Abstract
Protein remodeling at the cell–material interface is an important phenomenon that should be incorporated into the design of advanced biomaterials for tissue engineering. In this work, we address the relationship between fibronectin (FN) activity at the material interface and remodeling, including proteolytic cascades. To do so, we studied FN adsorption on two chemically similar substrates, poly(ethyl acrylate) (PEA) and poly(methyl acrylate) (PMA), which resulted in different distribution and conformation of the protein at the material interface: FN organized spontaneously upon adsorption on PEA into physiological-like fibrils, through a process called material-driven FN fibrillogenesis. The amount of adsorbed FN and its conformation were investigated in two different coating concentrations (2 and 20 μg/mL). Since FN activity at the material interface determines the initial cellular response, we followed the formation of focal adhesions (vinculin) and subsequent cell signaling by focal adhesion kinase (FAK) expression and its phosphorylation (pFAK). More detailed studies were performed to get further insights into integrin binding by crosslinking and extraction followed by immunofluorescence, as well as protein and gene expression for α5 and αv. To correlate cell adhesion with matrix degradation, gene expression and activity (zymography) of matrix metalloproteinases (MMPs) were measured. Overall, we demonstrated that the material-driven FN fibrillogenesis triggers proteolytic activity: MMP activity was higher on the material-driven FN fibrils, as a compensatory mechanism to the inability of cells to reorganize this FN network.
Keywords: cellular biology, extracellular matrix, molecular biology, tissue engineering
Introduction
Understanding and triggering the interaction of cells with synthetic materials are of fundamental importance in many biotechnological applications: biosensors, microfluidics, drug and protein delivery, tissue engineering, and regenerative medicine. Cell adhesion takes place through an intermediate layer of matrix proteins previously adsorbed on the material surface, coming from either the physiological fluids in vivo or culture medium in vitro, such as fibronectin (FN), vitronectin, and fibrinogen, representing the soluble matrix proteins in the blood.1–3 The activity of a protein after adsorption on a synthetic surface (i.e., the concentration, distribution, and motility of the adsorbed protein layer) plays a fundamental role in the biofunctionality of a synthetic material and is of paramount importance to understand the biological response of a substrate.4 Cells primarily interact with these proteins via integrins, a family of transmembrane receptors that link the extracellular matrix (ECM) to the actin cytoskeleton.5 Integrins play an important role in cell adhesion. After ligand binding with ECM proteins, integrins cluster into focal adhesions that contain structural proteins5,6 and signaling molecules (e.g., focal adhesion kinase [FAK], a nonreceptor protein tyrosine kinase). Cell migration, proliferation, and differentiation are triggered by FAK activation, which works as an integrator.7
Similar to the natural interactions of cells with the extracellular matrix, the cell–material interaction is a dynamic and complex bidirectional process.3,8 Cells receive inputs from the surrounding ECM and process it to secrete and remodel their matrix.1,5,9,10 Protein remodeling at the cell–material interface is an important factor to direct cell behavior on biomaterials for regenerative medicine and tissue engineering. When a biomaterial is implanted in vivo, adjacent cells do not always interact properly with its synthetic surface, most likely due to the lack of an ECM. The proteolytic remodeling of matrix proteins at the biomaterial's interface has only recently received attention.11–14 It is well known that cells tend to rearrange adsorbed matrix proteins at the material interface. We have shown that the activation of proteolytic routes is sensitive to surface chemistry,15 but there is still a lack of information about the organization and dynamics of the ECM at the cell–material interface. The main mechanism for ECM degradation and removal is the proteolytic cleavage of ECM components.16,17 The major enzymes that degrade ECM are matrix metalloproteinases (MMPs). This is a family of zinc-dependent endopeptidases that are involved in the degradation of ECM proteins together with other proteinases such as urokinase, plasmin, thrombin, and tissue plasminogen activator. MMPs can be either secreted or bound to the cell membrane in combination with different receptors and integrins.18 MMPs are known to play a role in pathological situations (cancer) but also in physiological situations (development) and in activating signaling molecules to restore homeostasis.11,17,19
Poly(ethyl acrylate) (PEA) is a synthetic material able to drive the organization of FN into physiological-like protein (nano) networks, a process that we have called material-driven FN fibrillogenesis.20–24 Here we address the ability of cells to remodel the FN fibrils assembled upon adsorption. Our overarching hypothesis is that the state (in terms of conformation, organization, and strength of interaction) of the layer of proteins adsorbed at the material interface will fine-tune the ability of cells to reorganize and degrade the ECM. To explore that idea, we used substrates of slightly different surface chemistry, PEA and poly(methyl acrylate) (PMA), on which very different conformations of FN occur upon simple adsorption from a protein solution. FN molecules remain globular and isolated on PMA, but material-driven FN fibrillogenesis occurs on PEA.20
Materials and Methods
Preparation of samples
Polymers were synthesized by radical polymerization of methyl acrylate and ethyl acrylate (Sigma-Aldrich, Steinheim, Germany). The initiator was benzoin at 1 wt% and 0.35 wt% respectively (98% pure, Scharlau, Barcelona, Spain). Afterwards, the samples were dried to constant weight. Spin-coating was used to coat glass coverslips and produce thin films of these polymers (Brewer Science, Rolla, MO). Polymer solutions were made in toluene with 6% PEA or 2.5% PMA. Samples for 24- and 6-well assays were centrifuged for 30 sec at 27 and 56 g, respectively. Finally, samples were dried at 60°C in vacuum.
Atomic force microscopy
A multimode device (NanoScope IIIa with Nanoscope 5.30r2 software, Veeco, Manchester, United Kingdom) operating in tapping mode 5%–10% below the resonance frequency was used for atomic force microscopy (AFM) experiments. Silicon cantilevers with stiffness 2.8 N/m and resonance frequency of 75 kHz (Veeco). Drive amplitude was 600 mV and the amplitude setpoint Asp (1.8 V) was kept at 80% of the free amplitude, maintaining the drive amplitude at 600 mV.
