagagggggaaggagagcacatagcgaacccatctatcctttggatattgtgga aaacaaacatttagaaagtgaagtcaatcccatcctgtattcaccatgatcgccctatacagaaatcacacaaagtctcttattgaggtaattatg ggccactgccttagcgttccacagaaaatcttgtttggcctacccaggatttataggggaagtgaaatcattaaataactagttaacctctgttt ggcaaaaagagtaagtaattactgaaaaggggagaaagaaaaaaa>>4aaaaaaaaaccctgcctttcacaaacagcacagatagcatc ctgaagggatttacacaatacaccaacg85tttgttctgtacccagagttgttaaaataattgtgatgacaggtgtctaggtaccactttcttaat ggttttcaatatccattgatttattattgtggctccccccacttacatctctagctctaagaaggggctaagaaaagagattttgcaccaaaggct gattctttattcctgggctgaaaagaactaataaaaataatcactcg84gcttttaagcagaaagcttccatttacagtatttcacccttacccccc ctccaagccccttgttatctaacataatggctagaatacatttccg83tctctccccttaataacttc>>3atg4<<cattaacatcctcttcacac atgcgcgtgcgtgcgcg78cg77cg76cg75cg74cg73cg72cacacacacacacacacacacagaggcaaacgcgcgcgcacccagcaaa aggcggaggtgggggggaaagaggcagcagaaatctcagctgtacttctagc3<<ccttttaaaaagtttgctctgtaaattggaatgag gtcagatttggagcttctcattgcacgcggagattattattgcatcgggttccaagccaatgggaaggacgggggaggggcttggcacgag gaagcgttagttacagcggctgattggctgtctgagaaaggataaagaagcgcgaggcggaattggggtctgctctaagctgcagccaga gaaacagagtgaggggaagagggccgtctccctcccggtttccagttcttgcacgctgtttcttagagagtctgcagtgggggaactctgcc ggtaaccagctccccttcttgcaggagggagggagaaacatacatttattcatgccggtctgttgcatgcaagcttcttggcttcctaccttgca acaaaataattgcaccaactcctcagcgccgattccgcccacagagagtcccggagccagagtcgctttggctttgcactgcaggaaaggg acttaggcgctagagacgatgtcgctttcctgagctaccgcgagctctcgtgaactgcaatcgactgcttcagggaaaggggtgggggaaa gacttgccccggaggcggcgagaaacttgcgtttggaagatactccggctaccaacgtttggagaaactcctcgccgcggccgactccag cctcggtgcccgcagggagcacttcagcggtgagggaggacggagggcctcggggactctaggttggcggcgggaggcggccgcccc tggcccgcgcgccgctcaggggaccctggtctccgccccggggagcccgcagggcccggcgaccgcgccgcgagcgtgtgagcgcgc gtgggcgcccggcaag:3′ |
Bold indicates CpG sites.
Bold and underline indicate CpG sites of amplicons 3 and 4 exhibiting differential methylation.
>>3: Start of amplicon 3; >>4: Start of amplicon 4; 3<<: End of amplicon 3; 4<<: End of amplicon 4.
Examination of methylation levels of individual CpG residues constituting the two DMRs revealed that changes in methylation levels of all CpG residues from GD12 to GD13 were in the same direction as that of the cognate DMR (Table 3). For DMR3, seven contiguous CpG residues (CpGs 72–78) demonstrated decreases of 30–50% in methylation levels, whereas the remaining nine residues showed relatively little change (<10%). Methylation levels in DMR4 increased from GD12 to GD13 for all three CpG residues – CpGs 83–85 (Table 3) – with the most pronounced increases being in CpGs 83 (40%) and 85 (30%); CpG 84 exhibited a 20% increase. These data provide support for the hypothesis that the decrease in Sox4 mRNA levels observed in the secondary palate from GD12 to GD13 (Table 1) was functionally correlated with the pronounced increases in methylation of CpG83 and CpG85. The methylation status of CpG residues 40–53 and 86–89 (Figure 4) could not be analyzed in this study, because these regions were presumably too degenerate to be amplified by PCR after bisulfite modification, a problem frequently encountered in bisulfite sequencing [35].
Functional analysis of DMR4, CpG83 & CpG85 on Sox4 promoter activity
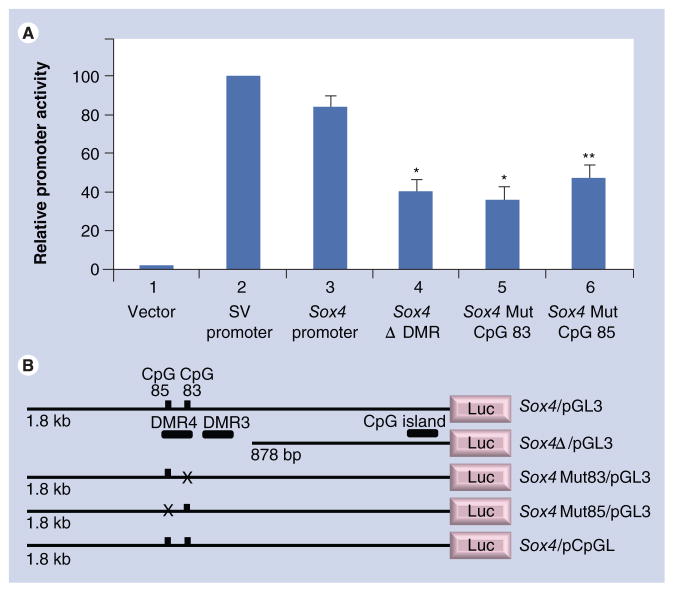
To determine if Sox4 DMR4 (containing CpGs 83 and 85) had any functional role in regulating gene expression, various deletion and mutant constructs were generated in the pGL3 (Promega) vector. Promoter activity was determined by transfection analysis in MEMM cells (Figure 5). The normalized promoter activity of the Sox4/pGL3 construct, which contains the 1.8-kb Sox4 5′ flanking region, was approximately 84% when compared with that of the positive control, pGL3-SV40 promoter (Figure 5A; #3 versus #2). The negative control (pGL3-Basic vector) exhibited negligible (1.5%) activity (Figure 5A; #1). The significant promoter activity of the Sox4/pGL3 construct suggested that it harbored the Sox4 promoter, which was most likely located within the CpG island, immediately flanking the gene (Figure 4). When an approximately 854-bp fragment containing both Sox4 DMRs was removed from the 5′ end of Sox4/pGL3 (Sox4Δ/pGL3), promoter activity dropped significantly to 40% (Figure 5A; #4). This decrease in promoter activity could be attributed to the loss of critical CpG residues residing in either or both DMRs and/or other sequence elements contained within the deleted region.
