Abstract
Several host genetic factors play an important role in susceptibility to human immunodeficiency virus type 1 (HIV-1) infection and in its progression to acquired immune deficiency syndrome (AIDS). The interleukin-18 (IL-18) is a multifunctional proinflammatory cytokine that regulates immune responses and plays a pathogenic role in HIV-1 infection by enhancing viral replication. Single nucleotide polymorphisms (SNPs) in the IL-18 gene promoter region may lead to altered transcriptional activity and IL-18 production, and may account for variation in the risk of HIV-1 infection. We have investigated the association between IL-18 promoter polymorphism −607C>A and HIV-1 infection through a case-control study of 500 patients with HIV-1/AIDS and an equal number of age and sex matched controls in a north Indian population. Genotyping using sequence specific primer-polymerase chain reaction (SSP-PCR) showed a statistically significant reduced risk of HIV-1 infection for the A>A genotype [odds ratio (OR) = 0.57, 95% confidence interval (95% CI) = 0.33–0.98, p = 0.040], but not for the C>A genotype (OR = 0.87, 95% CI = 0.66–1.14, p = 0.321). We concluded that the −607A allele of the IL-18 gene promoter polymorphism may play a protective role against the progression of HIV-1 infection in this population.
Keywords: Human immnunodeficiency virus type 1 (HIV-1), Acquired immune deficiency syndrome (AIDS), IL-18 gene, Single nucleotide polymorphisms (SNPs), Sequence specific primer-polymerase chain reaction (SSP-PCR)
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) infection results in progressive deterioration of the immune system and in acquired immune deficiency syndrome (AIDS). However, the rate at which the HIV-1 infection progresses to AIDS varies considerably between individuals, some progressing rapidly after primary infection, while others remain asymptomatic with no evidence of immune dysfunction for over 15 years [1,2]. The reasons for the variation are multifactorial and may involve genetic, virological or immunological factors that function in different ways [3]. The role of host genetic variations has been intensively examined in different populations and in all major risk groups. These have revealed that genetic polymorphisms in genes for chemokine receptors or in those of their natural ligands are likely to influence susceptibility or resistance to HIV-1 infection as well as the subsequent rate of disease progression [3,4]. Genes for human leukocyte antigens (HLA) that regulate host immune response to infection have also been correlated with the clinical course of HIV-1 infection [5]. So far, in addition to polymorphic variants in genes encoding for HIV-1 co-receptors and their ligands and those of HLA alleles, some T helper type 1 (Th1) and type 2 (Th2) cytokine genes have also been implicated to influence the rate of disease progression in HIV-1 seropositive patients either positively or negatively [6,7].
Cytokines play a crucial role in regulation of the balance between Th1 and Th2 immune responsiveness. The Th1 cytokines promote cell-mediated immunity to intracellular pathogens including viruses and other intracellular pathogens, whereas Th2 cytokines enhance humoral immunity by up-regulating antibody production against extracellular pathogens [8,9]. Furthermore, several cytokine related studies confirmed their key immunomodulatory role within the immune system and cytokine genes have been targeted for association studies of susceptibility and development of several immune-related and infectious diseases [10–13].
The interleukin-18 (IL-18) gene encoding human IL-18 cytokine is located at chromosome 11q22.2-22.3 [14,15]. Three single nucleotide polymorphisms (SNPs) (−656G>T, −607C>A, −137G>C) within the IL-18 gene promoter have been localized. Of these, those at positions −137 and −607 influence the quantitative expression of the IL-18 protein. The −607C>A disrupts a potential cyclic adenosine monophosphate (cAMP) responsive element-binding protein (CREB) binding site and −137G>C abolishes the human histone 4 transcription factor-1 (H4TF-1)-binding site, reducing its transcription. Thus, these two polymorphisms and their haplotypes seem to account for differential IL-18 expression and changes in the production of this cytokine [15].
The multiple biological functions of the IL-18 gene are associated with a wide variety of allergic, inflammatory, autoimmune, cancerous and infectious diseases [16,17]. In the context of HIV-1 infection, the IL-18 cytokine also plays a pathogenic role by enhancing viral replication in monocytic cells and in T cells [18,19]. Moreover, clinical studies have demonstrated the involvement of IL-18 in the immunopathogenesis of HIV-1 infection [20–22]. However, most of these studies focused primarily on the association of the levels of IL-18 and the pathogenesis of HIV-1 infection. Several studies have examined the association of SNPs in the IL-18 gene with various other diseases [23,24], but that with the HIV-1 infection has not been fully explored. For this reason, we have evaluated the association of the IL-18 gene promoter polymorphism −607C>A with the HIV-1/AIDS infections. To the best of our knowledge, this is the first association study of the IL-18 gene polymorphism in North Indian HIV-1/AIDS patients.