Protein adsorption
FN (human plasma, Sigma, St. Louis, MO) solutions in phosphate-buffered saline (PBS; 2 and 20 μg/mL) were used for AFM and quantification studies. Standard denaturing conditions were employed to separate FN from the samples as explained elsewhere.23 Using a semidry transfer cell system (Biorad, Hercules, CA), proteins were transferred to a polyvinylidene difluoride membrane (GE Healthcare, Madrid, Spain), and blocking was done by immersion in 5% skimmed milk in PBS. A rabbit antihuman FN polyclonal antibody (Sigma, 1:500) was used in PBS/0.1% Tween-20/2% skimmed milk to incubate the blot for 1 h at room temperature and then washed with PBS/0.1% Tween-20. Subsequently, the blot was incubated in HRP-conjugated secondary antibody (GE Healthcare) diluted 1:20,000 (in PBS/0.1% Tween-20/2% skimmed milk). Prior to exposing the blot to X-ray, a chemiluminescence-based system was used (GE Healthcare). Calibration was done using several dilutions of the FN solution and quantified by custom-made image analysis software.23
Antibody assay for FN conformation
After rinsing (PBS) and blocking (bovine serum albumin [BSA]/Dulbecco's PBS [DPBS]), HFN7.1 antibody (Developmental Hybridoma, Inc., Iowa City, IA) was used to assess the availability of the flexible link that joins the 9th and 10th type III FN domains. Primary antibody incubation (1:4000) was done for 1 h at 37°C. Washing (0.5% Tween 20/DPBS) and subsequent incubation in alkaline phosphatase–conjugated secondary antibody (1:5000) was done at 37°C for 1 h. Finally, surfaces were incubated in 4-methylumbelliferyl phosphate (4-MUP; Sigma) for 45 min at 37°C. A plate reader (Victor III, PerkinElmer, Waltham, MA) was used to quantify the reaction with fluorescence at 365 nm/465 nm.
Cell culture
NIH3T3 cells (European Collection of Cell Cultures, ECACC) were used. Cells were maintained prior to seeding on FN-coated substrates, in Dulbecco's modified Eagle's medium (1% penicillin-streptomycin and 10% calf serum). Cells were passaged twice a week using standard protocols. Material samples were coated with FN at concentration 2–20 μg/mL. Then, 2.6×104 (24-well) or 3×105 (6-well) cells were seeded and maintained at 37°C with 5% CO2 for 3 h. All cultures were realized in absence of calf serum except for integrin binding and gene expression (first hour without). Each experiment was performed in triplicate.
Immunofluorescence (vinculin)
After 4 h of culture, NIH3T3 cells were washed in DPBS (Gibco) and fixed in 10% formalin solution (Sigma) at 4°C. To permeabilize, cells were incubated for 5 min with 103 g/L sucrose, 2.92 g/L NaCl, 0.6 g/L MgCl2, 4.76 g/L HEPES buffer, 5 mL/L Triton X-100, pH 7.2. After blocking (1% BSA/DPBS), cells were incubated with vinculin primary antibody (Sigma, 1:400) for 1 h at room temperature and then rinsed in 0.5% Tween-20/DPBS. Cy3-conjugated secondary antibody (Invitrogen, Paisley, United Kingdom; 1:200) and BODIPY FL phallacidin (Invitrogen, 1:100) were dissolved together in 1% BSA/DPBS cells were then incubated with the mixture 1 h at room temperature. After washing, samples were mounted in Vectashield containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Visualization was done with a Nikon Eclipse 80i (Amsterdam, The Netherlands) fluorescent microscope.
Integrin binding
Bound integrins to FN were visualized via immunofluorescence following cross-linking and lysis of cellular components after 1.5 and 4 h. Samples were rinsed and integrin-FN pairs crosslinked for 20 min using ice-cold DTSSP (1.0 mM in DPBS/2 mM dextrose). The cross-linker not involved in the reaction was neutralized (50 mM Tris in DPBS) for 15 min and afterwards cells were removed in 0.1% SDS/350 mg/mL phenylmethylsulfonyl fluoride in DPBS. After rinsing and blocking (5% fetal bovine serum/0.1% Tween-20) for 1 h, anti–α5 integrin antibody was used to stain bound integrins (Millipore, Darmstadt, Germany; 1:400) for 1 h at 37°C. A Cy3-conjugated antibody (Jackson Research, Newmarket Suffolk, United Kingdom; 1:200) was used for visualization using a Nikon Eclipse 80i fluorescent microscope.
Protein expression analysis
Protein extraction was done by lysing cells using RIPA buffer (50 mM Tris-HCl pH 7.4, 1% nonidet p-40, 0.25% sodium deoxycolate, 150 mM NaCl, and 1 mM EDTA) supplemented with protease inhibitor cocktail tablets (Roche, Madrid, Spain) after 4 h of culture. Microcon YM-30 Centrifugal Filters units (Millipore) were use to concentrate the lysates that were afterwards separated in 7%–10% SDS PAGE under denaturing conditions. Standard Western blot was used (described above) for FAK, pFAK, and α5. The same amount of protein was charged in every case as measured with NanoDrop (ThermoScientific, Waltham, MA). Blots were incubated independently for primary antibody, FAK (Abcam, Cambridge, United Kingdom; 400 ng/mL), pFAK (Abcam; 1 μg/mL), and α5 integrin (Santa Cruz Biotechnology, Santa Cruz, CA; 1:100). HRP-linked secondary antibody was used with different dilutions: 1:25,000 for FAK, 1:10,000 for pFAK, and 1:150,000 for α5 integrin. Supersignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL) was used before X-ray exposition.
Gene expression analysis
Gene expression (mRNA) of α5 and αv integrins, MMP2, and MMP9 was analyzed after 4 h and 1 d of culture. RNAeasy Mini Kit (Qiagen) was used for RNA extraction, which was quantified using NanoDrop measurements (ThermoScientific). Afterwards, 1 μg was used as a template for SuperScript III RT (Invitrogen) and oligo(dT)12–18 (Invitrogen) for amplification. PCR experiments were done using Ampli Taq Gold 360 DNA polymerase (Invitrogen) Oligonucleotides are shown in Table 1. Independent experiments were done in triplicate.