Figure 5. Functional analysis of CpG83 and CpG85 on Sox4 promoter activity.
Various Sox4 constructs in luciferase reporter vectors were transfected into murine embryonic maxillary mesenchymal cells. (A) Values in the y-axis represent normalized luciferase activities. The Sox4 reporter activities of all constructs were compared with that of the SV40 promoter (column #2; taken as 100%). Plasmids used for transfection included: (#1) pGL3 vector; (#2) SV40/pGL3; (#3) Sox4/pGL3; (#4) Sox4Δ/pGL3; (#5) Sox4Mut83/pGL3; and 6) Sox4Mut85/pGL3. Note the significant decrease in promoter activities of the last three constructs (#4–#6) when compared with that of Sox4/pGL3 (#3). (B) Indicates the salient features of the Sox4 plasmid constructs utilized. The location of the two differentially methylated regions and the CpG island is indicated in Sox4/pGL3, which harbors the 1.8-kb 5′ upstream region of Sox4. X denotes mutated CpG sites. Sox4/pCpGL was used for methylation analysis and harbors the 1.8-kb 5′ upstream region of Sox4 (only CpGs 83 and 85 are indicated in the figure). Transfections were repeated four times using duplicates and values are presented as mean ± standard deviation.
*p< 0.0001; **p < 0.0002.
DMR: Differentially methylated region; Mut: Mutation; SV: Simian virus 40.
To assess if CpGs 83 and/or 85, specifically, could contribute to Sox4 expression, a single nucleotide change was introduced in these two residues to generate Sox4Mut83/pGL3 and Sox4Mut85/pGL3, respectively (Figure 5B). When the mutant constructs were transfected into MEMM cells, promoter activity decreased to 36% for CpGMut83 (Figure 5A; #5) and 47% for CpGMut85 (Figure 5A; #6), compared with the unmutated construct (Figure 5A; #3). These results suggest that CpGs 83 and 85 contribute significantly to Sox4 expression, with the caveat that the substitution mutations did not create novel binding sites for proteins inhibiting promoter function.
Methylation analysis of CpGs 83–85
Sox4/pCpGL, containing the 1.8-kb 5′ flanking promoter fragment, was methylated in vitro using SssI methylase. Methylation of Sox4/pCpGL was assessed by digestion with AclI and BstUI (Figure 6A), which recognize the sequences AACGTT and CGCG, respectively (CpG85 is one of two sites targeted by AclI in the vector). As expected, digestion of the unmethylated plasmid with AclI released an insert of 1.2 kb size (lane 5), whereas digestion with BstUI released multiple (>10) fragments ranging in size from 10 to 356 bp (lane 3). None of these bands were apparent after digestion of the methylated plasmid (lanes 2 and 4), indicating complete methylation of CpG sites within the approximately 1.8 kb 5′ upstream regions of Sox4. Methylated Sox4/pCpGL was then transfected into MEMM cells and its promoter activity compared with the unmethylated Sox4/pCpGL construct. Methylation completely abolished promoter activity (Figure 6B), reducing it to the level of the empty pCpGL-Basic vector. These data clearly indicate that CpG residues 83 and 85 constitute a critical functional component, regulating the Sox4 promoter.
Figure 6. Effect of methylation of the 5′ upstream region on Sox4 promoter activity.
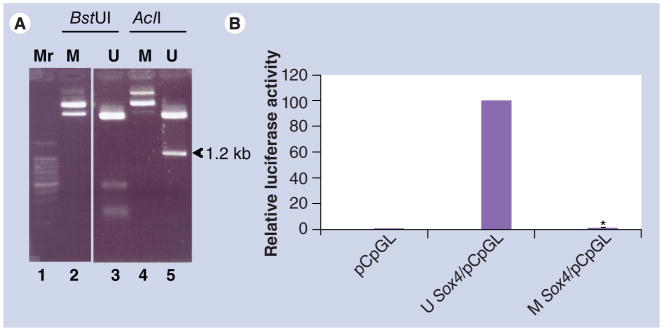
(A) Sox4/pCpGL harboring the 1.8-kb 5′ upstream region of Sox4 (see Figure 5B) was methylated in vitro using SssI methylase. Methylation of the plasmid was assessed by digesting the unmethylated and methylated plasmids with BstUI and AclI. The methylated plasmids were resistant to digestion (lanes 2 and 4), whereas the unmethylated plasmids (lanes 3 and 5) released expected digestion products. CpG85 is one of only two sites in Sox4/pCpGL that can be digested by AclI. The figure clearly indicates that CpG85 is fully methylated, as evidenced by the absence of the 1.2-kb digestion product from the methylated plasmid (lane 4) when compared with that derived from digestion of the unmethylated plasmid (lane 5). (B) Normalized promoter activities of the methylated and unmethylated plasmids after transfection into murine embryonic maxillary mesenchymal cells are presented. The relative promoter activity of the unmethylated plasmid was set at 100%. The experiment was repeated four times using duplicates and values are presented as mean ± standard deviation (*p < 0.0001).
