Abstract
Autism is a neurodevelopmental disorder of unknown origin that manifests in early childhood. Autism spectrum disorders (ASDs) refer to a broader group of neurobiological conditions, pervasive developmental disorders. Despite several arguments for a strong genetic contribution, the molecular basis in most cases remains unexplained. Several studies have reported an association between ASDs and mutations in the mitochondrial DNA (mtDNA) molecule. In order to confirm these causative relationship, we screened 21 individuals with idiopathic ASDs for a number of the most common mtDNA mutations. We identified two patients with candidate mutations: m.6852G>A that produces an amino acid change of glycine to serine in the MT-CO1 gene and m.8033A>G (Ile→Val) in the MT-CO2 gene. Overall, these findings support the notion that mitochondrial mutations are associated with ASDs. Additional studies are needed to further define the role of mitochondrial defects in the pathogenesis of autism.
Keywords: Autism spectrum disorders (ASDs), Genetic testing, Mitochondrial DNA (mtDNA) mutations
INTRODUCTION
Autism spectrum disorders (ASDs) are a heterogeneous group of neurodevelopmental disorders characterized by impaired reciprocal social interaction, lack of communication, isolated interests and repetitive or stereotyped behaviors [1]. It is now estimated that about one out of every 150 children is affected with ASDs [2]. Most cases are idiopathic, although there is increasing evidence that ASDs have an important genetic component with aetiological heterogeneity [3]. Some cases of autism have been associated with several different organic conditions including mitochondrial dysfunction [4–6], however, very few individuals with mitochondrial DNA (mtDNA) mutations have been found [7–10]. Pons et al.[7] reported two ASDs patients with the 3243A>G mutation and postulated that ASDs, with or without additional neurological features, can be an early presentation of the 3243A>G mutation and can be a prominent clinical manifestation of mtDNA depletion. One study of 810 patients with ASDs identified two individuals (0.2%) with the same mutation (3243A>G) [8]. Graf et al.[9] described a family with different neurological disorders associated with the mtDNA 8363G>A mutation; the phenotype of one child in the family was consistent with ASDs. Several variants of probable or unclear pathogenicity (3397A>G, 4295A>G, 3394T>C, 10394C>T, 11809T>C and 11984T>C) were detected in 25 ASDs patients [10]. Another study of 129 individuals with Asperger syndrome and 138 mothers of individuals with Asperger syndrome searched for the 3243A>G mutation, but no such mutation was found [11]. According to the Álvarez-Iglesias et al. “Therefore there is a certain amount of evidence that seems to suggest a role of mtDNA variants in ASDs; however, this evidence is weak and/or has not been replicated yet in different independent cohorts of patients.” [12]. To further investigate the hypothesis of an aetiological link between ASDs and mitochondrial disorders, we screened 21 autistic children for mtDNA mutations.
MATERIALS AND METHODS
Subjects. The patients included in this study were 21 autistic children (18 males and 3 females) ranged in age from 2 to 11 years. The diagnosis of ASDs was made by child neuropsychiatrists based on the criteria established in the American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) [1]. All patients are of Bulgarian ethnicity. The children were referred to our clinic for genetic and/or metabolic evaluation of autism but not specifically for evaluation for mitochondrial disease. All patients had an evaluation by neurologists, psychologists and clinical geneticists. Laboratory tests including standard karyotyping, fragile-X molecular testing. Molecular karyotyping by microarray comparative genomic hybridization was performed. Children with genetic anomalies potentially causative of ASDs were excluded from the study.
The research protocol was approved by The Ethics Committee for Research Investigations to the Medical University, Sofia, Bulgaria. Informed consent was obtained from the guardians of all patients.
Methods. Genomic high molecular weight DNA samples were extracted from peripheral blood lymphocytes, using a GENO-M6 automated system (Geno Vision, Geno M™-6; Olerup GmbH, Vienna, Austria). Polymerase chain reaction (PCR)-sequencing based technique(SBT) was used for detection of single nucleotide changes along the mtDNA. The mitochondrial genome was amplified regionally with an approximate fragments length of 1500 bp. Direct cycle sequencing with fluorescently labeled and terminating nucleotides was performed (BigDye™ Terminator Cycle sequencing Kit; Applied Biosystems, Foster City, CA, USA). Each DNA sequence variant was evaluated for pathogenicity by search in the MITOMAP and mtDB-Human Mitochondrial Genome databases [13,14]. The method allowed target identification of the most common pathogenic mutation points via the mitochondrial genome of the patients. Moreover it was possible to detect additional variants through the sequence as polymorphisms, silent and non synonymous mutations.