MATERIALS AND METHODS
In this case control study 500 patients with HIV-1/AIDS and an equal number of sex- and age-matched control groups were recruited from the Department of Internal Medicine, Post-Graduate Institute of Medical Education and Research at Chandigarh, India. Cases and controls were from the same geographical area and same ethnic backgrounds. Of the 500 cases, 174 were females and 326 males with a mean age of 35.4 ± 7.9, while of the 500 controls, 183 were females and 317 males with a mean age of 36.2 ± 9.8 years.
The research protocols were reviewed and approved by the ethics committee of the institute. Written informed consent was obtained from all the participating individuals. Information about age, sex, occupation and other relevant clinical data was gathered from the immunological cards of the HIV-1/AIDS patients. Peripheral blood (2–3 mL) was drawn and collected in an EDTA-coated tube for genomic DNA extraction. The samples were kept at −80°C until used for DNA isolation. Genomic DNA was extracted from peripheral blood leukocytes by sodium dodecyl sulphate lysis and proteinase K digestion followed by standard phenol-chloroform methods as previously described [15] and the extracts were subjected to a polymerase chain reaction (PCR).
Genotyping of the IL-18 −607C>A SNP was performed by (sequence specific primer)-SSP-PCR analysis [14]. Briefly, a common reverse primer 5′-TAA CCT CAT TCA GGA CTT CC-3′ and two sequence specific forward primers 5′-GTT GCA GAA AGT GTA AAA ATT ATT AC-3′ and 5′-GTT GCA GAA AGT GTA AAA ATT ATT AA-3′ were used. An amplification product of 196 bp was detected. A control forward primer 5′-CTT TGC TAT CAT TCC AGG AA-3′ was used to amplify a 301 bp fragment covering the polymorphic site as an internal positive amplification control.
The reaction was performed in a final volume of 25 μL consisting of 2.5 μL of 10X PCR buffer, 0.25 μL of 10 mmol of dNTP, 1.5 μL of 25 mM MgCl2, 2 μL of genomic DNA, and 0.5 U of Taq polymerase. For every reaction mixture, 0.5 μL of the −607C allele-specific or the −607A allele-specific forward primer and 0.5 μL of the common reverse primer were included. The 0.15 μL of internal positive control primer was added to reaction mixture. The PCR products (Figure 1) were visualized by ethidium bromide stained electrophoresis on 2% agarose gel. Two separate PCRs were carried out for every individual DNA sample.
Figure 1.
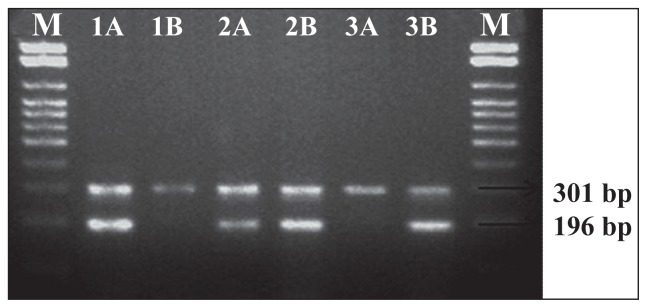
A 2% agarose gel electrophoresis of PCR products of the IL-18 gene at the position −607C>A. The 301 bp bands represent the PCR internal control amplified using the forward control primer and the common reverse primer. The 196 bp bands represents the PCR amplification products of the alleles C and A amplified using the sequence specific primers with the common reverse primers. M: 100 bp DNA marker; lanes 1A and 1B: wild CC genotype; lanes 2A and 2B: CA genotype, and lanes 3A and 3B: AA genotype.
Reactions were carried out in a Bio-Rad MyCycler (Bio-Rad Laboratories, Hercules, CA, USA). Denaturation for 2 min. at 94°C was performed as the first step. This was followed by seven cycles of 20 seconds at 94°C, 40 seconds at 64°C and 40 seconds at 72°C and 25 cycles of 20 seconds at 94°C, 40 seconds at 57°C and 40 seconds at 72°C. All PCR products were visualized under UV light on ethidium bromide-stained 2% agarose gels.