M: Methylated; Mr: Marker; U: Unmethylated.
Discussion
Deepening our understanding of factors that contribute to both normal and abnormal development of the secondary palate requires an appreciation of the molecular mechanisms governing regulation of gene expression during palatal ontogeny. Toward this end, mRNA profiling of developing (GD12–GD14) murine secondary palatal tissue identified a number of differentially expressed genes [14]. The present study focused on one of these genes – Sox4 – and examined one potential means by which its expression is regulated in tissue of the developing palate. Expression profiling suggested that the number of Sox4 transcripts decreased approximately twofold from GD12 to GD13 in the secondary palate, and that levels remained steady thereafter from GD13 to GD14 (Table 1) [14]. Sox4 encodes a transcription factor that is frequently overexpressed in a variety of cancers, and investigators have suggested that this implies a crucial role in cell proliferation, differentiation and/or apoptosis [15–25]. Members of the SOXC family (SOX4, SOX11 and SOX12) share molecular properties and functionally interact with each other [36]. Inactivation of Sox11 is known to result in CP, cleft lip and cleft lip with CP [37,38]. Indeed, when a mouse strain bearing a Sox4 mutation was bred in trans with another bearing a Sox4 deletion, 60% of offspring exhibited CP [39]. Furthermore, an extensive review of human linkage studies and mouse models of orofacial clefting led Juriloff and Harris to suggest that SOX4 is a strong candidate for human nonsyndromic cleft lip with CP [3]. SOX4 is vital for fetal development since Sox4−/− mice do not survive beyond E14 due to cardiac developmental defects [40]. Sox11−/− mice, however, die soon after birth, and Sox12−/− mice are viable and exhibit no obvious defects [41]. SOX4 has also been demonstrated to be essential for the development of the pancreas, bone, lymphocytes and the CNS [15,42,43]. It also regulates neuronal differentiation [44] and is required for the survival of neural and mesenchymal progenitors [38]. Collectively, these studies emphasize a role for Sox4 in cell fate decisions and provide a compelling rationale for examining the expression of Sox4 in the murine secondary palate [16]. In the current study, we focused on the questions of: whether Sox4 exhibits a differential spatiotemporal expression pattern in the developing secondary palate; and whether changes in DNA methylation of Sox4 are functionally related to the differential expression of Sox4 mRNA observed in the developing secondary palate. Genes that are regulated by DNA methylation during palatal development represent potential epigenetic targets for environmental factors that contribute to palatal clefting.
The palatal processes that give rise to the secondary palate arise bilaterally from the oral aspect of the maxillary prominence of the first branchial arch [8]. The morphological features of the murine palate begin to take visible shape at approximately GD11, and are more clearly defined on GD12, at which time the pair of processes are vertically positioned on either side of the tongue. On GD13, the palatal processes undergo a morphogenetic movement that brings them to a horizontal position, apposed to each other, above the tongue. On GD14, the MEE of each palatal process fuses medially along the anteroposterior axis to form the MES. Failure of the palatal processes to grow, reorient or fuse can result in CP. The precise mechanisms that contribute to mesenchymal confluency during palatal fusion are not clearly understood, but the general consensus is that this complex process involves epithelial–mesenchymal transition, apoptosis and cell migration [8]. The expression of Sox4 in branchial arch mesenchyme [41] and in the MEE just prior to palatal fusion (GD12–13.5; the current study), and its absence in the MES on GD14 (Hoser et al. [41] and this study), suggest that it may play a role in palatal MEE fusion. Our observations are consistent with the earlier study [41], when comparing equivalent developmental stages of the palate. Studies of SoxC single and double null mutants indicate that Sox4 participates in early functions and Sox11 in later functions in the survival of neural and mesenchymal precursors [45]. Collectively, these data are consistent with a role for members of the SOXC family, particularly SOX4, in both palatal mesenchymal as well as epithelial differentiation.
Sox4 is overexpressed in a variety of cancers [16–25] implying a central role in cell proliferation, differentiation and/or apoptosis, processes that also play significant roles in organogenesis, including that of the secondary palate. TGF-β signaling is known to increase Sox4 expression [17,19]. Indeed, SOX4 has been demonstrated to be part of a TGF-β–Sox4–Sox2 signaling axis required for maintaining stemness [17]. GD12–GD13 could therefore represent a critical juncture in palate development when diminution of Sox4 signaling is required for the differentiation of progenitor cells in the MEE. This potential role for Sox4 is compatible with the currently understood role TGF-β plays in palate development [14,26–28]. The importance of the TGF-β superfamily in palatal development is highlighted by the fact that inactivation or deletion of genes encoding family members such as TGF-βs, BMPs and their signaling mediators (Smads) results in a plethora of orofacial abnormalities [27,46,47].