Table 1.
Oligonucleotide Sequences Used for PCR Amplifications
| Gene | Sequence | Temperature (°C) | Reference |
|---|---|---|---|
| α5 integrin | F: GGA CGG AGT CAG TGT GCT G | 65 | 25 |
| R: GAA TCC GGG AGC CTT TGC TG | |||
| αv integrin | F: CAC CAG CAG TCA GAG ATG GA | 60 | 26 |
| R: GAA CAA TAG GCC CAA CGT CT | |||
| β-actin | F: TTC TAC AAT GAG CTG CGT GTG | 64 | M_007393.3 |
| R: GGG GTG TTG AAG GTC TCA AA | |||
| Gapdh | F: GTG TGA ACG GAT TTG GCC GT | 63 | NM_008084.2 |
| R: TTG ATG TTA GTG GGG TCT CG | |||
| MMP2 | F: GCC ATC CTT TCT CAA AGT TGT | 60 | 27 |
| R: GAG TTG GCA GTG CAA TAC CT | |||
| MMP9 | F: TGC CAC CAG GAA CAG G | 61 | 28 |
| R: ACA GCC AAC TAT GAC CAG |
Metalloproteinase activity analysis
Gelatin zymography was performed using 15 μL of supernatant after 4 and 24 h of culture. The samples were mixed with an equal volume of 2× loading buffer (2.5 mM Tris-HCl pH 6.8, 25% glycerol, 4% SDS, 0.01% bromophenol blue) and incubated for 10 min at room temperature before loading to 10% gelatin-ready zymogram gel (Biorad, 1611167). Gels were run at 50 V for 4 h at 4°C. The gels were incubated in 2.5% Triton X-100 for renaturation for 30 min. They were then equilibrated in fresh developing buffer (50 mM Tris base, 200 mM NaCl, 5 mM anhydride CaCl, 0.02%, 30% brij-35) and incubated at 37°C overnight. Gels were stained with 0.5% Coomassie R-250 (40% methanol, 10% acetic acid) for 2 h and finally destained with destaining buffer (40% methanol, 10% acetic acid) for 1 h. Areas of protease activity appear as clear bands against a dark blue background where the protease had digested the gelatin substrate.
Statistical analysis
All experiments were performed in triplicate unless otherwise noted. Data are described as mean±standard error. SYSTAT 8.0 (SPSS) was used to for one-way ANOVA, and when differences were significant (95% confidence level), pair-wise comparisons were done using a Tukey post hoc test.
Results
Fibronectin adsorption
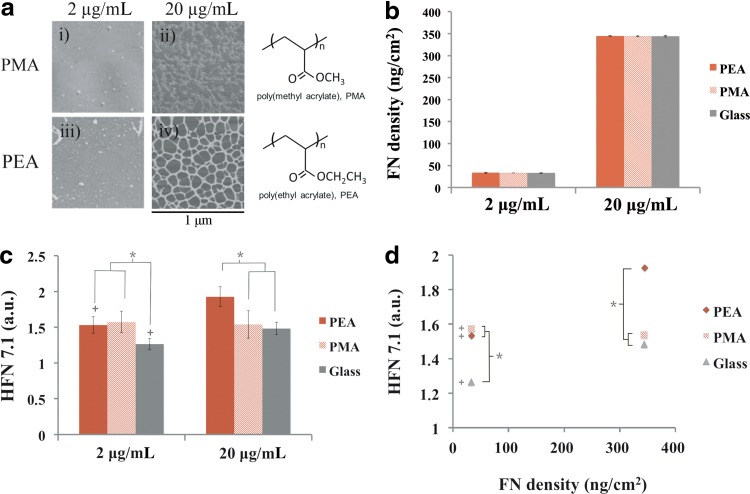
The molecular distribution of FN on the different substrates was previously studied by us using AFM.22 Figure 1a includes the organization of FN on PEA and PMA after adsorption from solutions of different concentrations (2 and 20 μg/mL). For each substrate, FN organization and distribution on the surface depends on the concentration of the initial solution from which the protein is adsorbed. Adsorption from a FN solution of concentration 2 μg/mL leads to isolated FN molecules on the material surface. As the concentration of the FN solution increases, the formation of a FN network occurs on PEA (e.g., material-driven FN fibrillogenesis) but not on PMA.22
FIG. 1.
Fibronectin adsorption and activity on the different substrates. (a) PEA and PMA chemical structure and fibronectin distribution on the different conditions as observed by the phase magnitude in AFM. The protein was adsorbed for 10 min from different solutions of 2 and 20 μg/mL concentration. (b) FN surface density after adsorption from two solutions with concentrations of 2 and 20 μg/mL. (c) Monoclonal antibody binding for HFN7.1 on the different substrates after FN adsorption from two solution of concentrations 2 and 20 μg/mL. (d) Activity of the adsorbed FN on the different substrates obtained by plotting the monoclonal antibody binding for HFN7.1 relative to the FN surface density calculated in (b). +There is a statistically significant difference between the conditions of 2 and 20 μg/mL; *conditions with a significant difference (P≤0.05). PEA, poly(ethyl acrylate); PMA, poly(methyl acrylate); AFM, atomic force microscopy; FN, fibronectin.
The surface density of adsorbed FN was quantified indirectly from the amount of protein remaining nonadsorbed in the supernatant using Western blot.23 Figure 1b shows the surface density of adsorbed FN from solutions of concentration 2 and 20 μg/mL. As expected, the amount of adsorbed FN was higher as the concentration of the protein solution increased. In contrast, there was no significant difference for the amount of adsorbed FN between material substrates at all FN concentrations, which remained constant and approximately 47 and 340 ng/cm2 for the 2 and 20 μg/mL FN solutions, respectively.