Statistical analysis
Results were analyzed with SPSS software, version 11 for windows 5 (SPSS Inc., Chicago, IL, USA). Hardy-Weinberg equilibrium between observed and expected genotype distributions was determined by means of the Pearson Chi-square (χ2) test. For the assessment of risk factors, ORs with 95% CIs were calculated according to Epi Info Version 3.5.1 2008; Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA. The frequencies of genotypes and alleles were calculated by manual counting. The Pearson χ2 test was used to calculate the significance of the differences in frequency of genotypes and alleles between the patients and control subjects. p Values of <0.05 were considered to be statistically significant.
RESULTS
The demographic profile of HIV-1/AIDS patients and healthy controls in the study group is summarized in Table 1. Regarding gender distribution, there was no significant difference between cases and controls. It was found that the proportion of males was 65.2 and 63.4% in cases and controls, respectively, whereas for females it was 34.8 and 36.6% in cases and controls, respectively. The mean age [± standard deviation (SD) in years] was 35.4 ± 7.9 years for cases and 36.2 ± 9.8 years for controls. There was no significant mean age difference between the cases and the controls (p = 0.180).
Table 1.
Demographic information on patients and controls.
| Parameters | HIV-1/AIDS Patients (n = 500) | Healthy Controls (n = 500) | p Value |
|---|---|---|---|
|
| |||
| Gender : n (%) | |||
| • Males | 326 (65.2) | 317 (63.4) | 0.553 |
| • Females | 174 (34.8) | 183 (36.6) | |
|
| |||
| Age (years) mean ± SD | 35.39 ± 7.90 | 36.15 ± 9.75 | 0.180 |
Staging of HIV infection was done by the physicians as per the World Health Organization (WHO) staging guidelines. Accordingly, all the four stages of HIV/AIDS were found in the cases. Of all the 500 seropositive cases, 74 (14.85%) were at stage I, 143 (28.6%) at stage II, 113 (22.6%) at stage III and 170 (34%) were at stage IV (AIDS) of the disease. The mean counts of CD4+-T cells at stages I, II, III, and IV were 331, 268, 206, and 113, respectively.
In all, 500 HIV-1/AIDS patients and an equal number of controls were studied for −607C>A IL-18 promoter polymorphism. As shown in Figure 1, there were C/C, C/A and A/A genotypes at position −607. The PCR products of homozygous individuals showed a 301 bp DNA segment and those of heterozygous individuals showed 196 and 301 bp fragments.
The genotypic distributions and allelic frequencies of −607C>A are presented in Table 2. The genotype distributions are in agreement with Hardy-Weinberg equilibrium. A significant increase in frequency of the wild C/C genotype was observed in patients when compared to control subjects (43.0 vs. 38.4%, p = 0.040). The frequency of the C/A genotype was slightly higher in controls (52.8%) than in cases (51.4%) (p = 0.321) and that of the A/A genotype was significantly lower (p = 0.040) in patients (5.6%) than in controls (8.8%). The frequencies of the C and A alleles were 68.7 and 31.3% in cases and 64.8 and 35.2% in controls (p = 0.071), respectively.
Table 2.
Genotype and allele distribution of IL-18 gene in patients and controls.
| IL-18 −607(C/A) | Patients (n = 500) | Controls (n = 500) | OR (95% CI) | p Value |
|---|---|---|---|---|
| Genotype | n (%) | n (%) | ||
| CC | 215 (43.0) | 192 (38.4) | 1.0 Reference | |
| CA | 257 (51.4) | 264 (52.8) | 0.87 (0.66–1.14) | 0.321 |
| AA | 28 (5.6) | 44 (8.8) | 0.57 (0.33–0.98) | 0.040 |
| Allelic | ||||
| C allele | 687 (68.7) | 648 (64.8) | 1.0 Reference | |
| A allele | 313 (31.3) | 352 (35.2) | 0.84 (0.69–1.01) | 0.071 |
OR: odds ratio [OR was calculated by Epi-info Version 3.5.1. (Centers for Disease Control and Prevention, Atlanta, GA, USA); 95% CI: 95% confidence interval; a p value of ≤0.05 was considered significant.