Evidence for a specific role for TGF-β1 and TGF-β3 in palatal MEE differentiation has recently been presented. While TGF-β1 contributes to cell-cycle arrest in the MEE, TGF-β3 is necessary for MEE apoptosis and subsequent epithelial–mesenchymal transition [48,49]. The expression profile of Sox4 in the developing palate parallels that observed for TGF-β1 – i.e., expression in the MEE epithelium decreases significantly prior to MES formation, and is completely absent in the MES. Moreover, TGF-β is known to play a significant role in palate mesenchymal growth, eliciting changes in orofacial cell proliferation, as well as synthesis and remodeling of the extracellular matrix [50–53]. If, therefore, as has been shown elsewhere [17,19], Sox4 is a target for TGF-β signaling in the secondary palate, then a role for Sox4 in both MEE differentiation as well as mesenchymal growth can be considered.
The pattern of Sox4 expression in the developing secondary palate is suggestive of a migratory phenotype – that is, Sox4-expressing cells follow a defined path from the MEE on GD12 to the rugae on GD14 (Figure 1). While our study does not confirm that the spatiotemporal change in Sox4 expression is due to cells migrating from the MEE to the rugae, it nevertheless is supported by evidence that palatal MEE cells are indeed migratory [54]. BMP7 and FGF signaling contribute to the formation of rugae [55,56], which serve as Shh signaling centers that mediate the extension of the palate in an anteroposterior direction [55,57]. Indeed, Fgf10 KO mice exhibit loss of rugae and CP [56]. The anterior extension of the palate is driven by the temporal formation of rugae signaling centers that integrate Fgf10 and Bmp4 signaling to induce epithelial Shh expression [57].
Canonical Wnt signaling also contributes to palate formation. Ablation of Wnt signaling in the oral epithelium blocks rugae formation and induces CP [58]. Interestingly, SOX4 can also regulate Wnt signaling by increasing stability of β-catenin, thereby acting as a Wnt signaling agonist [59,60]. β-catenin is abundantly expressed in the rugae epithelium [58], and elimination of β-catenin expression in the palate epithelium results in loss of SHH expression and CP [58]. These data suggest an obligatory role for canonical Wnt signaling in rugae development. Sox4, like Shh, is also expressed in the rugae epithelium on GD14 and not in the underlying mesenchyme (Figure 3B). These data allow consideration of the hypothesis that, during development of the palate, Sox4 acts to integrate the aforementioned signaling systems. This is illustrated in Figure 7.
Figure 7. A model for SOX4 regulation in the rugae epithelium during secondary palate development.
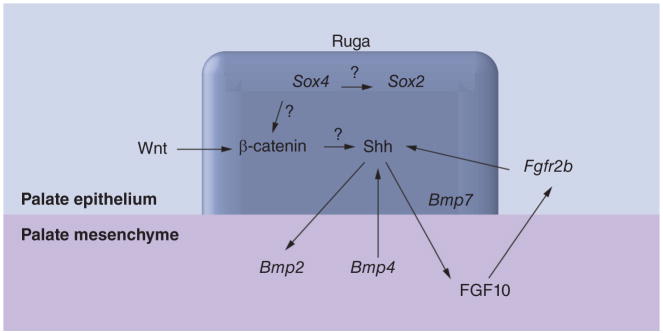
A model illustrating how SOX4 potentially integrates diverse developmental signals in the rugae during secondary palate development is presented. The ruga (boxed) is the Shh signaling center. Shh signaling facilitates palate development through epithelial–mesenchymal cross-talk involving Wnt, FGF and BMP signaling pathways. SOX4 acts to increase the stability of β-catenin, thereby acting as a Wnt agonist, and/or activates SOX2, a pluripotent factor, via binding to the Sox2 enhancer. The latter is consistent with the rugae being a niche for neural stem cells. ‘?’ denotes interactions whose underlying mechanisms are unknown. All other interactions are from published literature [9,17,55–61].
Interestingly, adult rodent rugae harbor neural crest-related stem cells that are positive for neural crest stem cell markers (Nestin, SOX2 and p75) and for the ‘four factors’ (SOX2, KLF4, c-MYC and OCT4) required for reprogramming somatic cells into pluripotent cells [61]. Sox4 expression in the rugae could thus be directed towards maintaining the stem cell fate of neural progenitor cells through induction of Sox2 (see Figure 7). The demonstration that Sox2 induction occurs through the binding of SOX4 to the 3′ flanking enhancer of Sox2 [17] lends support to this premise. Indeed, SOX4 and SOX11 are both required for the survival of neural and mesenchymal progenitors [38,45]. A role for SOX4 in maintaining the survival of neural and mesenchymal progenitors is consistent both with its transient expression in the MEE as well as in maintaining a neural stem cell niche in the rugae.
