Abstract
The use of photolabile protecting groups is a versatile and well-established means of synthesizing high complexity microarrays of biopolymers, such as nucleic acids and peptides, for high-throughput analysis. The synthesis takes place in a photochemical reaction cell which positions the microarray substrate at the focus of the optical system delivering the light and which can be connected to a fluidics system which delivers appropriate reagents to the surface in synchrony with the light exposure. Here we describe a novel photochemical reaction cell which allows for the simultaneous synthesis of microarrays on two substrates. The reaction cell positions both substrates within the limited depth-of-focus of the optical system while maintaining the necessary reagent flow conditions. The resulting microarrays are mirror images of each other but otherwise essentially identical. The new reaction cell doubles the throughput of microarray synthesis without increasing the consumption of reagents. In addition, a secondary flow chamber behind the reaction cell can be filled with an absorbent and index-matching fluid to eliminate reflections from light exiting the reaction cell assembly, greatly reducing unintended light exposure that reduces the sequence fidelity of the microarray probes.
Microarrays are versatile and widely used analytical tools with the capacity to simultaneously detect several hundred thousand to millions of different biomolecules simultaneously. Microarrays can be made by presynthesizing the probe molecule and spotting it on a surface using appropriate tethering chemistry, but modern microarrays are made with in situ methods in which the biomolecules are synthesized directly on the substrate from their monomer components, which allows for high probe densities, high uniformity, and high reproducibility.
Light-directed in situ synthesis of microarrays derives from the photolithographic technology used in the semiconductor industry in combination with combinatorial chemistry based on the selective removal of photolabile protecting groups. The technology was first commercialized by Affymetrix, which used the photolabile MeNPOC group on the 5′ end of DNA phosphoramidites to synthesize high-density DNA microarrays for genomics applications.1 The synthesis technology was improved with the use of optical systems incorporating digital micromirror devices (DMD) to replace physical masks in the patterning of light on the microarray substrate, as well as by the use of the NPPOC photolabile group, which has significantly improved photodeprotection yield.2−7 This maskless array synthesis (MAS) technology, originally used for DNA microarray synthesis has also been extended for the synthesis of RNA, aptamer,8 and peptide microarrays.9−13
In situ microarray synthesis is robust and efficient in comparison with spotted synthesis; however, the total synthesis time and the consumption of solvents and reagents are still a significant economic constraint. In addition, the light-directed chemistry is sensitive to stray light in the system, which leads to unintended photodeprotection which degrades the sequence fidelity of the microarray probes.7,14 Here we present an improved microfluidic photochemical reaction cell for use in light-directed synthesis that addresses both of these concerns. This reaction cell places two microarray substrates within the depth-of-focus plane of the optical system, so that two microarrays are synthesized simultaneously using the same reagents. The microarrays thus synthesized are mirror images of each other but otherwise essentially identical. The microarrays can be used independently but may have additional utility as matched pairs for experiments that would benefit from very close data comparisons; the quality of in situ synthesized microarrays, however, is very high and in most common applications, variations in quality between microarrays synthesized at different times are not experimentally relevant. In addition, the reaction cell assembly has a secondary chamber that can be filled with a light-absorbing and index-matching fluid to eliminate reflections that are a primary source of sequence error in light-directed synthesis.
Materials and Methods
Photochemical Reaction Cell Concept and Assembly
The reaction cell needs to position the two microarray substrates at the focal plane of the optical system. There is some tolerance to this positioning: the depth of focus of the imaging optics. The imaging optics are a 1:1 Offner relay system,15,16 an off-axis conjugate system composed of two spherical concentric mirrors, primary and secondary. The system was designed with a numerical aperture (NA) of 0.08 to result in a resolving power of 2.7 μm. This resolving power is sufficient since it is significantly smaller than the size of individual mirrors of the digital micromirror device (DMD), 13 μm × 13 μm, separated by a 0.7 μm gap and is similar or better than those of most available microarray scanners. A low value of numerical aperture lowers the cost of the primary mirror but, more importantly, reduces the amount of scattered light originating from dust and imperfections in the optical system, which is proportional to NA2. Unintended photodeprotection, from scattering, diffraction, and local flare, is the largest source of sequence error in light-directed microarray synthesis.7 The depth of focus is intrinsically limited by diffraction to <∼λ/NA2, ∼60 μm, but in practice, the positioning of the microarray substrates in the focal plane is somewhat less restricted due to limited resolution of microarray scanners. Therefore, the primary optical constraint in the simultaneous light-directed synthesis of microarray pairs is that the two substrates must be within ∼60–100 μm of each other, depending on the scanner resolution.