RESULTS
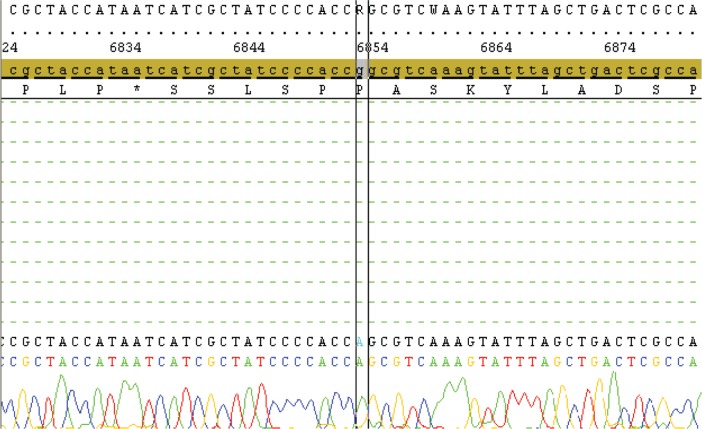
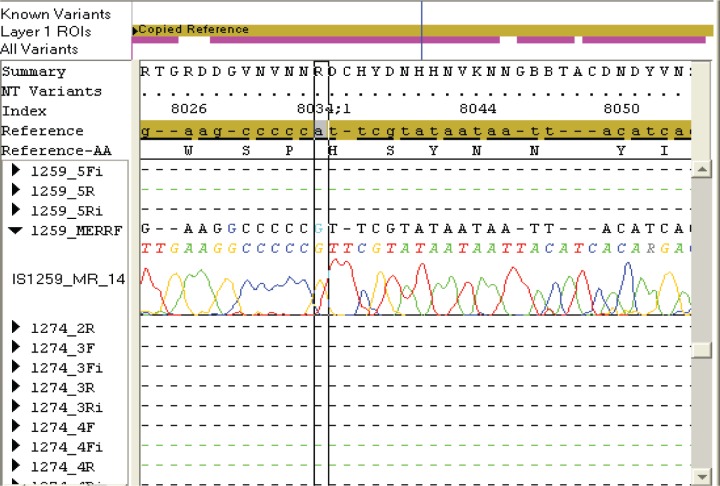
Table 1 summarizes some of the clinical data and mtDNA testing results of the ASDs patients in the present study. Molecular genetic studies were performed by isolation of total DNA from blood by standard procedures, screening for the mtDNA mutations by PCR-SBT analysis and subsequent sequencing of the whole mitochondrial genome. Sixteen of the 21 children carried reported polymorphisms and/or silent variants and two patients carried mtDNA mutations of likely pathogenicity (Table 1). In one patient, a G>A transition in protein-coding regions (COI, subunit of cytochrome c oxidase) was detected: m.6852 G>A, p.(Gly317Ser) (Figure 1). A candidate mutation m.8033A>G that produces an amino acid change p.(Ile 150Val) in the mitochondrial gene (MT-CO2) coding for complex IV that has not been reported before, was observed in one boy with autism, moderate mental retardation and hypertrichosis (Figure 2). These mutations were present in homoplasmy in the patient’s DNA. Normal blood lactate levels were found in the two children carrying mtDNA mutations.
Table 1.
Clinical data and mitochondrial DNA testing results of 21 autistic patients.