DNA methylation is one mechanism by which differential Sox4 expression in developing palatal tissue could be regulated. CpG methylation profiling of a 1.8-kb 5′ upstream region of Sox4 in developing palatal tissue identified two DMRs (DMR3 and DMR4) with contrasting methylation patterns. Specifically, DMR4 had two CpG sites (CpGs 83 and 85) that exhibited increased methylation, while DMR3 had seven contiguous sites (CpGs 72–78) that demonstrated decreased methylation from GD12 to GD13 (Table 3). We hypothesized that the downregulation of Sox4 mRNA observed from GD12 and GD13 was due to increased methylation of CpG residues 83 and 85 in DMR4. Consistent with this interpretation was the finding that methylation levels of these two residues stabilized between GD13 and GD14, a period when mRNA expression remained constant. Transfection analysis using luciferase reporter vectors in cells derived from the developing palate (MEMM cells) indicated that the 1.8-kb region harbored the Sox4 promoter as it exhibited 84% relative promoter activity compared with a control (SV40) promoter (Figure 5A). Moreover, the Sox4 promoter is likely associated with a CpG island located immediately upstream of the ATG start site (Figure 4). Complete methylation of the 1.8-kb fragment eliminated promoter activity indicating that methylation represses gene expression (Figure 6B). CpG residues constituting the CpG island (Amplicon 1; Figure 4) remained unmethylated on all 3 GDs analyzed (GD12–GD14), consistent with Sox4 being expressed on all 3 days, albeit to varying degrees (Figures 1 & 4). However, not all of the promoter activity can be attributed to the CpG island, as removal of both distally located DMRs from the 1.8-kb fragment (Sox4Δ/pGL3; Figure 5A) reduced promoter activity from 84 to 40% (Figure 5A). This suggests that either or both DMRs might be required for Sox4 expression in the palate and that elements distal to the CpG island contribute significantly to promoter activity. When mutant versions of CpG83 and 85 were used in transfection assays, promoter activity was significantly reduced to 36 and 47%, respectively. Indeed, the loss in promoter activity observed for each mutated CpG site was comparable to that lost when both DMRs were deleted (Sox4Δ/pGL3). This finding emphasizes the importance of CpGs 83 and 85 to Sox4 expression in palatal tissues, and suggests that they are likely mediated by a cooperative, rather than an additive, mechanism.
CpGs 83 and 85 are localized in low-density CpG regions (Box 1) located outside the CpG island (which spans CpGs 13–51). Their role in Sox4 regulation is consistent with the emerging view that methylation changes occurring in CpG-poor regions, located distal to the promoter, may have potential tissue-specific regulatory function [62]. Irizarry et al. observed differential methylation in CpG residues lying at the outer periphery of CpG islands called ‘CpG shores’ that dictate tissue specificity [63]. These findings support the notion that differential expression of Sox4 in developing palatal tissue from GD12 to GD13 can be attributed to changes in methylation levels of CpGs 83 and 85, residues located distant from the CpG island in the Sox4 promoter. A search for potential (vertebrate) transcription factors binding to CpGs 83–85 revealed that CpG85 was a preferred target for HNF (or Forkhead box) proteins (Table 4), whereas CpG83 is a potential target for ISGF3, ELF-1, SP1 and NF-AT proteins. Foxc2 and Foxe1 have been identified as candidate genes for human syndromic and nonsyndromic palatal clefting, highlighting the importance of HNF proteins to palate development and the potentially critical role of CpG85 [64,65]. We therefore demonstrate, for the first time, that the developmental expression of Sox4 in the secondary palate is regulated via temporal changes in DNA methylation occurring at critical CpG residues located distal to the putative promoter.
Table 4.
Putative transcription factor binding sites spanning CpG83 and CpG85.
| CpG | Sequence† | Strand | Transcription factor | Factor name |
|---|---|---|---|---|
| 85 | GTTTGTTCTT | (+) | HNF1A (TCF1) | Hepatocyte nuclear factor 1 homeobox A (transcription factor 1) |
| HNF1B (TCF2) | Hepatocyte nuclear factor 1 homeobox B (transcription factor 2) | |||
| HNF1C | – | |||
|
| ||||
| GTTTGTTTT | (+) | HNF-3 (FOXM1) | Hepatocyte nuclear factor 3 (forkhead box M1) | |
| HNF3A (FOXA1) | Hepatocyte nuclear factor 3A (forkhead box A1) | |||
| HNF3B (FOXA2) | Hepatocyte nuclear factor 3B (forkhead box A2) | |||
|
| ||||
| 83 | GGAA | (−) | ISGF3 (IRF9) | Interferon-stimulated gene factor 3 (interferon regulatory factor 9) |
| NF-AT2 | Nuclear factor of activated T cells 2 | |||
| NF-AT3 | Nuclear factor of activated T cells 3 | |||
| NF-AT4 | Nuclear factor of activated T cells 4 | |||
| ELF-1 | E74-like factor 1 | |||
|
| ||||
| GTCTCTCCCCTT | (+) | SP1 | Specificity protein 1 | |
Sequence mismatch underlined. CG sites (or parts thereof) in bold. Transcription factor binding sites (vertebrates) identified using Patch and AliBaba2.1 programs [102].
Overall, the expression pattern of Sox4 in the developing palate suggests a number of potential functional roles, including contributions to MEE fusion, palatal growth and interaction with rugae signaling centers, or maintenance of a putative neural stem cell niche. The detection of Sox11 expression at sites where the palatal shelves fuse [36,41] adds another level of complexity to the role of SOX4. The redundant expression of SOXC proteins indicates that a definitive answer to the role of Sox4 in facilitating palate morphogenesis will come from conditional knockouts of Sox4 and Sox11 in the palates of mice. It is noteworthy that Sox4 and Sox11 null mice are apparently normal until GD8.5, but die on GD14 and soon after birth, respectively [38]. This indicates that SoxC genes function in embryonic processes that occur after E8.5, consistent with the timeframe for secondary palate development (GD12–GD14). SOX4 is also situated at a nodal point where it can integrate several developmentally important pathways such as the TGF-β-, Wnt- and Hippo-signaling pathways.