For accounting of the risk assessment the genotypic data was analyzed with Epi Info software. The results showed a significantly reduced risk of HIV-1 infection with the homozygous variant −607A>A genotype when it was computed against healthy controls [odds ratio (OR) = 0.57, 95% confidence interval (95% CI) = 0.33–0.98, p = 0.040]). Conversely, a statistically insignificant reduced risk of HIV-1 infection was observed with the heterozygous −607C>A genotype (OR = 0.87, 95% CI = 0.66–1.14, p = 0.321). Likewise, a statistically insignificant association of HIV-1 infection was observed with the −607A allele (OR = 0.84, 95% CI = 0.69–1.01, p = 0.071).
The genotypic and allelic frequencies of −607C>A between only seropositive for HIV-1 and seropositive patients with disease are presented in Table 3. Of the 500 seropositive patients with HIV-1, 330 (66%) were patients at stages I–III without AIDS and the remaining 170 (34%) were at the AIDS stage. The frequency of the wild C/C genotype in seropositive patients without AIDS disease was 130 (26%) as compared to 85 (17%) in HIV-1 patients at at the AIDS stage. The frequency of the heterozygous C/A genotype was 178 (35.6%) and 79 (15.8%) for seropositive patients without AIDS and those at the AIDS stage, respectively. The frequency of the mutant genotype A/A was 4.4% (22) in HIV-1 patients without AIDS disease and 1.2% (six) in patients with AIDS. A statistically insignificant reduced risk of HIV infection was observed with the C/A genotype of IL-18-607 (OR = 0.68, 95% CI = 0.47–1.01, p = 0.057). Moreover, a statistically insignificant reduced risk of HIV infection was observed with the homozygous variant A/A genotype (OR = 0.42, 95% CI = 0.14–1.14, p = 0.098).
Table 3.
Genotype and allele distribution of the IL-18 gene in HIV-1 seropositive patients.
| Genotype | HIV-1 Patients With AIDSa (n = 170) | HIV-1 Patients Without AIDsa (n = 330) | OR (95% CI) | p Value |
|---|---|---|---|---|
| CC | 85 | 130 | 1.0 Reference | |
| CA | 79 | 178 | 0.68 (0.46–1.01) | 0.057 |
| AA | 6 | 22 | 0.42 (0.14–1.14) | 0.098 |
| C | 249 | 438 | 1.0 Reference | |
| A | 91 | 222 | 0.72 (0.53–0.97) | 0.031 |
Staging of HIV/AIDS was done by the physicians as per the World Health Organization (WHO) clinical staging of HIV disease in adults and adolescents.
DISCUSSION
Cytokines are important immunomodulatory molecules in immune response against pathogenic micro-organisms and also have an important role in the setting of disorders affecting the immune system. Cytokine genes are highly polymorphic and genetic variations that can alter the expression of cytokine genes could have evident pathological consequences, potentially leading to a number of chronic diseases, increased risk of infection, and altered outcome of acute disorders [8,10]. With regard to HIV-1, cytokines have been identified as important tools toward better understanding of the profound interactions occurring between HIV-1 and the human immune system. Moreover, cytokine genes have been implicated to influence the rate of disease progression in HIV-1 seropositive patients either positively or negatively [6,7,26,27].
Interleukin-18 is a pleiotropic proinflammatory cytokine that is recognized as an important regulator of innate and acquired immune responses [28]. It is mainly produced by monocytes/macrophages and many other immune and non immune cells [17,29]. By virtue of its nature, IL-18 plays a key role in induction of both Th1 and Th2 cytokines and, thereby, modulates their immune responses. Interleukin-18 possesses broad and potent immunomodulatory properties. It is therefore not surprising that it appears essential to host defenses against a variety of infections. The involvement of IL-18 has been identified in several inflammatory and autoimmune diseases, in a variety of cancers, and in the context of numerous infectious diseases [16,17]. Furthermore, it has been reported that IL-18 is not only one of several inflammatory markers that increases during HIV-1 infection, but may also be involved in the immunopathogenesis of this disease [22].
The expression of the IL-18 gene seems to be regulated at the transcriptional level by the two SNPs at positions −607C>A and −137G>C in the promoter of this gene. The C>A substitution at position −607 disrupts the binding site of a potential cAMP-responsive element binding protein and the G>C substitution at position −137 abolishes the H4TF-1 nuclear factor-binding site. The −607C and −137G alleles have been associated with higher transcription activity than the −607A and −137C alleles, respectively [15]. Moreover, investigations carried out to determine the association of SNPs in the IL-18 gene and various types of diseases have also shown the influence of the −607C/A promoter polymorphism on quantitative expression of the IL-18 protein. According to these studies, subjects with −607C/C and C/A genotypes have had significantly higher IL-18 concentrations than those with the −607A/A genotype [23,24].
