Abstract
Understanding early stage renal malfunctions with regard to the glomerular filtration processes is essential for nephropathological prescreening strategies and intervention at an early stage. Mass spectrometry imaging (MSI) in combination with histopathology can provide an universal analytical approach. Proteomic and lipidomic aspects of glomerular biocompositions were applied for micro-structural differentiation in healthy rat kidney samples. Usability of commonly used tissue embedding media and the compatibility of histological staining and fixation methods were of interest. It was demonstrated that ultra-thin tissue samples (500 nm, 1 and 10 μm) can be used for lipid and peptide-based differentiation at the glomerular resolution level in formalin-fixed tissue samples in combination with preceding histological staining for correlating optical and molecular mass images.
Keywords: Glomerular microstructures, Lipids, Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (MS) imaging, Nephropathy, Proteins
INTRODUCTION
A combination of histopathology and mass spectrometry imaging (MSI) offers comprehensive information regarding structure, molecular composition and pathological information in tissue samples. Mass spectrometry imaging is a rapidly developing technique using spatially resolved mass spectrometry (MS) techniques to simultaneously trace distributions of hundreds of biomolecules directly from tissue samples using essentially the same technology. Peptides, proteins, pharmaceuticals and metabolites can also be analyzed but without a label and without prior knowledge. With MSI, molecular peak information is correlated to the underlying tissue architecture and a virtual image is rebuilt with respect to the intensity of each molecular species to understand the distribution of differential signals.
Renal dysfunction has a high demand for early stage identification to initiate healing processes. Recent investigations proved nephropathy is associated with insulin resistance leading to malfunctions of the glomerular filtration [1], manifested in potential glomerular basement membrane modifications [2]. Lipid phosphatase levels, promoting podocyte apoptosis leading to diabetic nephropathy, are up-regulated before histological changes are observed [1]; moreover, the membrane contains protein complexes associated with cholesterol binding which alters the lipid environment [2]. Lipid accumulations have been reported for diabetic kidney diseases [3] and oxidized phosphatidylcholine species seem to be associated with renal dysfunction [4]. Recently tubuli-related phosphatidylcholine classes identified by MSI were correlated to immunoglobulin A nephropathy [5].
Thus, for visualizing renopathological microstructures, MSI is a promising tool, allowing further discoveries of yet unknown disease-related analytes. Special challenges regarding ion suppression effects during the matrix-assisted laser desorption/ionization (MALDI) process were observed whilst obtaining lipidomic information using MSI. Consequently, special attention had to be paid to sample preparation methods, regarding washing procedures, matrix application and protein denaturation, making adaptation to the particular analytical question mandatory [6].
MATERIALS AND METHODS
Chemicals were of analytical grade (Sigma-Aldrich, St. Louis, MO, USA). Water (ddH2O) was purified in a Simplicity system (Millipore, Billerica, MA, USA). Frozen rat kidney tissue (formalin-fixed, unfixed) embedded in paraffin (FFPE), optimal cutting temperature (OCT) compound (Sakura Finetek, Tokyo, Japan) or sucrose was sliced to 10, 5 and 0.5 μm using a cryo-microtome (Leica, Wetzlar, Germany) and mounted on indium-tin oxide coated glass slides (Sigma-Aldrich). The OCT compound and sucrose were removed by washing the samples five times (4°C) for 45 seconds with 200 μL ddH2O/cm2. A xylene bath followed by a descending ethanol gradient (100.0, 96.0, 70.0, 50.0, 0.0% in ddH2O) removed the paraffin. All samples were washed three times with 200 μL 70.0% ethanol/cm2 for 45 seconds (4°C) and vacuum dried for 15 min. before the MALDI matrix application. Samples were hematoxylin/eosin (H/E) stained before MSI treatment.
The MALDI matrices [α-cyano-4-hydroxycinnamic acid (HCCA), sinapinic acid, 2,5-dihydroxybenzoic acid] were dissolved in solvents containing 50.0 or 70.0% acetonitrile or ethanol in 0.1% aqueous trifluoroacetic acid. Matrix deposition was performed using a ChIP-1000 (Shimadzu Biotech Kratos Analytical). For protein identification, tissue was trypsinized before matrix deposition by depositing enzyme directly on pre defined tissue spots [1 ng/μL in 50 mM ammonium bicarbonate, 0.1% Rapigest (Waters, Manchester, Greater Manchester, UK)]. Samples were incubated in a humidified atmosphere overnight at 37°C before desiccation in vacuum and heat treatment (85°C).
Mass spectrometry imaging experiments were performed using a MALDI-TOF (time of flight) instrument (AXIMA TOF2; Shimadzu Biotech Kratos Analytical, Manchester, Greater Manchester, UK; 337nm nitrogen laser, 20Hz) and a MALDI-QqTOF instrument (Synapt HDMS, Waters; 355nm Nd-YAG laser, 200Hz). The optical diameter of 60 μm was reduced to an operational resolution of 35 μm by over sampling.