| Patient Number | Sex-Age (y:moths) | Non Synonymous Mutations | Silent Variants | Polymorphisms | Lactate (mmol/L)a | Clinical Data |
|---|---|---|---|---|---|---|
| 1 | M-2:7 | C7028T; G9055A; A9494G; T9698C | 3.24 | Autism, moderate mental retardation, strabismus | ||
| 2 | F-3:2 | A4188G; C7028T | 2.30 | Autism, mental retardation, dysmorphic face, skin hyperpigmentation | ||
| 3 | M-3:4 | T3197C; G8581A; G9477A; G9554A | 2.61 | Autism, mental retardation | ||
| 4 | M-4:6 | 7.88 | Autism, relative macrocephaly | |||
| 5 | M-4:2 | C6371T; C7028T | 1.36 | Autism, mild mental retardation, family history of psychiatric disorders | ||
| 6 | M-7:8 | C7028T; T9698C; G9055A | 1.99 | Autism, cognitive regression | ||
| 7 | M-3:9 | A8348G | 5.24 | Autism, moderate mental retardation, hyperactivity | ||
| 8 | M-4:10 | G3834A; T8602C | 2.94 | Autism | ||
| 9 | M-11:0 | A476G; A8718G G8994A | 1.03 | Autism, moderate mental retardation, microcephaly | ||
| 10 | F-6:4 | 6852G>A (MT-COI gene); Gly317Ser (complex IV) | 6809A>G (complex I) | 2.07 | Autism, mild mental retardation | |
| 11 | M-4:10 | T3197C; G8994A | 2.01 | Autism, moderate mental retardation | ||
| 12 | M-5:11 | G8269A; G8557A; T4216C; G5460A | 1.27 | Autism, strabismus | ||
| 13 | M-6:8 | 6464C>A (complex I); 7810C>T (complex II) | T4216C; C7028T; A12612G | 4.82 | Autism, dolichocephaly, dymorphic face | |
| 14 | M-5:5 | 3.11 | Autism, borderline intelligence, hyper-trichosis | |||
| 15 | F-3:4 | T4336C | 1.75 | Autism, mental retardation, dolichocephaly | ||
| 16 | M-6:7 | C6371T; A6791G C7028T; T8503C | 2.10 | Atypical autism, severe mental retardation, dysmorphic face | ||
| 17 | M-2:8 | G3591A; A4310G | 1.60 | Autism, mild mental retardation, muscle hypotonic, bilateral inguinal hernia | ||
| 18 | M-11:0 | 8033A>G (MT-CO2 gene); Ile150Val (complex IV) | 1.83 | Autism, moderate mental retardation, hypertrichosis, dysmorphic face | ||
| 19 | M-8:9 | 1.10 | Autism, moderate mental retardation, microcephaly | |||
| 20 | M-2:0 | A8805G | 5.64 | Autism, mild mental retardation | ||
| 21 | M-2:7 | 2.66 | Autism, mild mental retardation, dysmorphic face |
Normal range for blood lactate: 1.10–2.40 mmol/L.
Figure 1.
Sequencing-based identification of the complex IV mutation: m.6852G>A, p.(Gly317Ser).
Figure 2.
Sequencing-based identification of the complex IV mutation: m.8033A>G, p.(Ile150Val).
DISCUSSION
The autism spectrum disorders are clinically and etiologically heterogeneous developmental disorders of the brain. Many causes of autism have been proposed, but understanding of the etiology of ASDs is incomplete. The brain is strongly dependent on the ATP production of the mitochondrion, and the high energy demands of the brain and nervous system make them particularly vulnerable to impaired energy production. Recent epidemiological studies indicate that the incidence of mitochondrial disorders in children with autism could be 550 to 770 times higher than that of the general population [15]. All these facts have led several researchers to explore mitochondrial respiratory chain disorders as potential risk factors in ASDs. Multiple studies have confirmed the role of mitochondrial dysfunctions in ASDs [4–6,15,16]. For most individuals with defects of oxidative phosphorylation, the diagnosis is made through the determination of the activities of the enzyme complexes of the electron transport chain but an underlying mitochondrial mutation (including mtDNA depletion, mutations and deletions) can rarely be identified [7–10,17]. The present study aimed to explore the roles of mtDNA mutations in ASDs and find more evidence to support the relationship between these two disorders. In an ongoing survey, we screened 21 autistic children for mtDNA mutations. Sequencing of mtDNA showed a number of polymorphisms and two mutations, presented in the homoplasmic state, which have never been reported before in autistic patients (Table 1). Patient 10 had the G6852A mutation that replaces glycine with serine in protein-coding regions (COI) documented in blood (Figure 1). This mutation was reported previously by Taylor et al.[18]. They examined colonic crypt stem cells derived from a 75-year-old patient undergoing resection for a colonic tumor and found the G6852A mutation in the heteroplasmic (60.0%) state. Probably, the variability of phenotypes associated with this mutation, is according to the existence of environmental or genetic factors in addition to the mtDNA mutation that contribute to the development of clinical presentations. We also identified the p.(Ile→Val) substitution (m.8033A>G) in protein COII, subunit of cytochrome c oxidase (Figure 2) in a patient with autism, moderate mental retardation, hypertrichosis and dysmorphic face. To the best of our knowledge, the same mutation has not been previously observed in a patient with autistic or non autistic behavior and mitochondrial cytopathies. Hair abnormalities and pigmented skin eruptions belong to the broad spectrum of presenting symptoms of mitochondrial disease and 10.0% of the children with mitochondrial disorders develop specific cutaneous manifestations [19]. Hypertri-chosis is one of the most frequent skin disorders associated with mitochondrial dysfunction, however, the relationship between cutaneous manifestations in our patient and the A8033G mutation is speculative.