A secondary constraint is imposed by reagent delivery. A larger reaction cell volume requires larger flow rates of solvents and reagents, the consumption of which scales with cell volume. Since our original reaction cell (for synthesizing microarrays on a single surface) had a depth of 70 μm and worked well with a standard oligonucleotide synthesizer (Expedite 8909), we took this value as a starting point. Thus, the reaction cell should consist of two standard microarray substrates (75 mm × 25 mm × 1 mm) separated by a uniform gap of ∼70 μm. The microarray substrates form the entrance and exit windows for the ultraviolet light used in the synthesis. Reagents need to be introduced into this gap and to uniformly flow across the surface before exiting. We used these criteria to design and built the reaction cell shown in Figure 1. The reaction cell assembly consists of a black anodized aluminum support block, a quartz block, the two microarray substrates, two gaskets, and a clamping frame and screws to hold the parts together. Reagent delivery tubes attach to the underside of the quartz block and connect to the oligonucleotide synthesizer.
Figure 1.
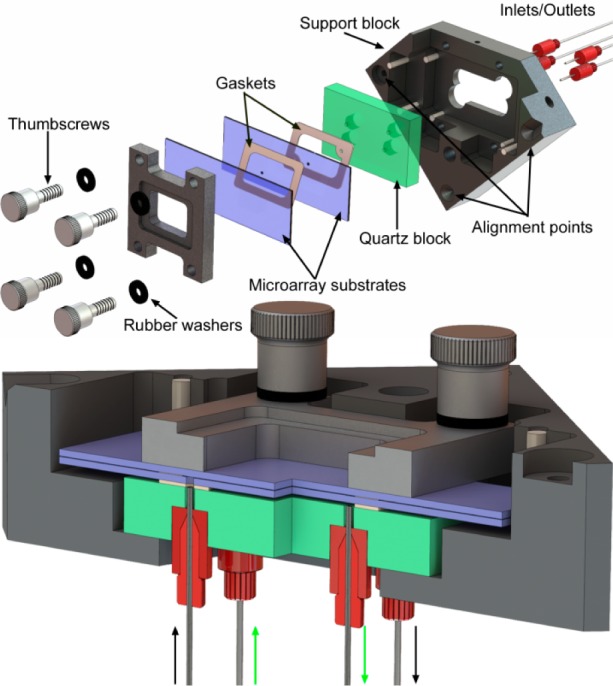
Exploded and section view of reaction cell assembly. The reaction cell is formed by two microarray substrates (75 mm × 25 mm × 1 mm) separated by a 50 μm PTFE gasket. Reagents enter and exit the cell via two 0.9 mm holes through the lower substrate. These holes are coupled to the inlets/outlets via an additional 250 μm thick FFKM gasket separating the lower substrate from the quartz block. The lower gasket forms a chamber that can be independently filled with a light-absorbing and index-matching fluid to reduce reflections from both quartz surfaces and from the back surface of the lower substrate. The thickness of the upper and lower gaskets in the section view have been exaggerated by a factor of 2 for visual clarity.
The support block forms the rigid structure for the assembly of the reaction cell and allows for the reaction cell to be precisely positioned in the focal plane. Three alignment points make contact with ball-tipped, high-precision adjustment screws (Newport AJS127-0.5H) in the optical system. After initial adjustment of the screws, the reaction cell assembly can be quickly and reproducibly positioned. The support blocks hold a quartz block. The quartz block has four 0.8 mm through-holes (two inlets, two outlets) that are countersunk on the back side to accommodate microfluidics ports. The microfluidics ports (IDEX 6-32 Coned NanoPort Assemblies) were turned on a lathe to reduce their diameter to 6.4 mm and attached within each countersunk hole with common cyanoacrylate adhesive. The front and back surfaces of the quartz block were machined to a surface parallelism error of <30 arc sec and polished to an optical flatness of λ/4 (Mindrum Precision). During reaction cell assembly, the lower gasket is placed on the quartz surface. This gasket forms the lower chamber, which can be filled via two of the fluidics ports. Prior to microarray synthesis, this chamber can be filled with an index-matching and light absorbing fluid to prevent light reflections from light exiting the reaction chamber. In the legacy reaction cell design, an antireflective coating on the back surface of the quartz block can reduce the back reflection to a minimum of about 0.25% when new, but this value is typically larger, ∼1%, due to the presence of dust, chemical films, and scratches. This 0.25–1% value is sufficient to make this unintended light exposure the largest source of error after diffraction, but unlike diffraction, the error is not confined primarily to the gaps between microarray features.7 An alternative strategy to reduce back reflections is to fill the lower chamber with an index-matching fluid with dissolved chromophores which absorb the light exiting the reaction chamber and which either convert the light to heat or Stokes shift it beyond the absorption band of the light-labile group.