Executive summary.
Spatiotemporal expression of Sox4 in the developing murine secondary palate
A prior study from our laboratory using an mRNA profiling strategy identified several differentially expressed genes, including Sox4, in the developing (gestational day [GD]12–GD14) secondary palate. Sox4 expression decreased twofold from GD12 to GD13 and remained steady thereafter (i.e., GD13–GD14). The aims of this study were to examine the spatiotemporal expression profile of Sox4 in the development of the secondary palate and to determine whether changes in DNA methylation of Sox4 were functionally related to the differential expression of Sox4 mRNA observed in the developing secondary palate.
Sox4 expression on GD12 was visible in the medial edge epithelium (MEE) of the palatal processes. From GD13 to GD13.5, Sox4 expression was predominantly observed in the rugae and its expression in the MEE began to dissipate. On GD14, upon palatal fusion, Sox4 expression was localized only in the rugae with no traces of expression in the medial edge seam.
Sectional in situ hybridization of Sox4 expression in the developing murine secondary palate
Coronal sections of secondary palates indicated intense Sox4 expression in the inferior margins of the presumptive MEE on both GD12 and GD13.
Sagittal sections of GD14 secondary palates indicated that Sox4 expression in the secondary palate was confined to the epithelial thickenings of the palatal rugae.
Sox4 mRNA levels during development of the murine secondary palate
Gene-expression profiling indicated that Sox4 mRNA levels decreased twofold from GD12 to GD13 in the secondary palate, and expression remained unchanged in the tissue from GD13 to GD14. We hypothesized that this pattern of expression was due to increased methylation of the 5′ flanking/promoter regions of Sox4 in the secondary palate from GD12 to GD13, followed by methylation levels remaining relatively stable between GD13 and GD14.
CpG methylation profiling of the Sox4 5′ flanking region
Approximately 1.8 kb of the 5′ flanking region of the Sox4 gene was analyzed by PCR amplification of bisulfite-modified DNA. Six different amplicons (amplicons 1–6) were generated spanning the 5′ flanking region.
Amplicons 3 and 4 exhibited prominent differences in methylation levels between GD12 and GD13, but not between GD13 and GD14. However, methylation levels of amplicon 3 decreased by 19%, whereas that of amplicon 4 increased by 30% between GD12 and GD13.
Amplicons 1–6 exhibited no significant differences in methylation levels between GD13 and GD14, suggesting that methylation levels had stabilized by GD13.
Amplicons 3 and 4 were termed differentially methylated regions 3 and 4 (DMR3 and DMR4), respectively. We hypothesized that the increase in methylation of CpG residues in DMR4 from GD12 to GD13 could account for the decrease in Sox4 mRNA levels observed during this period.
DMR4 contained only three CpG residues (CpGs 83–85). Methylation levels of all three CpGs increased from GD12 to GD13, with the most pronounced increases being in CpGs 83 (40%) and 85 (30%).
Functional analysis of DMR4, CpG83 & CpG85 on Sox4 promoter activity
Various deletion and mutant Sox4 constructs were generated and promoter activity was determined by transfection analysis in murine embryonic maxillary mesenchymal cells.
The normalized promoter activity of a construct carrying 1.8 kb of the murine Sox4 5′ flanking region was approximately 84% when compared with that of a positive control (SV40 promoter).
Removal of a region containing both DMR3 and DMR4 significantly reduced promoter activity by 44%.
Transfection analysis utilizing mutant CpG constructs indicated that CpGs 83 and 85 of DMR4 contribute significantly to Sox4 expression in the palate.
Methylation analysis of CpGs 83–85
In vitro methylation of the approximately 1.8 kb 5′ upstream region of Sox4 completely abolished promoter activity.
Discussion
Sox4 is a TGF-β-regulated transcription factor.
The decrease in Sox4 mRNA levels from GD12 to GD13 during secondary palate development correlates well with the increase in methylation of a CpG-poor region (DMR4) localized upstream of the putative Sox4 promoter.
Changes in the methylation levels of CpGs 83 and 85 in DMR4 appear critical for Sox4 expression in the secondary palate.
The spatiotemporal expression pattern of Sox4 in the developing palate suggests a role for Sox4 in mediating palatal fusion, palatal extension, and/or the maintenance of the neural stem cell niche in the rugae.
Sox4 is situated at a nodal point where it can integrate several developmental pathways critical for secondary palate development, such as the TGF-β, Wnt and Hippo signaling pathways.
Footnotes
Disclaimer The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIH.