The two patients carrying mtDNA mutations had normal blood lactate levels (Table 1), however, normal lactate levels do not exclude mitochondrial disease. Biochemical markers may be abnormal only during illness and variations in blood collection techniques are well known to cause differences in lactate values. Only a few studies evaluated autistic children who had a normal lactate for mitochondrial disorders [7,9,20,21]. Using only lactate as a screening test for mitochondrial dysfunction may miss some individuals with mitochondrial diseases.
The phenotypic presentation of mitochondrial disorders in ASDs is quite broad. Some investigators described autistic patients with mitochondrial dysfunction as not different from the general ASDs population [10,16,22]. Other investigators found that children with ASDs and mitochondrial disorders demonstrated significant neurological (developmental regression, seizures, motor delay) and/or gastrointestinal (reflux, constipation) problems [10,23]. One of our patients (patient 10, Table 1) did not have any major clinical features that distinguished her from typical autism. The patient with the A8033G mutation had a dysmorphic face and hypertrichosis and might be classified as having a syndromic autism. The substantial clinical heterogeneity of individuals with cooccurring autism and mtDMA mutations and lack of classical features associated to mitochondrial diseases constitute a challenge for clinicians to diagnose these patients.
The presence of mtDNA mutations observed in this study, performed on ASDs children, may indicate an etiological role of the mitochondrial dysfunction in autistic patients. The pathogenetic link is that all of these mutations impair mitochondrial protein synthesis, however, additional studies are needed to investigate whether the mitochondrial dysfunction in children with autism is primary or secondary. A plausible hypothesis is that the mitochondrial dysfunction can lead to reduced synaptic neurotransmitter release in GABAergic (neurons whose primary neurotransmitter is GAMMA-AMINO BUTYRIC) neurons during a specific developmental period between 12 and 30 months of age [24,25]. However, an imbalance in the excitatory and inhibitory neurotransmitter systems reported in ASDs could also contribute to secondary mitochondrial dysfunction through an inhibition of mitochondrial b-oxidation [26]. Mitochondrial dysfunction has been associated with other behavioral abnormalities besides autism, such as schizophrenia and depression, which are frequent psychiatric conditions in relatives of children with autism [7,27].
Our study has some limitations. First, the number of individuals in whom mtDNA mutations were assessed was relatively small, and further studies in a larger sample would be needed to determine the association between ASDs and the results of mtDNA analyses reported herein. Second, only a panel of the most common mutations in mtDNA has been tested and, therefore, a mutation elsewhere in the mitochondrial genome cannot be ruled out. Third, we tested only blood samples, and lack of detectable mutation in one tissue does not exclude the presence of a mutation in other tissues. Fourth, the fact that diagnosed variants are non synonymous and rare does not guarantee their pathogenic status. Additional analyses of mtDNA genomes could supply new findings about the etiological role of rare mutations.
CONCLUSIONS
Despite of the mentioned limitations, our study suggests that mtDNA mutations may be an additional patho-genetic factor for a subset of individuals with autism. It is important for ASDs children with mitochondrial disorders to be identified early in life and to start a proper treatment with antioxidants and mitochondrial cofactors. Some ASDs patients who have a mitochondrial dysfunction can be phenotypically indistinguishable from children with idiopathic autism. In order to reach an early diagnosis, all children with ASDs should be screened for mitochondrial disorders. Further studies are needed to prove the clinical significance of the present findings and to understand how mitochondrial defects may contribute to autism.