To evaluate the availability of integrin binding domains after FN adsorption, enzyme-linked immunosorbent assay with a monoclonal antibody was used (Fig. 1c). This is a well-established method to probe for structural or conformational changes in adsorbed proteins.29,30 HFN7.1 (monoclonal antibody) was directed against the flexible linker between the 9th and 10th type III repeats of FN, a probe for cell adhesion and integrin binding.31 Adsorption from the 2 μg/mL solution resulted in the same FN activity on PEA and PMA. Further increase of the concentration of the protein solution (20 μg/mL) provided higher activity of FN on PEA, but not on PMA. It is interesting to note that the amount of protein on a surface and its biological activity are two independent parameters. In our case, after the amount of protein on the surface was increased ∼10-fold, the activity measured by HFN 7.1 binding was almost the same, which revealed the role of protein–protein interactions in hiding the availability of the integrin binding sequence of FN. Since the same amount of FN is adsorbed on every material for each one of the adsorbing solutions, Figure 1d represents the activity of the protein (availability of cell adhesion domains) as a function of the protein surface density, which revealed higher activity of FN on PEA than PMA for the highest FN density.
Cell adhesion
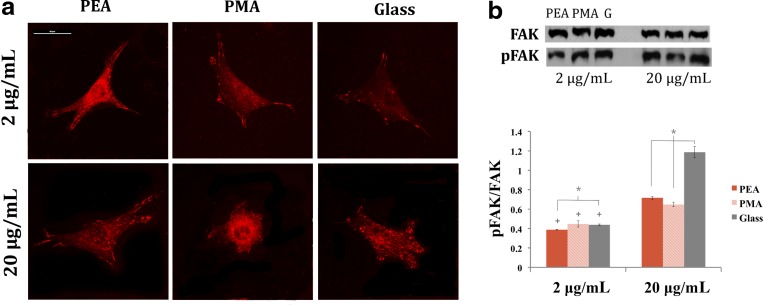
To gain insights into the mechanisms controlling matrix remodeling on material-driven FN networks, we began by examining the organization of proteins involved in the formation of focal adhesion complexes by immunofluorescence. Figure 2 shows the distribution of vinculin in cells adhering on the different substrates. Poorly developed focal adhesions were observed on both PEA and PMA for the 2 μg/mL FN coating. However, well-defined focal plaques formed on PEA, but not on PMA, for the 20 μg/mL FN coating. Likewise, although the formation of F-actin fibers on PEA and PMA already occurred at the lowest FN concentration, more prominent F-actin cables terminating in well-developed focal adhesion complexes were found on PEA at the highest FN concentration (Fig. 2).
FIG. 2.
Cell adhesion and focal adhesion kinase (FAK) phosphorylation after 4 h on both substrates. (a) Distribution of focal adhesion protein vinculin and its incorporation into focal contact plaques. Scale bar=50 μm. (b) Representative Western blot bands for FAK and pFAK with quantification by image analysis of Western blot bands. The error bars represent the standard deviation of three independent experiments. +There is a statistically significant difference between FN concentrations (2 and 20 μg/mL). *Conditions with a significant difference (P≤0.05).
FAK localizes to focal adhesions to activate multiple signaling pathways that regulate cell migration, survival, proliferation, and differentiation.7 The phosphorylation of FAK at Y-397, the autophosphorylation site and a binding site for Src and PI-3 kinases,32 is shown in Figure 2. It remained the same for PEA and PMA at the lower FN concentration, and it increased at the highest FN concentration (20 μg/mL). Moreover, higher levels of pFAK were obtained on PEA than PMA, revealing enhanced signaling from the substrate-assembled FN networks.
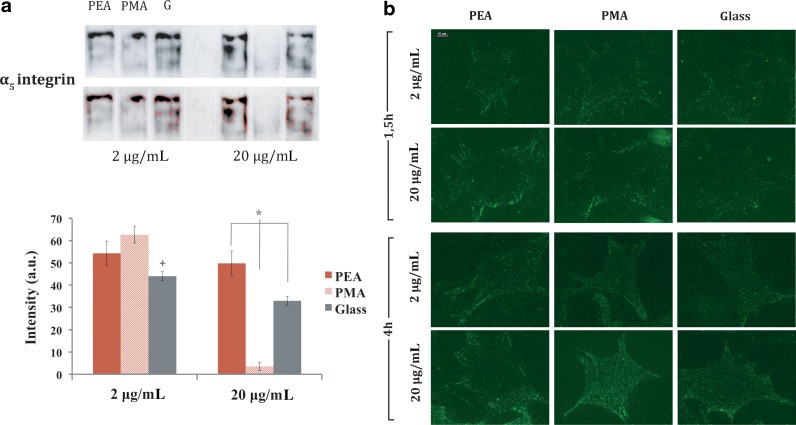
To gain insights into the adhesion mechanism on these synthetic FN matrices assembled at the material interface, we examined expression and binding of α5β1 integrin to the adsorbed FN since this receptor provides the primary adhesion mechanism in this cell model.33 First, we examined α5 integrin expression by Western blot (Fig. 3a). No difference was found on PEA between the two FN concentrations used, with a similar expression level on PMA with FN at 2 μg/mL. Surprisingly, very low integrin expression was found on PMA after increasing the concentration of the adsorbing solution to 20 μg/mL. Afterwards, integrin binding to FN-coated materials was analyzed via immunostaining following cross-linking of bound integrins to FN and extraction of cellular components (Fig. 3b). No significant differences were found among the material surfaces regardless of FN concentration, demonstrating that the substrate-assembled FN network does not alter integrin binding in the presence of serum.22
FIG. 3.
α5 integrin expression after 4 h on the different substrates. (a) Representative Western blot bands for α5 integrin and quantification by image analysis of Western blot bands. The expected band for this antibody might be quite wide according to the manufacturer, which is why we developed in-house software to detect the band based on the intensity threshold criterion. Error bars represent the standard deviation of three independent experiments. +Statistically significant difference between FN concentrations (2 and 20 μg/mL). *Conditions with a significant difference. (b) α5 integrins bound to FN are observed after crosslinking and extraction of cellular components. Scale bar=10 μm (P≤0.05).