Ethical conduct of research The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure This work was supported in part by NIH grants DE018215, HD053509 and P20 RR017702 from the COBRE program of the National Center for Research Resources and NIGMS, and by a grant from the Cleft Palate Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: * of interest, ** of considerable interest
- 1.Yazdy MM, Honein MA, Rasmussen SA, Frias J. Priorities for future public health research in orofacial clefts. Cleft Palate Craniofac J. 2007;44(4):351–357. doi: 10.1597/06-233.1. [DOI] [PubMed] [Google Scholar]
- 2.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301(2):309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- ▪3.Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82(2):63–77. doi: 10.1002/bdra.20430. Comprehensive review that identifies Sox4 as a candidate gene for human nonsyndromic cleft lip with cleft palate. [DOI] [PubMed] [Google Scholar]
- 4.Lidral AC, Moreno LM. Progress toward discerning the genetics of cleft lip. Curr Opin Pediatr. 2005;17(6):731–739. doi: 10.1097/01.mop.0000185138.65820.7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71(10):1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- 7.Vieira AR. Unraveling human cleft lip and palate research. J Dent Res. 2008;87(2):119–125. doi: 10.1177/154405910808700202. [DOI] [PubMed] [Google Scholar]
- 8.Greene RM, Pisano MM. Palate morphogenesis: current understanding and future directions. Birth Defects Res C Embryo Today. 2010;90(2):133–154. doi: 10.1002/bdrc.20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He F, Xiong W, Wang Y, et al. Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev Biol. 2010;347(1):109–121. doi: 10.1016/j.ydbio.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trainor PA. Craniofacial birth defects: the role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A(12):2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marazita ML. Genetic etiologies of facial clefting. In: Mooney MP, Siegel MI, editors. Understanding Craniofacial Anomalies: The Etiopathogenesis of Craniosynostoses and Facial Clefting. Wiley-Liss; NY, USA: 2002. pp. 147–161. [Google Scholar]
- 12.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Kartiko S, Finnell RH. Importance of gene–environment interactions in the etiology of selected birth defects. Clin Genet. 2009;75(5):409–423. doi: 10.1111/j.1399-0004.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- ▪14.Mukhopadhyay P, Greene RM, Pisano MM. Expression profiling of TGFβ superfamily genes in developing orofacial tissue. Birth Defects Res A Clin Mol Teratol. 2006;76(7):528–543. doi: 10.1002/bdra.20276. Identifies Sox4 as a differentially expressed gene during murine palatogenesis and is a precursor for the current work. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42(3):425–428. doi: 10.1016/j.biocel.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno CS. The sex-determining region Y-Box 4 and homeobox C6 transcriptional networks in prostate cancer progression. Crosstalk with the Wnt, Notch and PI3K pathways. Am J Pathol. 2010;176(2):518–527. doi: 10.2353/ajpath.2010.090657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪17.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5(5):504–514. doi: 10.1016/j.stem.2009.08.018. Identifies a TGF-β–Sox4–Sox2 signaling pathway in gliomas. [DOI] [PubMed] [Google Scholar]
- 18.Jafarnejad SM, Ardekani GS, Ghaffari M, Li G. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1187-y. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruebel KH, Leontovich AA, Tanizaki Y, et al. Effects of TGF β1 on gene expression in the HP75 human pituitary tumor cell line identified by gene expression profiling. Endocrine. 2008;33(1):62–76. doi: 10.1007/s12020-008-9060-3. [DOI] [PubMed] [Google Scholar]
- 20.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastases. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10(2):203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69(23):9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen CL, Christensen LL, Thorsen K, et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100(3):511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina PP, Castillo SD, Blanco S, et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet. 2009;18(7):1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 25.de Bont JM, Kros JM, Passier MM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncol. 2008;10(5):648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwata J, Parada C, Chai Y. The mechanism of TGF-β signaling during palate development. Oral Dis. 2011;17(8):733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139(2):231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng L, Bian Z, Torensma R, Von den Hoff JW. Biological mechanisms in palatogenesis and cleft palate. J Dent Res. 2009;88(1):22–33. doi: 10.1177/0022034508327868. [DOI] [PubMed] [Google Scholar]
- 29.Warner DR, Smith HS, Webb CL, Greene RM, Pisano MM. Expression of Wnts in the developing murine secondary palate. Int J Dev Biol. 2009;53(7):1105–1112. doi: 10.1387/ijdb.082578dw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay P, Brock G, Pihur V, Webb C, Pisano MM, Greene RM. Developmental microRNA expression profiling of murine embryonic orofacial tissue. Birth Defects Res A Clin Mol Teratol. 2010;88(7):511–534. doi: 10.1002/bdra.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aranyi T, Tusnady GE. BiSearch: ePCR tool for native or bisulfite-treated genomic template. Methods Mol Biol. 2007;402:385–402. doi: 10.1007/978-1-59745-528-2_20. [DOI] [PubMed] [Google Scholar]