REFERENCES
- 1.Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) American Psychiatric Association; Washington DC: American Psychiatric Association; 1994. [Google Scholar]
- 2.Newschaffer CJ, Croen LA, Daniels J, Giarelli E, Grether JK, Levy SE, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gargus JJ, Imtiaz F. Mitochondrial energy-deficient endophenotype in autism. Am J Biochem Biotech. 2008;4(2):198–207. [Google Scholar]
- 5.Rossignol DA, Bradstreet JJ. Evidence of mitochondrial dysfunction in autism and implications for treatment. Am J Biochem Biotech. 2008;4(2):208–217. [Google Scholar]
- 6.Palmieri L, Persico AM. Mitochondrial dysfunction in autism spectrum disorders: cause or effect? Biochim Biophys Acta. 2010;1797(6–7):1130–1137. doi: 10.1016/j.bbabio.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. J Pediatr. 2004;144(1):81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Serajee FJ, Zhang H, Huq A. Prevalence of common mitochondrial point mutations in autism. Neuropediatrics. 2006;37(Suppl 1):S127. [Google Scholar]
- 9.Graf WD, Marin-Garcia J, Gao HG, Pizzo S, Naviaux RK, Markusic D, et al. Autism associated with the mitochondrial DNA G8363A transfer RNA(Lys) mutation. J Child Neurol. 2000;15(6):357–361. doi: 10.1177/088307380001500601. [DOI] [PubMed] [Google Scholar]
- 10.Weissman JR, Kelley RI, Bauman ML, Cohen BH, Murray KF, Mitchell RL, et al. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS One. 2008;3(11):e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent L, Lambert C, Pyle A, Elliott H, Wheelwright S, Baron-Cohen S, et al. The mitochondrial DNA A3243 A>G mutation must be an infrequent cause of Asperger syndrome. J Pediatr. 2006;149(2):280–281. doi: 10.1016/j.jpeds.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 12.Álvarez-Iglesias V, Mosquera-Miguel A, Cuscó I, Carracedo Á, Pérez-Jurado LA, Salas A. Reassessing the role of mitochondrial DNA mutations in autism spectrum disorder. BMC Med Genet. 2011;12:50. doi: 10.1186/1471-2350-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.www.mitomap.org.
- 14.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34(Database issue):749–751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giulivi C, Zhang Y-F, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira G, Diogo L, Grazina M, Garcia P, Ataide A, Marques C, et al. Mitochondrial dysfunction in autism spectrum disorders: a population-based study. Dev Med Child Neurol. 2005;47(3):185–189. doi: 10.1017/s0012162205000332. [DOI] [PubMed] [Google Scholar]
- 17.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112(9):1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodemer C, Rötig A, Rustin P, Cormier V, Niaudet P, Saudubray JM, et al. Hair and skin disorders as signs of mitochondrial disease. Pediatrics. 1999;103(2):428–433. doi: 10.1542/peds.103.2.428. [DOI] [PubMed] [Google Scholar]
- 20.Ezugha H, Goldenthal M, Valencia I, Anderson CE, Legido A, Marks H. 5q14.3 deletion manifesting as mitochondrial disease and autism: case report. J Child Neurol. 2010;25(10):1232–1235. doi: 10.1177/0883073809361165. [DOI] [PubMed] [Google Scholar]
- 21.Tsao CY, Mendell JR. Autistic disorder in 2 children with mitochondrial disorders. J Child Neurol. 2007;22(9):1121–1123. doi: 10.1177/0883073807306266. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira G, Ataide A, Marques C, Miguel TS, Coutinho AM, Mota-Vieira L, et al. Epidemiology of autism spectrum disorder in Portugal: prevalence, clinical characterization, and medical conditions. Dev Med Child Neurol. 2007;49(10):726–733. doi: 10.1111/j.1469-8749.2007.00726.x. [DOI] [PubMed] [Google Scholar]
- 23.Filiano JJ, Goldenthal MJ, Rhodes CH, Marin-Garcia J. Mitochondrial dysfunction in patients with hypotonia, epilepsy, autism, and developmental delay: HEADD syndrome. J Child Neurol. 2002;17(6):435–439. doi: 10.1177/088307380201700607. [DOI] [PubMed] [Google Scholar]
- 24.Anderson MP, Hooker BS, Herbert MR. Bridging from cells to cognition in autism pathophysiology: biological pathways to defective brain function and plasticity. Am J Biochem Biotechnol. 2008;4(2):167–176. [Google Scholar]
- 25.Herlenius E, Lagercrantz H. Development of neurotransmitter systems during critical periods. Exp Neurol. 2004;190(Suppl 1):S8–S21. doi: 10.1016/j.expneurol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Pastural E, Ritchie S, Lu Y, Jin W, Kavianpour A, Khine Su-Myat K, et al. Novel plasma phospholipid biomarkers of autism: mitochondrial dysfunction as a putative causative mechanism. Prostaglandins Leukot Essent Fatty Acids. 2009;81(4):253–264. doi: 10.1016/j.plefa.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Shachar D. Mitochondrial dysfunction in schi-zophrenia: a possible linkage to dopamine. J Neurochem. 2002;83(6):1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]