Peptide and lipid identification was based on post source decay (PSD) and collision induced dissociation (CID) fragmentation in combination with database search [proteins: SwissProt (http://www.uniprot.org/help/uniprotkb); lipids: Lipid Maps (http://www.lipidmaps.org/data/structure/)]. Selected ion images were generated using BioMAP (Novartis, Basel, Switzerland).
RESULTS AND DISCUSSION
Histology and Mass Spectrometry Imaging. Integrating MSI in histopathology necessitates the combination of different requirements from pathology and MS. The primary demands for histology are the preservation of high spatial resolution for tissue structures, long lasting analyte preservation and low sample consumption. For MS, spatial resolution can usually be disregarded and analytes have to be mobilized instead of preserved. For MSI, additionally, histological information has to be retained and tissue disintegration has to be limited to a minimum. We present an approach arranging histological staining prior to MSI using ultra-thin tissue sections showing good molecular results comparable to microscopic images.
It was found that MALDI-TOF MSI analysis of H/E-stained tissue showed no limiting aspects concerning proteomic approaches. Protein digestion directly from the tissue and measuring peptides by MS could be achieved. Stain-related signals, however, were observed below m/z signals of 500, eventually leading to ion suppression effects for neutral lipids. Phospholipid classes were obtained unaffected from ultra-thin sucrose samples. Histological images of OCT embedded samples were often difficult to correlate to MSI results because of structural changes after thawing. Sucrose embedding in combination with cutting thickness of 1 μm was sufficient for IMS analysis, even reducing signal background and enhancing signal quality. The selected ultra-thin samples considerably improved signal quality whilst preserving spatial resolution.
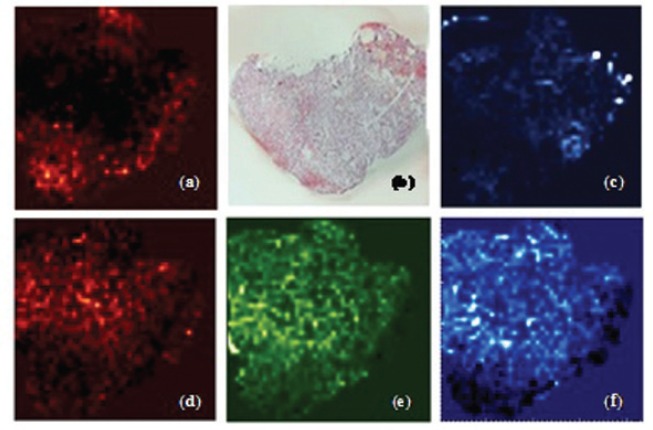
Lipid Differentiation. Lipid-based histological differentiation was performed using unfixed rat kidney samples embedded in sucrose and formalin-fixed tissue embedded in OCT Low embedding medium related signals and intensive species-related signals were obtained from both OCT and sucrose embedded samples. Several lipid classes could be localized, identified and characterized by fragmentation experiments. Unfixed tissue species revealed different distribution pattern phosphoatidylcholine species with varying alkyl chain lengths. The 1-tetradecanoyl-2-sn-glycero-3-phosphocholine, 1-hexadecanolyl-2-sn-glycero-3-phosphocholine and 1-octadecanoyl-2-sn-glycero-3-phosphocholine showed specifiable areas related to kidney structures, i.e., cortex, medulla and pelvis (Figure 1). The 1-hexadecyl-2-acetyl-sn-glycero-3-phosphocholine and 1-hexadecanoyl-sn-glycero-3-phosphocholine exhibited different localization as well as 1-O-(1Z-tetradecenyl)-2-(9Z-octadecenoyl)-snglycerol and its oxidized co-species. Besides 16 different phospholipids, diacylglycerophosphoethanolamine, N-(tetracosanoyl)-sphing-4-enine-1-phosphocholine and two glomeruli specific metabolites, cholesterol and squalene, were also discovered. Histological correlation revealed the potential to identify pathologically modified membrane structures in glomerular associated diseases.
Figure 1.
Tissue differentiation according to identified lipid species. Distribution of (a) diacylglycerol on (b) 5 μm OCT embedded unfixed kidney tissue; (c) oxidized diacylglycerol; (d) 1-tetradecanoyl-2-sn-glycero-3-phosphocholine; (e) 1-hexadecanolyl-2-sn-glycero-3-phosphocholine; (f) 1-octadecanoyl-2-sn-glycero-3-phosphocholine.