The lower gasket has two holes that align with two of holes in the quartz block. These holes couple the corresponding fluidics ports to the microarray synthesis cell. This gasket is made from 250 μm thick Chemraz 584 perfluoroelastomer (FFKM), cut to shape with a laser cutter (Spirit GX). The microarray synthesis cell is a chamber consisting of two glass substrates separated by a very thin gasket. This chamber is accessed via two 1 mm holes, in the lower substrates, which align with the holes in the lower gasket.
The thickness of the upper gasket determines the depth of the photochemical reaction cell and therefore needs to be ∼70 μm thick, chemically resistant and sufficiently elastic to form a seal for the duration of the synthesis, up to ∼12 h for an array of 70mers. These requirements are quite exceptional and we were unable to find any references to such thin gaskets in the scientific or engineering literature. A perfluoroelastomer, such as Chemraz, would likely work, but the manufacturer is unable to make them thinner than 250 μm. We tried expanded polytetrafluoroethylene (ePTFE), which is commonly used in gasket applications due to its chemical resistance and ability to compress to form a seal, but found seepage through the gasket, presumably due to its porous nature. In the end we found that the common PTFE tape used for plumbing applications works well. This tape is made from unsintered PTFE and is therefore sufficiently compressible to form a seal but not porous. PTFE tape is made in many thicknesses and densities, which allowed for some experimentation. We initially used ∼100 μm (120 μm uncompressed) PTFE with a density of ∼1.4 g/cm3 (Gasoila yellow tape), sintered PTFE has a density of about 2 g/cm3, but found some loss of focus when microarrays were scanned at a resolution of 2.5 μm. Another problem with the 100 μm gap were indications that reagents were flowing in a channel through the center of the reaction cell rather than sweeping the whole surface. This was particularly apparent with the helium drying step, which was not capable of fully removing solvent from the corners of the reaction cell. Switching to thinner and lower density PTFE tape (Gasoila Industrial Strength SD, ∼0.7 g/cm3) gave a thickness of ∼50 μm under compression. With this thickness, both of the paired arrays produce sharp scans with resolution limited only by the 2.5 μm pixel size of the scanner and both reagent and helium flow sweep uniformly across the entire surface of both substrates. The 50 μm PTFE gaskets are also formed with a laser cutter. Because of their thinness, they are too delicate to be reusable but can be made quickly and inexpensively.
Microarray Synthesis and Hybridization
Schott Nexterion Glass D slides functionalized with N-(3-triethoxysilylpropyl)-4-hydroxybutryamide (Gelest SIT8189.5). The arrays with holes were drilled with a 0.9 mm diamond bit and washed and rinsed in an ultrasonic bath prior to functionalization. The slides were loaded in a metal staining rack and completely covered with a 500 mL of a solution of 10 g of the silane in 95:5 (v/v) ethanol–water and 1 mL of acetic acid. The slides were gently agitated for 4 h and then rinsed twice for 20 min with gentle agitation in the same solution but without the silane. The slides were then drained and cured overnight in a preheated vacuum oven (120 °C). After cooling to room temperature, the slides were stored in a desiccator cabinet until use. Microarrays were synthesized directly on the slides using a maskless array synthesizer, which consists of an optical imaging system that used a digital micromirror device to deliver patterned ultraviolet light near 365 nm to the synthesis surface. Microarray layout and oligonucleotide sequences are determined by selective removal of the NPPOC photocleavable 5′-OH protecting group. Reagent delivery and light exposures are synchronized and controlled by a computer. The chemistry is similar to that used in conventional solid-phase oligonucleotide synthesis. The primary modification is the use of NPPOC phosphoramidites. Upon absorption of a UV photon, and in the presence of a weak organic base, e.g., 1% (m/v) imidazole in DMSO, the NPPOC group comes off, leaving a 5′-terminal hydroxyl which is able to react with an activated phosphoramidite in the next cycle. The DNA sequences on the microarrays in this project were synthesized with a light exposure dose of 4.5 J/cm2, with coupling time of 40 s at monomer concentrations of 30 mM. After synthesis, the microarrays were deprotected in 1:1 (v/v) ethylenediamine in ethanol for 2 h at room temperature, washed twice with distilled water, dried with argon, and stored in a desiccator until hybridization.