Taking into account the findings of Giedraitis et al.[15], Sakai et al. [23] and Xu et al. [24], 500 HIV-1 patients and an equal number of healthy control subjects were studied for the IL-18 −607C>A gene promoter polymorphism to assess its possible association with the risk of HIV-1 infection. Analysis of the present data demonstrated an association between the IL-18 −607C/A polymorphism and risk of HIV-1 infection in North Indians. In the present study, statistically significant reduced risk of HIV-1 infection was observed with the −607A/A genotype, as compared to healthy controls (OR = 0.57, 95% CI = 0.33–0.98, p = 0.040). On the other hand, a statistically insignificant reduced risk of HIV-1 infection was observed with the heterozygous −607C/A genotype (OR = 0.87, 95% CI = 0.66–1.14, p = 0.321).
As to the effect on HIV-1 infection, increased IL-18 concentrations in HIV-1 patients are likely to play an important role in the disease progression and related clinical conditions [30]. Interleukin-18 has also been shown to induce HIV-1 replication in acutely or chronically infected human monocytic and T cell lines, suggesting its pathogenic role in HIV-1 pathogenesis [18,19,31]. Furthermore, Stylianou et al. [22] suggested the involvement of IL-18 in augmenting the expression of the HIV-1 co-receptor CXCR4 in peripheral blood monocytic cells (PBMCs) from HIV-1-infected patients. Such up-regulation of CXCR4 by IL-18 seems to facilitate viral entry and replication, indicating a further role of the cytokine in the disease progression. Moreover, Wiercinska-Drapalo et al. [21] demonstrated the presence of an association between plasma level of IL-18 with viral load and disease progression in HIV-1-infected patients. In another instance, Song et al.[32] demonstrated the presence of overall positive correlation between serum IL-18 and HIV-1 viral load accompanied by negative correlation between serum IL-18 and CD4+-T cell count.
The allelic variants of the IL-18 gene −607C>A promoter polymorphism have been shown to influence the transcriptional activity and subsequently a level of IL-18 production. The IL-18 −607C/C and C/A genotypes represent the potential to produce high levels of IL-18, while, the A/A genotype is associated with low transcription activity and low production of this proinflammatory cytokine [23,24]. In connection with HIV-1/AIDS, Segat et al.[33] demonstrated the association between the IL-18 gene −607C>A promoter polymorphism and risk of HIV-1 infection in seropositive Brazilian children. Moreover, high concentrations of circulating IL-18 have been correlated with enhanced HIV-1 replication and accelerated progression of the HIV-1 infection towards AIDS [21,30].
The results of the current study revealed the occurrence of the −607 A/A genotype at statistically significant lower frequency in the HIV-1 patients than healthy controls (p = 0.040). This suggests that subjects possessing the −607 A/A genotype may be at reduced risk of HIV-1 infection. Furthermore, the existence of the A allele in the heterozygous −607 C/A genotype may also confer reduced risk against disease progression. However, as the IL-18 gene −607 C>A promoter polymorphism was the only SNP examined in this study, we cannot rule out the possibility that genetic variants from other promoter positions of this gene may also be participating in disease progression of HIV-1 infection.
The findings of the present study provide additional evidence regarding the possible involvement of host genetic variations in the immunopathogenesis of HIV-1 infection. It was found that the −607 A allele in the promoter of IL-18 gene may be playing a protective role against the progression of HIV-1 infection to AIDS. The −607 A allele is assumed to results in attenuated transcription activity and lower IL-18 cytokine production that in turn may be involved in slowing down viral replication. As a result, HIV-1 patients with the −607 A/A and C/A genotypes may progress to AIDS at slower rate than those with the C/C genotype. However, the results of this study need to be further evaluated by undertaking investigations correlating IL-18 gene promoter polymorphisms with serum levels of IL-18 in HIV-1/AIDS patients for precise determination of the role IL-18 plays in HIV-1 infection. Likewise, how the −607 A allele interacts with other possible genetic factors in influencing the pathogenesis of HIV-1 infection requires further study.
ACKNOWLEDGMENTS
The present study was approved by the Ethics Committee of the Post-Graduate Institute of Medical Education and Research at Chandigarh, India. The authors wish to thank the staff of Dr. Ajay Wancho (Department of Internal Medicine, Post-Graduate Institute of Medical Education and Research at Chandigarh, India) for the provision of the blood samples and for the demographic and clinical data collected in this study.