Gene expressions for α5 and αv integrins were obtained by reverse-transcription PCR (Fig. 4). α5 expression (Fig. 4b) decreases with time but increases with FN concentration. At the lowest FN concentration (2 μg/mL), α5 expression was higher on PEA than PMA (Fig. 4b), as it was, but with less difference, at the 20 μg/mL FN concentration. In addition, the opposite trend was found for αv expression, which remained higher on PMA than PEA at both FN coatings (Fig. 4c).
FIG. 4.
Integrin adhesion quantified by gene expression of α5 and αv integrins. (a) Representative bands of reverse-transcription (RT)-PCR for α5 and αv integrins after 4 h and 1 day of culture on the different substrates (PEA, PMA, glass; unlabeled bands follow the same pattern). Gapdh was included as a constitutive gene. (b, c) Quantification of gene expression for α5 and αv integrins on the different substrates. The intensity of each band was related to the level of Gapdh under the same conditions, and represented as fold change relative to the values obtained for 2 μg/mL on glass. Error bars are represented by the standard deviation of three different experiments. *Conditions with a significant difference for every protein concentration and culture time (P≤0.05).
Matrix degradation
Seeking to understand the relationship between FN adsorption, cell adhesion, and matrix remodeling, we next examined MMP2, MMP9, and MMP13 at both protein activity and gene expression levels. Figure 5 shows gene expression for MMP2 and MMP9 after 4 h and 1 day. The MMP2 level (Fig. 5b) was lower on PEA than PMA at the highest concentration of the FN coating (20 μg/mL), that is, on the material-assembled FN fibrils. Only at the shorter time (4 h) and lower FN concentration (2 μg/mL) was MMP2 expression similar on PEA and PMA. The opposite trend was found for MMP9 expression (Fig. 5c): similar levels were found on both PEA and PMA at every time but at 4 h at the lowest FN concentration (2 μg/mL).
FIG. 5.
Matrix degradation quantified by gene expression of MMP2 and MMP9. (a) Representative bands of RT-PCR for MMP2 and MMP9 after 4 h and 1 day of culture on the different substrates (PEA, PMA, glass; unlabeled bands follow the same pattern). Gapdh was included as a constitutive gene. (b, c) Quantification of gene expression for MMP2 and MMP9 on the different substrates. The intensity of each band was related to the level of Gapdh under the same conditions, and represented as fold change relative to the values obtained for 2 μg/mL on glass. Error bars represent the standard deviation of three different experiments. *Conditions with a significant difference (P≤0.05). MMP, matrix metalloproteinase.
Further insights in matrix degradation can be obtained by investigating MMP activity by gelatin zymography (Fig. 6). Both the pro-form of the protein and the active one (MMP2) were highly dependent on the underlying material surface and the observation time, with higher level of activity on PEA than PMA (Fig. 6c, d). Increasing the concentration of the FN solution did not significantly alter the activity of the protein. (Fig. 6c, d). The activity of MMP9 and its forms (Fig. 6b) could only be observed after 1 day of culture, while the pro-MMP13 behaved as MMP2 for the higher concentration of the FN solution (Fig. 6e). That is to say, MMP13 was more active on cells seeded on the material-driven FN network after 1 day.
FIG. 6.
Matrix degradation quantified by enzymatic activity of MMP2 and MMP9. (a) Representative bands of gelatin zymography for pro-MMP9, MMP9+TIMP-1, pro-MMP9, MMP9, pro-MMP2, MMP2, and pro-MMP13 after 4 h and 1 day of culture on the different substrates (PEA, PMA, Glass). (b) Amplification of representative bands of gelatin zymography for pro-MMP9, MMP9+TIMP-1, pro-MMP9, MMP9, and pro-MMP13 with the different substrates and FN adsorption at 1 day of culture. (c, d, e) Quantification of MMP2, pro-MMP2, and pro-MMP13 activity on the different substrates. Error bars are represented by the standard deviation of three different experiments. *Conditions with a significant difference. TIMP, tissue inhibitors of metalloproteinase (P≤0.05).
Discussion
Material-based research direct cellular responses has demanded significant efforts to engineer materials that summarize the characteristics of the ECM, such as the presence of adhesive and degradable sequences.34,35 Nevertheless, several attempts have been made to reconstitute the network structure and bioactivity of FN fibrillar matrices in a cell-free way. The structure of FN has to be changed in order to promote fibril assembly, which is why unfolding and denaturing agents as well as mechanical forces application have been used.36–39 We have previously shown that FN adsorption on PEA induces conformational changes in the molecule facilitating FN-FN interactions and driving FN fibril assembly.20–24 It is important to note that this material-driven assembly of FN on PEA occurs in a physiological-like way because it involves the 70 kDa amino-terminal regions of the molecule as in vivo.40,41
We have previously shown the biological activity of this material-driven FN network in terms of cell adhesion, signaling, cytoskeleton organization, matrix reorganization, and cell differentiation.20–24 Here we address the link between material-driven FN assembly and matrix protein remodeling (degradation) at the cell–material interface, which we hypothesized must be related to integrin-mediated adhesion through the conformation of the adsorbed protein. We investigated two polymer surfaces that consist in a vinyl chain with a slightly different (one carbon) side group: PEA and PMA.22 These surfaces adsorbed the same amount of FN and showed similar wettability (Fig. 1b). However, the conformation and distribution of the protein following passive adsorption onto these surfaces are completely different (Fig. 1a). FN was self-organized into fibrils on PEA (Fig. 1a-iv) as the so-called material-driven fibrillogenesis, whereas individual globule-like molecules or small molecular aggregates were present on PMA (Fig. 1a-iii). FN fibril formation on PEA was dependent on the FN solution concentration, since lower concentrations (e.g., 2 μg/mL) result in dispersed adsorbed molecules.22
The amount of FN adsorbed on every surface (PEA, PMA) was approximately the same for both concentrations of the protein solution (Fig. 1b). However, the different distribution of the protein revealed by AFM (Fig. 1a)—dispersed globular aggregates on PMA versus a network of assembled FN fibrils on PEA—also involved different availability of the integrin binding domain in FN: Figure 1c shows significantly higher signal for the HFN7.1 antibody against the flexible linker joining the III9 and III10 domains of FN.31 Moreover, this higher activity of FN on PEA was very much dependent on the concentration of the protein solution, and it was drastically diminished upon adsorption from the 2 μg/mL FN solution (Fig. 1c). To fully assess these results, we have included in Figure 1d a representation for the availability of HFN7.1 antibody versus the amount of FN adsorbed on every surface. This representation clearly reveals the synergistic effect of the material-driven FN fibrillogenesis on FN activity, and it disregards any influence of the total amount of adsorbed FN in a comparison of results between PEA and PMA. The higher availability of cell adhesion domains for the material-driven FN fibrils on PEA supports previous results for the biological activity of this surface in terms of cell adhesion and differentiation.20–24