Microarrays were hybridized in an adhesive chamber (SecureSeal SA200, Grace Biolabs) with a solution consisting of 0.3 pmol of 5′-Cy5-labeled probe and 200 μg of acetylated BSA in 400 μL of MES buffer (100 mM MES, 1 M NaCl, 20 mM EDTA, 0.01% Tween-20). After 2 h of rotation at 42 °C, the chamber was removed and the microarrays were vigorously washed in a 50 mL centrifuge tube with 30 mL of nonstringent wash buffer (SSPE; 0.9 M NaCl, 0.06 M phosphate, 6 mM EDTA, 0.01% Tween-20) for 2 min and then with stringent wash buffer (100 mM MES, 0.1 M NaCl, 0.01% Tween-20) for 1 min. The microarrays were then dipped for a few seconds in a final wash buffer (0.1× SSC) and then dried with a microarray centrifuge. Arrays were scanned with a Molecular Devices GenePix 4400A at a resolution of 2.5 μm.
Detection and Suppression of Reflected Light
To test the possibility of eliminating reflected light reaching the synthesis area, a small piece of radiochromic film (Far West Technology, FWT-60-20f), with a 2 mm punched hole, was placed in the reaction cell. A 9.5 mm metal disk with a 1 mm pinhole (Edmund Optics, 39730) was aligned over the hole in the film to serve as a physical mask. The entire reaction cell assembly was tilted by ∼7° to move the reflection spot away from the mask hole. The lower chamber was filled with either DMSO (control) or UV absorbers dissolved in DMSO or dichloromethane. The UV absorbers (beta carotene, 9-methylanthracene, and riboflavin) were chosen for high extinction coefficients near 365 nm, high Stokes shift, low fluorescence quantum yield, and solubility in DMSO. The synthesis cell was exposed using all mirrors, with an exposure of 60 J/cm2 (80 mW/cm2 for 750 s).
Results and Discussion
Synthesis of Mirror-Image Microarrays
Simultaneous synthesis of mirror-image microarrays in this microfluidic photochemical reaction chamber produces high-quality microarrays with little additional cost or effort beyond those of the single microarray synthesis of the legacy method. The primary concern with this method is that both arrays are in focus. To test the image quality of paired microarrays, we initially synthesized simple microarrays of 30mers (GTC ATC ATC ATG AAC CAC CCT GGT CTT TTT), hybridized them with labeled complementary oligonucleotides and scanned them at high resolution. The results of one such experiment is shown in Figure 2. The top row shows pixel-level close-ups from both of the arrays. Each white square corresponds to a microarray feature synthesized with a single DMD mirror. In both close-ups, the features are individually resolved, and the 0.7 μm gap between features are also clearly visible. The middle row shows plots of the scan image intensity along a horizontal line through the center of each of the pixel-level close-ups. The intensity drops by ∼1000-fold between the center of hybridized pixels and unhybridized pixels, which is a typical signal/background for this type of microarray. The gap between immediately adjacent hybridized pixels is visible as a drop in intensity of about 20%. This interstitial intensity is due to the limited resolution of the scanner (2.5 μm), which leads to image pixels that derive most of their intensity from the adjacent bright microarray features. Diffraction also contributes significantly to intensity in gaps between microarray features, about 40% of the intensity of adjacent features when both features are exposed, and about 20% of the intensity of an adjacent feature when only one of the features is exposed.7 The vertical sawtooth pattern probably originates from signal latency during rastering by the scanner. The microarrays are fully resolved within the constraints of scanner resolution and diffraction. The bottom row of Figure 2 shows 3-D surface intensity plots of the same close-ups. From the perspective of common microarray use, the each of the mirror image microarrays from the pair can be used as an individual microarray, but in some experimental contexts requiring close comparisons, matched pairs might be used to increase confidence in the comparison.
Figure 2.