- 33.Singh S, Greene RM, Pisano MM. Arsenate-induced apoptosis in murine embryonic maxillary mesenchymal cells via mitochondrial-mediated oxidative injury. Birth Defects Res A Clin Mol Teratol. 2010;88(1):25–34. doi: 10.1002/bdra.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klug M, Rehli M. Functional analysis of promoter CpG Methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1(3):127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 35.Seelan RS, Pisano MM, Greene RM, Casanova MF, Parthasarathy R. Differential methylation of the gene encoding myo-inositol synthase (Isyna1) in rat tissues. Epigenomics. 2011;3(1):111–124. doi: 10.2217/epi.10.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ▪36.Dy P, Penzo-Méndez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins – Sox4, Sox11 and Sox12 – exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36(9):3101–3117. doi: 10.1093/nar/gkn162. Compares the expression patterns of the three SoxC genes and the DNA binding properties of the cognate proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24(15):6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattaram P, Penzo-Méndez A, Sock E, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldsworthy M, Hugill A, Freeman H, et al. Role of the transcription factor Sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008;57(8):2234–2244. doi: 10.2337/db07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83(10):986–994. doi: 10.1161/01.res.83.10.986. [DOI] [PubMed] [Google Scholar]
- 41.Hoser M, Potzner MR, Koch JMC, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcriptional factors. Mol Cell Biol. 2008;28(15):4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nissen-Meyer LS, Jemtland R, Gautvik VT, et al. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120(Pt 16):2785–2795. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 43.Hoser M, Baader SL, Bosl MR, et al. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27(20):5495–5505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potzner MR, Griffel C, Lütjen-Drecoll E, Bösl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27(15):5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thein DC, Thalhammer JM, Hartwig AC, et al. The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J Neurochem. 2010;115(1):131–141. doi: 10.1111/j.1471-4159.2010.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay P, Webb CL, Warner DR, Greene RM, Pisano MM. BMP signaling dynamics in embryonic orofacial tissue. J Cell Physiol. 2008;216(3):771–779. doi: 10.1002/jcp.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parada C, Chai Y. Roles of BMP signaling pathway in lip and palate development. Front Oral Biol. 2012;16:60–70. doi: 10.1159/000337617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iordanskaia T, Nawshad A. Mechanisms of transforming growth factor β induced cell cycle arrest in palate development. J Cell Physiol. 2011;226(5):1415–1424. doi: 10.1002/jcp.22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu W, Ruest LB, Svoboda KK. Regulation of epithelial-mesenchymal transition in palatal fusion. Exp Biol Med (Maywood) 2009;234(5):483–491. doi: 10.3181/0812-MR-365. [DOI] [PubMed] [Google Scholar]
- 50.Iwata J, Parada C, Chai Y. The mechanism of TGF-β signaling during palate development. Oral Dis. 2011;17(8):733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.d'Amaro R, Scheidegger R, Blumer S, et al. Putative functions of extracellular matrix glycoproteins in secondary palate morphogenesis. Front Physiol. 2012;3:377. doi: 10.3389/fphys.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwata J, Hacia JG, Suzuki A, Sanchez-Lara PA, Urata M, Chai Y. Modulation of noncanonical TGF-β signaling prevents cleft palate in Tgfbr2 mutant mice. J Clin Invest. 2012;122(3):873–885. doi: 10.1172/JCI61498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Oliveira Demarchi AC, Zambuzzi WF, Paiva KB, et al. Development of secondary palate requires strict regulation of ECM remodeling: sequential distribution of RECK, MMP-2, MMP-3, and MMP-9. Cell Tissue Res. 2010;340(1):61–69. doi: 10.1007/s00441-010-0931-6. [DOI] [PubMed] [Google Scholar]
- 54.Carette MJ, Ferguson MW. The fate of medial edge epithelial cells during palatal fusion in vitro: an analysis by DiI labeling and confocal microscopy. Development. 1992;114(2):379–388. doi: 10.1242/dev.114.2.379. [DOI] [PubMed] [Google Scholar]
- 55.Pantalacci S, Prochazka J, Martin A, et al. Patterning of palatal rugae through sequential addition reveals an anterior/posterior boundary in palatal development. BMC Dev Biol. 2008;8:116. doi: 10.1186/1471-213X-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porntaveetus T, Oommen S, Sharpe PT, Ohazama A. Expression of Fgf signalling pathway related genes during palatal rugae development in the mouse. Gene Expr Patterns. 2010;10(4–5):193–198. doi: 10.1016/j.gep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Welsh IC, O'Brien TP. Signaling integration in the rugae growth zone directs sequential SHH signaling center formation during the rostral outgrowth of the palate. Dev Biol. 2009;336(1):53–67. doi: 10.1016/j.ydbio.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C, Fisher AV, Yin Y, et al. The inductive role of Wnt-β-Catenin signaling in the formation of oral apparatus. Dev Biol. 2011;356(1):40–50. doi: 10.1016/j.ydbio.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinner D, Kordich JJ, Spence JR, et al. Sox17 and Sox4 differentially regulate β-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27(22):7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee AK, Ahn SG, Yoon JH, Kim SA. Sox4 stimulates β-catenin activity through induction of CK2. Oncol Rep. 2011;25(2):559–565. doi: 10.3892/or.2010.1091. [DOI] [PubMed] [Google Scholar]
- ▪61.Widera D, Zander C, Heidbreder M, et al. Adult palatum as a novel source of neural crest-related stem cells. Stem Cells. 2009;27(8):1899–1910. doi: 10.1002/stem.104. Demonstrates the presence of neural crest-related stem cells in adult palatal rugae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stadler MB, Murr R, Burger L, et al. NA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- ▪63.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. Demonstrates that tissue-specific regulatory regions are located outside CpG islands and promoters, in regions termed ‘CpG island shores’. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nikopensius T, Kempa I, Ambrozaitytė L, et al. Variation in FGF1, FOXE1, and TIMP2 genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2011;91(4):218–225. doi: 10.1002/bdra.20791. [DOI] [PubMed] [Google Scholar]
- 65.Vreeburg M, Heitink MV, Damstra RJ, Moog U, van Geel M, van Steensel MA. Lymphedema-distichiasis syndrome: a distinct type of primary lymphedema caused by mutations in the FOXC2 gene. Int J Dermatol. 2008;47(Suppl 1):S52–S55. doi: 10.1111/j.1365-4632.2008.03962.x. [DOI] [PubMed] [Google Scholar]
Websites
- 101.EMBOSS explorer cpgplot. www.bioinformatics.nl/cgi-bin/emboss/cpgplot
- 102.gene-regulation.com. www.gene-regulation.com/pub/programs.html