Differences in the availability of FN adhesion domains after adsorption on PEA and PMA from a solution of concentration 20 μg/mL influence the initial cell–material interaction, in terms of integrin expression, focal adhesion formation, and F-actin cytoskeleton development. We examined binding of α5β1 integrin to the adsorbed FN since this receptor provides the primary adhesion mechanism in this cell model.33 Immunofluorescence staining following crosslinking of bound integrins to FN and extraction of cellular components revealed no significant differences in integrin binding among FN on different surfaces. To enhance the visualization of integrin clusters, this experiment was performed in the presence of serum, which is known to contain large amounts of FN and vitronectin that might perturb the effect of the provisional FN matrix previously assembled at the material surface. This is why we investigated protein and gene expression in absence of any additional serum, to enable focusing on the sole effect of FN at the material interface. α5 integrin expression was higher for cells seeded on the material-driven FN network (PEA) than on the dispersed globular-like FN matrix adsorbed on PMA (Fig. 3a). The same result was obtained at the gene level for the α5 receptor (Fig. 4a). The opposite trend was found for αv expression, which was enhanced on PMA compared with PEA. These results reveal that α5 is the main receptor involved during the initial cell interaction with the material-driven FN network, as happens for the interaction of cells with natural physiological matrices.33 α5β1 binding has been related to the simultaneous availability of the synergy and RGD sequences within FN.33 In contrast, when FN is adsorbed as discrete aggregates on PMA, the availability of the whole integrin binding site of FN is reduced (Fig. 1) and cell adhesion mostly occurs through the αv receptor, which only needs the exposition of the RGD sequence.
After adhesion, cells reorganize the adsorbed layer of FN at the material interface and then secrete matrix proteins.9 In this way, cells assemble FN into a network of fibrils. Along the way, FN experience particular conformational changes, that can be limited after adsorption on the material surface.40,41 This may explain the role of the properties of material surfaces (e.g., wettability) in FN matrix formation.9,42 We have previously shown that cell-mediated FN reorganization does not occur on the material-driven FN network assembled on PEA, due to the high strength of interaction between FN fibrils and the underlying surface chemistry.43 The enhanced initial biological response of the material-driven FN network (Figs. 2–4) can be explained as a consequence of the following phenomena occurring at the cell–material interface after FN assembly into physiological-like fibrils: (i) the availability of the integrin binding sequence (Fig. 1); (ii) α5 integrin expression, which leads to enhanced focal adhesion formation and cytoskeleton development (Figs. 2, 3); and (iii) enhanced phosphorylation of FAK (Fig. 2).
Matrix protein dynamics is significantly altered on material-driven FN fibrils, since cells are not able to reorganize the underlying matrix of proteins at the material interface.43 Reorganization processes at the material interface have been thought to direct the ability of cells to secrete new matrix, so that FN is only secreted after protein reorganization: cell movements, in the range of the size of the focal adhesion plaques, must take place for matrix deposition to occur normally.21 This has been clearly shown for model hydrophilic and hydrophobic surfaces.9,42,44,45 However, matrix dynamics involve not only the organization of new secreted matrix, but also proteolytic degradation (remodeling) to remove the excess ECM. The relationship between the biocompatibility of a material and matrix remodeling at the cell–material interface remains to be elucidated. The importance of matrix remodeling has been recognized in the design of new biomaterials that include MMP-sensitive sequences.46–48
We hypothesize that the material-driven FN network must also lead to a different proteolytic activity at the material interface, compared with a similar chemistry on which the protein is not organized (Fig. 1). The effect of material chemistry on the proteolytic activity of cells has been barely addressed so far. Expression of MMP2 and MMP9 has been observed in cells cultured on tissue culture polystyrene. There are only a few studies that address the effect of synthetic materials on MMP expression and activity.14,49–52 On model surfaces with controlled wettability and surface chemistry, the activation of proteolytic routes occurs in an MMP-dependent way. It has been previously suggested a direct relationship between MMP9 and FN activity at the material interface,15 which we also found in our experiments (Fig. 6b): FN activity was higher on the assembled FN fibrils (Fig. 1), as was the qualitative interpretation of the band for MMP9 activity in Figure 6b (without quantification). Moreover, MMP2 and MMP13 activities (zymography) were much higher on the material-assembled FN matrix, after 4 h and 1 day (Fig. 6). MMP2 has FN type II repeats inserted into the catalytic domain,11 and it has been found to cleave vitronectin and FN in vivo, leading to enhanced cell migration.11,12 Degradation of the assembled fibrils at the material interface would enhance the exposure of adhesion sites, which would support enhanced integrin (α5) expression and focal adhesion formation in this system. It also supports the proposed relationship between FN activity, the ability to reorganize the underlying layer of proteins at the material interface, and proteolytic cascades: higher MMP activity is required to remodel the provisional matrix when cells are not able to reorganize this layer of proteins at the material interface, as in fact occurs with the material-driven FN network. It is precisely this initial proteolytic activity (after 4 h) that enhanced cellular behavior on the FN fibrils assembled on PEA. This result supports the hypothesis that cells need to rearrange the initial layer of proteins at the material interface, and that this is very much related to the biocompatibility of materials: when the protein–material interaction is so strong that reorganization cannot occur, proteolytic cascades are enhanced to degrade FN, seeking to start microscale movements at the material interface to direct cell function.