REFERENCES
- 1.Lifson A, Rutherford G, Jaffe J. The natural history of human immunodeficiency virus infection. J Infect Dis. 1988;158(6):1360–1367. doi: 10.1093/infdis/158.6.1360. [DOI] [PubMed] [Google Scholar]
- 2.Pantaleo G, Fauci AS. Immunopathogenesis of HIV-1 infection. Ann Rev Microbiol. 1996;50:825–854. doi: 10.1146/annurev.micro.50.1.825. [DOI] [PubMed] [Google Scholar]
- 3.Hogan CM, Hammer SM. Host determinants in HIV infection and disease. Part 1: cellular and humoral immune responses. Ann Intern Med. 2001;134(9, Pt. 1):761–776. doi: 10.7326/0003-4819-134-9_part_1-200105010-00013. [DOI] [PubMed] [Google Scholar]
- 4.Kaslow RA, Dorak T, Tang J. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191(Suppl 1):S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 5.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 6.Singh P, Gurvinder K, Gaurav S, Narinder KM. Immunogenetic basis of HIV-1 infection, transmission and disease progression. Vaccine. 2008;26(24):2966–2980. doi: 10.1016/j.vaccine.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee K. Host genetic factors in susceptibility to HIV-1 infection and progression to AIDS. J Genet. 2010;89(1):109–116. doi: 10.1007/s12041-010-0003-4. [DOI] [PubMed] [Google Scholar]
- 8.Lucey DR, Clerici M, Shearer GM. Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clin Microbiol Rev. 1996;9(4):532–562. doi: 10.1128/cmr.9.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 10.Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, Oksenberg J, McNicholl J, Pociot F, Hardt C, D’Alfonso S. Cytokine gene polymorphism in human disease: on-line databases, supplement 1. Genes Immun. 2001;2(2):61–70. doi: 10.1038/sj.gene.6363733. [DOI] [PubMed] [Google Scholar]
- 11.Dahlman I, Eaves IA, Kosoy R, Morrison VA, Heward J, Gough SC, Allahabadia A, Franklyn JA, Tuomilehto J, Tuomilehto-Wolf E, Cucca F, Guja C, Ionescu-Tirgoviste C, Stevens H, Carr P, Nutland S, McKinney P, Shield JP, Wang W, Cordell HJ, Walker N, Todd JA, Concannon P. Parameters for reliable results in genetic association studies in common diseases. Nat Genet. 2002;30(2):149–150. doi: 10.1038/ng825. [DOI] [PubMed] [Google Scholar]
- 12.O’Shea JJ, Ma A, Lipsky P. Cytokines and autoimmunity. Nat Rev Immunol. 2002;2(1):37–45. doi: 10.1038/nri702. [DOI] [PubMed] [Google Scholar]
- 13.Hill AV. Immunogenetics and genomics. Lancet. 2001;357(9273):2037–2041. doi: 10.1016/S0140-6736(00)05117-5. [DOI] [PubMed] [Google Scholar]
- 14.Nolan KF, Greaves DR, Waldmann H. The human interleukin 18 gene IL-18 maps to 11q22.2-q22.3, closely linked to the DRD2 gene locus and distinct from mapped IDDM loci. Genomics. 1998;51(1):161–163. doi: 10.1006/geno.1998.5336. [DOI] [PubMed] [Google Scholar]
- 15.Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112(1–2):146–152. doi: 10.1016/s0165-5728(00)00407-0. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 17.Gracie JA, Robertson SE, McInnes IB. Interleukin-18. J Leukocyte Biol. 2003;73(2):213–224. doi: 10.1189/jlb.0602313. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro L, Puren AJ, Barton HA, Novick D, Peskind RL, Shenkar R, Gu Y, Su MS-S, Dinarello CA. Interleukin 18 stimulates HIV type 1 in monocytic cells. Proc Natl Acad Sci USA. 1998;95(21):12550–12555. doi: 10.1073/pnas.95.21.12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein SA, Klebba C, Kauschat D, Pape M, Ozmen L, Hoelzer D, Ottmann OG, Kalina U. Interleukin-18 stimulates HIV-1 replication in a T-cell line. Eur Cytokine Netw. 2000;11(1):47–52. [PubMed] [Google Scholar]