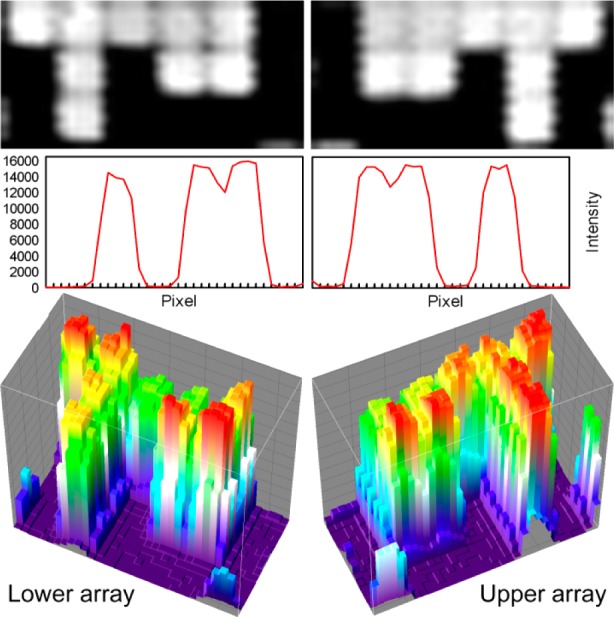
Scanned images and pixel intensities from two mirror-image microarrays synthesized simultaneously. Figures on the left are from the lower substrate (closest to quartz block in Figure 1), and those on the right are from the upper substrate. Top row: 3 × 6 array of features from the center of a 1024 × 768 array, scanned at 2.5 μm. Each features measures 13 μm × 13 μm and are separated by a 0.7 μm gap. Middle row: Intensity profiles of lines drawn horizontally through the close-ups above. Lower row: 3D surface intensity plots of the same close-ups.
Blocking Reflections
The use of a light-absorbing fluid in the lower chamber resulted in the complete blockage of reflected light. Initial trials with 9-methylanthracence and riboflavin in DMSO were only partially successful due to incomplete absorption of violet light from the mercury lamp. Most of the photodeprotection of NPPOC results from the 365 nm line, but the mercury lines at 405 and 436 nm are also transmitted through the optical system and result in measurable deprotection. Beta carotene was able to completely absorb the incident light and prevent any reflection. Beta carotene is insufficiently soluble in DMSO but is highly soluble in dichloromethane,17 which also has an index of refraction similar to that of glass. Figure 3 shows the effect of 5.5 mM beta carotene in dichloromethane. The control experiment (left film) has DMSO in the lower chamber and clearly shows the reflection from the light transmitted through the 1 mm pinhole as a round exposed spot on the lower right-hand side. Another reflection is also apparent on the left side of the circle; this originates from transmission outside the pinhole disk that is not entirely absorbed by the radiochromic film. The film on the right shows that the beta carotene solution completely suppresses the reflections.
Figure 3.
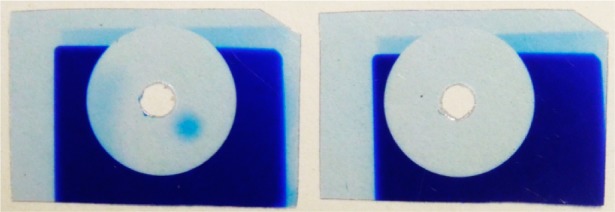
Visualization of light reflected into the synthesis chamber from the back surface of the quartz block and the complete suppression thereof using a light-absorbing fluid in the lower chamber. A 9.5 mm metal disk with a 1 mm diameter pinhole was used to mask radiochromic film in the synthesis chamber. The pinhole was aligned with a 2 mm hole in the film to allow the passage of light (60 J/cm2), and the reaction cell assembly was tilted 7° to direct the reflection away from the hole. With the secondary chamber filled with a nonabsorbent fluid (left), there is a clear reflection to the lower right of the hole. When the secondary chamber is filled with a light-absorbing fluid, the reflection is completely suppressed (right).
There are four principle sources of unintended photodeprotection: (1) global scattering, (2) edge scattering, (3) local flare (which includes reflections), and (4) diffraction.7 Global scattering from imperfections and dust in the optical system is relatively small and results in a contrast ratio of better than 1/2500. Edge scattering originates primarily from the edges of the micromirrors and has a similar magnitude as global scattering. Diffraction is an intrinsic limitation of all imaging systems and results in partial exposure (∼20%) of the area of the synthesis surface corresponding to the gaps between mirrors. Local scattering is primarily due to reflections of light exiting the reaction block but also includes scattering from bubbles in the exposure solvent. Bubbles can be eliminated by using appropriate fluidics protocols, primarily the use of helium as the blanket gas and adequate flushing of the reaction cell with exposure solvent before exposure. Reflection and diffraction remain alone as the largest sources of unintended exposure, each contributing approximately 1–2% of incident light. The use of an effective light absorber in the lower chamber, as demonstrated here, therefore reduces unintended exposure by approximately 50%. Diffraction remains as a large source of unintended exposure, but because the intensity is mostly confined to the gaps between microarray features (“spots”), it does not strongly affect the sequence fidelity within the features.