Conclusions
Material-driven FN fibrillogenesis provides physiological-like FN fibrils upon simple adsorption on specific material chemistries. We have shown that FN assembly results in protein conformation and distribution at the material interface that enhanced the availability of the integrin-binding sequence. Thus cell adhesion occurs primarily through the specific α5β1 receptor occupancy, with enhanced focal adhesion formation and FAK signaling. Furthermore, we have shown that MMP activity is also enhanced on the material-driven FN fibrils, through a mechanism that is dependent on the ability of cells to reorganize the adjacent layer of proteins at the cell/material interface.
Abbreviations
- AFM
atomic force microscopy
- BSA
bovine serum albumin
- DPBS
Dulbecco's PBS
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- FN
fibronectin
- MMP
matrix metalloproteinase
- 4-MUP
4-methylumbelliferyl phosphate
- PBS
phosphate-buffered saline
- PEA
poly(ethyl acrylate)
- pFAK
phosphorylation of FAK
- PMA
poly(methyl acrylate)
Acknowledgments
We acknowledge the financial support of the Spanish Ministry of Science and Innovation through the project MAT2012-38359-C03-01/03, the support of CIBER-BBN, and the fellowship EEBB-I-13-07652.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Grinnell F. Focal adhesion sites and the removal of substratum-bound fibronectin. J Cell Biol. 1986;103:2697–2706. doi: 10.1083/jcb.103.6.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarz US. Bischofs IB. Physical determinants of cell organization in soft media. Med Eng Phys. 2005;27:763–772. doi: 10.1016/j.medengphy.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Sipe J. Tissue engineering and reparative medicine. Reparative Medicine: Growing Tissues and Organs. Ann NY Acad Sci. 2002;961:1–9. doi: 10.1111/j.1749-6632.2002.tb03040.x. [DOI] [PubMed] [Google Scholar]
- 4.Anselme K. Bigerelle M. Noel B, et al. Qualitative and quantitative study of human osteoblast adhesion on materials with various surface roughnesses. J Biomed Mater Res. 2000;49:155–166. doi: 10.1002/(sici)1097-4636(200002)49:2<155::aid-jbm2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 5.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 6.Garcia AJ. Get a grip: integrins in cell-biomaterial interactions. Biomaterials. 2005;26:7525–7529. doi: 10.1016/j.biomaterials.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 7.Mitra SK. Hanson DA. Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 8.Griffith LG. Naughton G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 9.Altankov G. Groth T. Reorganization of substratum-bound fibronectin on hydrophilic and hydrophobic materials is related to biocompatibility. J Mater Sci. 1994;5:732–737. [Google Scholar]
- 10.Avnur Z. Geiger B. The removal of extracellular fibronectin from areas of cell-substrate contact. Cell. 1981;25:121–132. doi: 10.1016/0092-8674(81)90236-1. [DOI] [PubMed] [Google Scholar]
- 11.Page-McCaw A. Ewald AJ. Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenny HA. Kaur S. Coussens LM, et al. The initial steps of ovarian cancer cell metastasis are mediated by MMP-2 cleavage of vitronectin and fibronectin. J Clin Invest. 2008;118:1367–1379. doi: 10.1172/JCI33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang CM. Chien CS. Yao CC, et al. Mechanical strain induces collagenase-3 (MMP-13) expression in MC3T3-E1 osteoblastic cells. J Biol Chem. 2004;279:22158–22165. doi: 10.1074/jbc.M401343200. [DOI] [PubMed] [Google Scholar]
- 14.Wan R. Mo Y. Zhang X, et al. Matrix metalloproteinase-2 and -9 are induced differently by metal nanoparticles in human monocytes: The role of oxidative stress and protein tyrosine kinase activation. Toxicol Appl Pharmacol. 2008;233:276–285. doi: 10.1016/j.taap.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llopis-Hernandez V. Rico P. Ballester-Beltran J, et al. Role of surface chemistry in protein remodeling at the cell-material interface. PLoS One. 2011;6:e19610. doi: 10.1371/journal.pone.0019610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koblinski JE. Ahram M. Sloane BF. Unraveling the role of proteases in cancer. Clin Chim Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 17.Mohamed MM. Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 18.Buck MR. Karustis DG. Day NA, et al. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem J. 1992;282:273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodríguez D. Morrison CJ. Overall CM. Matrix metalloproteinases: What do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim Biophys Acta. 2010;1803:39–54. doi: 10.1016/j.bbamcr.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Brizuela Guerra N. Gonzalez-Garcia C. Llopis V, et al. Subtle variations in polymer chemistry modulate substrate stiffness and fibronectin activity. Soft Matter. 2010;6:4748–4755. [Google Scholar]
- 21.Gonzalez-Garcia C. Sousa SR. Moratal D, et al. Effect of nanoscale topography on fibronectin adsorption, focal adhesion size and matrix organisation. Colloids Surf B Biointerfaces. 2010;77:181–190. doi: 10.1016/j.colsurfb.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 22.Salmeron-Sanchez M. Rico P. Moratal D, et al. Role of material-driven fibronectin fibrillogenesis in cell differentiation. Biomaterials. 2011;32:2099–2105. doi: 10.1016/j.biomaterials.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 23.Rico P. Rodriguez Hernandez JC. Moratal D, et al. Substrate-induced assembly of fibronectin into networks: influence of surface chemistry and effect on osteoblast adhesion. Tissue Eng Part A. 2009;15:3271–3281. doi: 10.1089/ten.TEA.2009.0141. [DOI] [PubMed] [Google Scholar]
- 24.Gugutkov D. Gonzalez-Garcia C. Rodriguez Hernandez JC, et al. Biological activity of the substrate-induced fibronectin network: insight into the third dimension through electrospun fibers. Langmuir. 2009;25:10893–10900. doi: 10.1021/la9012203. [DOI] [PubMed] [Google Scholar]