- 20.Torre D, Speranza F, Martegani R, Pugliese A, Castelli F, Basilico C, Biondi G. Circulating levels of IL-18 in adult and paediatric patients with HIV-1 infection. AIDS. 2000;14(14):2211–2212. doi: 10.1097/00002030-200009290-00023. [DOI] [PubMed] [Google Scholar]
- 21.Wiercinska-Drapalo A, Jaroszewicz J, Flisiak R, Prokopowicz D. Plasma interleukin-18 is associated with viral load and disease progression in HIV-1-infected patients. Microbes Infect. 2004;6(14):1273–1277. doi: 10.1016/j.micinf.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Stylianou E, Bjerkeli V, Yndestad A, Heggelund L, Waehre T, Damas JK, Wachre T, Damas JK, Aukrust P, Froland SS. Raised serum levels of interleukin-18 is associated with disease progression and may contribute to virological treatment failure in HIV-1-infected patients. Clin Exp Immunol. 2003;132(3):462–466. doi: 10.1046/j.1365-2249.2003.02179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai K, Kita M, Sawai N, Shiomi S, Sumida Y, Kanemasa K, Mitsufuji S, Imamichi J, Yamaoka Y. Levels of interleukin-18 are markedly increased in Helicobacter pylori-infected gastric mucosa among patients with specific IL18 genotypes. J Infect Dis. 2008;197(12):1752–1761. doi: 10.1086/588196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Tin SK, Sivalingam SP, Thumboo J, Koh DR, Fong KY. Interleukin-18 promoter gene polymorphisms in Chinese patients with systemic lupus erythematosus: association with CC genotype at position –607. Ann Acad Med. 2007;36(2):91–95. [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. pp. E3–5. [Google Scholar]
- 26.Sobti RC, Nega B, Salih AM, Rupinder K, Seyed AH, Vijesh K, Ajay W. Polymorphisms of IL-6 174 G/C, IL-10 –592 C/A and risk of HIV/AIDS among North Indian population. Mol Cell Biochem. 2009;337(1–2):145–152. doi: 10.1007/s11010-009-0293-0. [DOI] [PubMed] [Google Scholar]
- 27.Sobti RC, Salih AM, Nega B, Seyed AH, Rupinder K, Vijesh K, Ajay W. Insights into the role of IL-12B and IFN-γ cytokine gene polymorphisms in HIV-1/AIDS infection. Folia Biologica (Praha) 2010;56(3):110–115. [PubMed] [Google Scholar]
- 28.Dinarello CA, Fantuzzi G. Interleukin-18 and host defense against infection. J Infect Dis. 2003;187(Suppl 2):S370–S384. doi: 10.1086/374751. [DOI] [PubMed] [Google Scholar]
- 29.Skurk T, Kolb H, Muller-Scholze S, Rohrig K, Hauner H, Herder C. The proatherogenic cytokine interleukin-18 is secreted by human adipocytes. Eur J Endocrinol. 2005;152(6):863–868. doi: 10.1530/eje.1.01897. [DOI] [PubMed] [Google Scholar]
- 30.Iannello A, Samarani S, Debbeche O, Tremblay C, Toma E, Boulassel M-R, Routy J-P, Ahmad A. Role of interleukin-18 in the development and pathogenesis of AIDS. AIDS Rev. 2009;11(3):115–125. [PubMed] [Google Scholar]
- 31.Pugliese A, Gennero L, Vidotto V, Speranza F, Tambini R, Torre D. Interleukin-18 enhances HIV-1 production in a human chronically-infected T cell line (H9-V) Cell Biochem Funct. 2002;20(4):333–337. doi: 10.1002/cbf.981. [DOI] [PubMed] [Google Scholar]
- 32.Song W, Wilson CM, Allen S, Wang C, Li Y, Kaslow RA, Tang J. Interleukin 18 and human immunodeficiency virus type I infection in adolescents and adults. Clin Exp Immunol. 2006;144(1):117–124. doi: 10.1111/j.1365-2249.2006.03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segat L, Bevilacqua D, Boniotto M, Arraes LC, de Souza PR, de Lima Filho JL, Crovella S. IL-18 gene promoter polymorphism is involved in HIV-1 infection in a Brazilian pediatric population. Immunogenetics. 2006;58(5–6):471–473. doi: 10.1007/s00251-006-0104-7. [DOI] [PubMed] [Google Scholar]