Conclusions
We have presented a method for doubling the efficiency of in situ, light directed microarray synthesis by assembling a reaction cell from two very closely spaced substrates. The method is straightforward, and we have adopted the method for routine synthesis of both DNA and RNA microarrays and for applications including gene expression and miRNA expression studies.18,19 For microarray applications requiring high sequence fidelity, the reaction cell assembly provides a chamber that can be used to completely suppress reflections.
Acknowledgments
Funding by the University of Vienna, the Faculty of Chemistry of the University of Vienna, the Austrian Science Fund (Grant FWF P23797), the Austrian Federal Ministry of Economy, Family and Youth, and the Austrian National Foundation for Research, Technology and Development is gratefully acknowledged. We thank John Wallace and Kurt Heinrich for providing assistance with material sourcing, specifications, and mechanical drawings from the original reaction cell and Walter Leuthner for machining the support block and prototypes of various components.
The authors declare no competing financial interest.
References
- Fodor S. P.; Read J. L.; Pirrung M. C.; Stryer L.; Lu A. T.; Solas D. Science 1991, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- Singh-Gasson S.; Green R.; Yue Y.; Nelson C.; Blattner F.; Sussman M.; Cerrina F. Nat. Biotechnol. 1999, 17, 974–978. [DOI] [PubMed] [Google Scholar]
- Beier M.; Hoheisel J. D. Nucleic Acids Res. 2000, 28, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A.; Stengele K.; Giegrich H.; Cornwell P.; Isham K.; Sachleben R.; Pfleiderer W.; Foote R. Tetrahedron 1997, 53, 4247–4264. [Google Scholar]
- Pirrung M. C.; Wang L.; Montague-Smith M. P. Org. Lett. 2001, 3, 1105–1108. [DOI] [PubMed] [Google Scholar]
- Agbavwe C.; Somoza M. M. PLoS ONE 2011, 6, e22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbavwe C.; Kim C.; Hong D. G.; Heinrich K.; Wang T.; Somoza M. M. J. Nanobiotechnol. 2011, 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen-van Hal N. L. W.; van der Putte P.; Hellmuth K.; Matysiak S.; Kretschy N.; Somoza M. M. Anal. Chem. 2013, 85, 5950–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan K. R. Org. Biomol. Chem. 2006, 4, 1857–1859. [DOI] [PubMed] [Google Scholar]
- Shin D.-S.; Lee K.-N.; Yoo B.-W.; Kim J.; Kim M.; Kim Y.-K.; Lee Y.-S. J. Comb. Chem. 2010, 12, 463–471. [DOI] [PubMed] [Google Scholar]
- Lackey J. G.; Mitra D.; Somoza M. M.; Cerrina F.; Damha M. J. J. Am. Chem. Soc. 2009, 131, 8496–8502. [DOI] [PubMed] [Google Scholar]
- Wang T.; Oehrlein S.; Somoza M. M.; Perez J. R. S.; Kershner R.; Cerrina F. Lab Chip 2011, 11, 1629–1637. [DOI] [PubMed] [Google Scholar]
- Lackey J. G.; Somoza M. M.; Mitra D.; Cerrina F.; Damha M. J. Chim. Oggi 2009, 27, 30–33. [Google Scholar]
- Garland P. B.; Serafinowski P. J. Nucleic Acids Res. 2002, 30, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offner A. Opt. Eng. 1975, 14, 130–132. [Google Scholar]
- Offner A. Photogr. Sci. Eng. 1979, 23, 374. [Google Scholar]
- Craft N. E.; Soares J. H. J. Agric. Food Chem. 1992, 40, 431–434. [Google Scholar]
- Holik A.-K.; Rohm B.; Somoza M. M.; Somoza V. Food Funct. 2013, 4, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Rohm B.; Holik A.-K.; Somoza M. M.; Pignitter M.; Zaunschirm M.; Ley J. P.; Krammer G. E.; Somoza V. Mol. Nutr. Food Res. 2013, 10.1002/mnfr.201200846. [DOI] [PubMed] [Google Scholar]