- 25.do Carmo Costa M. Bajanca F. Rodrigues A, et al. Ataxin-3 plays a role in mouse myogenic differentiation through regulation of integrin subunit levels. PLoS One. 2010;5:e11728. doi: 10.1371/journal.pone.0011728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veeravalli KK. Chetty C. Ponnala S, et al. MMP-9, uPAR and cathepsin B silencing downregulate integrins in human glioma xenograft cells in vitro and in vivo in nude mice. PLoS One. 2010;5:e11583. doi: 10.1371/journal.pone.0011583. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lee EK. Lee YS. Lee H, et al. 14-3-3 epsilon protein increases matrix metalloproteinase-2 gene expression via p38 MAPK signaling in NIH3T3 fibroblast cells. Exp Mol Med. 2009;41:453–461. doi: 10.3858/emm.2009.41.7.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao H. Miao Z. Hui-Chen, et al. Nucleophosmin gene mutations promote NIH3T3 cell migration and invasion through CXCR4 and MMPs. Exp Mol Pathol. 2011;90:38–44. doi: 10.1016/j.yexmp.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Ugaova T. Zamarron C. Veklich Y, et al. Conformational transitions in the cell-binding domain of fibronectin. Biochemistry. 1995;34:4457–4466. doi: 10.1021/bi00013a039. [DOI] [PubMed] [Google Scholar]
- 30.McClary KB. Ugarova T. Grainger DW. Modulating fibroblast adhesion, spreading, and proliferation using self-assembled monolayer films of alkylthiolates on gold. J Biomed Mater Res. 2000;50:428–439. doi: 10.1002/(sici)1097-4636(20000605)50:3<428::aid-jbm18>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 31.Schoen RC. Bentley KL. Klebe RJ. Monoclonal-antibody against human fibronectin which inhibits cell attachment. Hybridoma. 1992;1:99–108. doi: 10.1089/hyb.1.1982.1.99. [DOI] [PubMed] [Google Scholar]
- 32.Schaller M. Hilderbrand J. Shannon J, et al. Autophosphorylation of the focal adhesion kinase, Pp125(fak), directs Sh2 dependent binding of Pp60(src) Mol Cell Biol. 1994;14:1680–1688. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruoslahti E. Obrink B. Common principles in cell adhesion. Exp Cell Res. 1996;227:1–11. doi: 10.1006/excr.1996.0243. [DOI] [PubMed] [Google Scholar]
- 34.Lutolf MP. Gilbert PM. Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrie TA. Raynor JE. Dumbauld DW, et al. Multivalent integrin-specific ligands enhance tissue healing and biomaterial integration. Sci Transl Med. 2010;2:45ra60. doi: 10.1126/scitranslmed.3001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosher D. Johnson R. In vitro formation of disulfide-bonded fibronectin multimers. J Biol Chem. 1983;258:6595–6601. [PubMed] [Google Scholar]
- 37.Morla A. Zhang ZH. Ruoslahti E. Superfibronectin is a functionally distinct form of fibronectin. Nature. 1994;367:193–196. doi: 10.1038/367193a0. [DOI] [PubMed] [Google Scholar]
- 38.Baneyx G. Vogel V. Self-assembly of fibronectin into fibrillar networks underneath dipalmitoyl phosphatidylcholine monolayers: role of lipid matrix and tensile forces. Proc Natl Acad Sci USA. 1998;96:12518–12523. doi: 10.1073/pnas.96.22.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulmer J. Geiger B. Spatz JP. Force-induced fibronectin fibrillogenesis in vitro. Soft Matter. 2008;4:1998–2007. [Google Scholar]
- 40.Schwarzbauer JE. Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J Cell Biol. 1991;113:1463–1473. doi: 10.1083/jcb.113.6.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Y. Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Altankov G. Groth T. Fibronectin matrix formation and the biocompatibility of materials. J Mater Sci Mater Med. 1996;7:425–429. [Google Scholar]
- 43.Gugutkov D. Altankov G. Rodriguez Hernandez JC, et al. Fibronectin activity on substrates with controlled -OH density. J Biomed Mater Res Part A. 2010;92A:322–331. doi: 10.1002/jbm.a.32374. [DOI] [PubMed] [Google Scholar]
- 44.Altankov G. Grinnell F. Groth T. Studies on the biocompatibility of materials: Fibroblast reorganization of substratum-bound fibronectin on surfaces varying in wettability. J Biomed Mater Res. 1996;30:385–391. doi: 10.1002/(SICI)1097-4636(199603)30:3<385::AID-JBM13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 45.Altankov G. Groth T. Krasteva N, et al. Morphological evidence for a different fibronectin receptor organization and function during fibroblast adhesion on hydrophilic and hydrophobic glass substrata. J Biomater Sci Polym Ed. 1997;8:721–740. doi: 10.1163/156856297x00524. [DOI] [PubMed] [Google Scholar]
- 46.Lutolf MP. Lauer-Fields JL. Schmoekel HG, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics. Proc Natl Acad Sci USA. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelps EA. Landazuri N. Thule PM, et al. Bioartificial matrices for therapeutic vascularization. Proc Natl Acad Sci USA. 2010;107:1–6. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider RK. Puellen A. Kramann R, et al. The osteogenic differentiation of adult bone marrow and perinatal umbilical mesenchymal stem cells and matrix remodelling in three-dimensional collagen scaffolds. Biomaterials. 2010;31:467–480. doi: 10.1016/j.biomaterials.2009.09.059. [DOI] [PubMed] [Google Scholar]
- 49.Uchida M. Shima M. Shimoaka T, et al. Regulation of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) by bone resorptive factors in osteoblastic cells. J Cell Physiol. 2000;185:207–214. doi: 10.1002/1097-4652(200011)185:2<207::AID-JCP5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 50.Zambuzzi WF. Paiva KBS. Menezes R, et al. MMP-9 and CD68(+) cells are required for tissue remodeling in response to natural hydroxyapatite. J Mol Histol. 2009;40:301–309. doi: 10.1007/s10735-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 51.Chung AS. Waldeck H. Schmidt DR, et al. Monocyte inflammatory and matrix remodeling response modulated by grafted ECM-derived ligand concentration. J Biomed Mater Res Part A. 2009;91A:742–752. doi: 10.1002/jbm.a.32259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ducy P. Zhang R. Geoffroy V, et al. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]