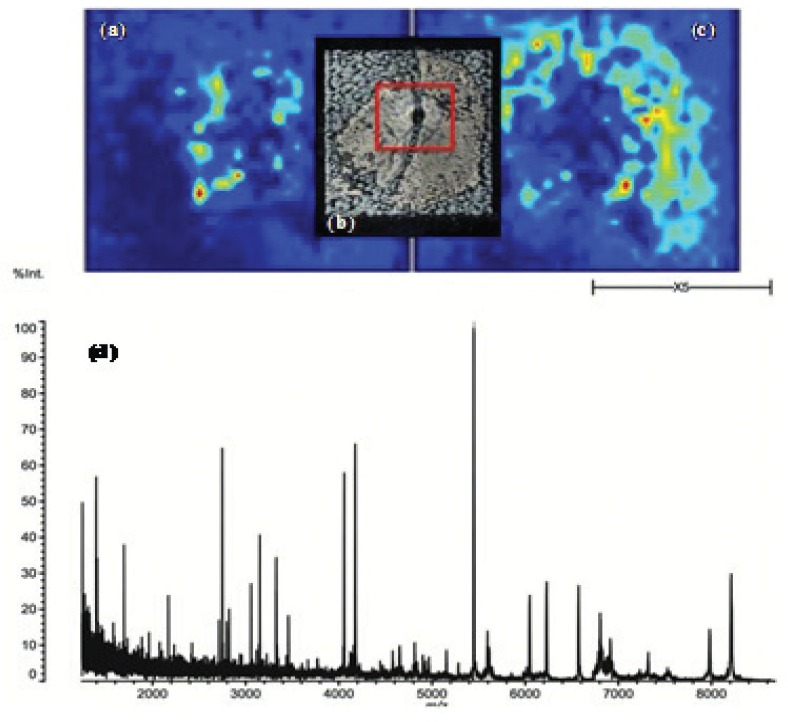
Protein Identification. To detect most of the proteins available in an ultra-thin sample, several washing steps were required for complete removal of analyte species which are much easier to ionize than proteins, e.g., lipids [7]. Our study showed that mass spectra quality for tissue analysis was enhanced by heat treatment before MSI experiments. Several tissue-related peptides could be identified by PSD and CID, revealing the possibility to distinguish pelvis and cortex, both tissue macrostructures. For all investigated embedding materials as well as fixation methods, it was possible to obtain sufficient signals up to m/z values of 10000 (Figure 2). The application of danaturation agents and heat treatment supports subsequent on-tissue enzymatic treatment for protein identification on fixed and embedded rat kidney. Highly cross-linked proteins (formalin) showed improved signal intensities for tryptic peptides after denaturation, which significantly supported protein identification based on peptide fragmentation. For protein identification in MSI approaches, peptide sequencing was indispensable because of protein co-localization and the non applicable approach of peptide mass fingerprinting. The presented protocol on ultra-thin tissue sections allowed the identification of Tubulin and Claudin-4, besides seven other peptide fragments on differentiated, but so far unspecific, tissue regions.
Figure 2.
Protein based differentiation of (a) medulla and (c) cortex in; (b) FFPE rat kidney tissue (10 μm) after HCCA application; (d) MALDI-TOF-MS profile of the marked sample area (red).
Perspectives. Combining histopathology and MSI for investigating structural membrane modifications associated with glomerular related nephropathology looks promising. Structural modifications and molecular changes can be detected and investigated very early in progression.
REFERENCES
- 1.Hyvönen ME, Saurus P, Wasik A, et al. Lipid phosphotase SHIP2 downregulates insulin signalling in podocytes. Mol Cell Endocrinol. 2010;328(1–2):70–79. doi: 10.1016/j.mce.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Kibel G, Heilhecker A, Von Bruchhausen F. Lipids associated with bovine kidney glomerular basement membranes. Biochem J. 1976;155(3):535–541. doi: 10.1042/bj1550535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Jiang T, Li J, et al. Regulation of renal lipid metabolism, lipid accumulation, and glomerulosclerosis in FVBdb/db mice with type 2 diabetes. Diabetes. 2005;54(8):2328–2335. doi: 10.2337/diabetes.54.8.2328. [DOI] [PubMed] [Google Scholar]
- 4.Jimi S, Uesugi N, Saku K, et al. Possible induction of renal dysfunction in patients with lecithin:cholesterol acyltransferase deficiency by oxidized phosphatidylcholine in glomeruli. Arterioscler Thromb Vasc Biol. 1999;19(3):794–801. doi: 10.1161/01.atv.19.3.794. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko Y, Obata Y, Nishino T, et al. Imaging mass spectrometry analysis reveals an altered lipid distribution pattern in the tubular areas of hyper-IgA murine kidneys. Exp Mol Pathol. 2011;91(2):614–621. doi: 10.1016/j.yexmp.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Kaletaş BK, van der Wiel IM, Stauber J, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9(10):2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 7.van Hove ER, Smith DF, Fornai L, et al. An alternative paper based tissue washing method for mass spectrometry imaging: localized washing and fragile tissue analysis. J Am Soc Mass Spectrom. 2011;22(10):1885–1890. doi: 10.1007/s13361-011-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]