Abstract
Oral leukoplakias are histopathologically-diagnosed as Candida leukoplakia or non-Candida leukoplakia by the presence or absence of hyphae in the superficial epithelium. Candida leukoplakia lesions have significantly increased malignant potential. Candida albicans is the most prevalent fungal species associated with oral leukoplakia and may contribute to malignant transformation of Candida leukoplakia. To date, no detailed population analysis of C. albicans isolates from oral leukoplakia patients has been undertaken. This study investigated whether specific C. albicans genotypes were associated with Candida leukoplakia and non-Candida leukoplakia in a cohort of Irish patients. Patients with histopathologically-defined Candida leukoplakia (n = 31) or non-Candida leukoplakia (n = 47) were screened for Candida species by culture of oral rinse and lesional swab samples. Selected C. albicans isolates from Candida leukoplakia patients (n = 25), non-Candida leukoplakia patients (n = 19) and oral carriage isolates from age and sex matched healthy subjects without leukoplakia (n = 34) were subjected to multilocus sequence typing (MLST) and ABC genotyping. MLST revealed that the clade distribution of C. albicans from both Candida leukoplakia and non-Candida leukoplakia lesions overlapped with the corresponding clade distributions of oral carriage isolates and global reference isolates from the MLST database indicating no enrichment of leukoplakia-associated clones. Oral leukoplakia isolates were significantly enriched with ABC genotype C (12/44, 27.3%), particularly Candida leukoplakia isolates (9/25, 36%), relative to oral carriage isolates (3/34, 8.8%). Genotype C oral leukoplakia isolates were distributed in MLST clades 1,3,4,5,8,9 and 15, whereas genotype C oral carriage isolates were distributed in MLST clades 4 and 11.
Introduction
Several Candida species cause opportunistic infections in humans, the most prevalent and pathogenic of which is Candida albicans. Candida albicans is also the most common Candida species isolated from the oral cavity as both a commensal and pathogenic organism in healthy individuals and those with underlying disease [1].
Oral leukoplakia is used as a general, clinical descriptive term to describe oral white patches or lesions of questionable risk, having excluded other known diseases that carry no increased risk for cancer [2]. These lesions can be further defined based on histopathological diagnosis or clinical history. Lehner [3] introduced the term “Candida leukoplakia” to describe chronic oral Candida infection presenting in the form of leukoplakia, for which increased malignant potential has been reported [4] compared to simple leukoplakia. Candida leukoplakia usually presents clinically as a well demarcated, rough, raised, white plaque-like lesion that cannot be rubbed off. The lesion may have an homogenous white surface, (homogenous leukoplakia), or in contrast a non-homogenous, nodular or speckled apperance, with an erythematous base. The commissure is the most common site affected, but it may also affect the palate and tongue. These lesions may be associated with angular cheilitis. Candida albicans is by far the most prevalent species associated with oral leukoplakia lesions [5].
Candida leukoplakia (CL) lesions are difficult to distinguish from non-Candida leukoplakias (NCLs) clinically, but the presence of invading Candida hyphae in the superficial layer of epithelium accompanied by infiltratration of polymorphic neutrophils histologically distinguish CL lesions [4], [6]. The role of Candida in inducing keratotic changes and cellular atypia is uncertain, and there is some controversy regarding the initiation of epithelial hyperplasia by Candida. Jepsen and Winther [7] suggested that Candida invades a pre-existing hyperplastic lesion, whereas in contrast Cawson and Lehner [4] proposed that Candida infection is the primary cause of CL. Holmstrup and Besserman [8] demonstrated reversion of non-homogenous CL to the homogenous type after the use of topical antifungal agents. McCullough et al. [9] reported a strong statistical association between increased oral yeast density, oral epithelial dysplasia and oral squamous cell carcinoma, with the degree of epithelial dysplasia correlating with increased oral yeast density. Based on these findings, the authors hypothesised that the progression of Candida leukoplakia to dysplasia is promoted by C. albicans. With NCL lesions, the inference is that Candida species isolated from these lesions are non-pathogenic, commensal yeast blastospore contaminants.
Detailed molecular epidemiological and population studies based on C. albicans isolates recovered from CL lesions are, to date, mostly lacking. This is most likely due to practical difficulties in obtaining sufficient isolates from such lesions for detailed studies due to the relative rarity of the condition and the requirement for biopsy and histopathological investigation. Furthermore, histopathological diagnosis of such lesions has previously been quite vague. A previous report based on biotyping experiments suggested that C. albicans isolates recovered from oral leukoplakia lesions in 17 separate patients may be genetically different to those which typically colonise the normal oral mucosa [5]. In contrast, another study that used PCR-based fingerprinting methods using two interrepeat primer combinations and a minisatellite-specific primer failed to find clonal restrictions among 38 C. albicans strains recovered from 17 patients with CL [10]. However, in this study not all lesions were histopathologically confirmed as CL.
Multilocus sequence typing (MLST) of C. albicans has been used widely to assess the genetic relatedness of isolates recovered from disparate geographic locations and for C. albicans population analysis [11], [12], [13]. MLST is a DNA-based method that examines nucleotide sequence variation in seven housekeeping genes and is highly reproducible between different laboratories. A curated, internet-based C. albicans MLST database (http://calbicans.mlst.net) has facilitated the comparison of isolates from a wide variety of anatomical sites and geographic locations. Candida albicans isolates can also be ABC genotyped based on the presence or absence of an intron in 25S rDNA [14]. Although this method has previously been described as a helpful confirmatory test in instances where isolates differ by one or more single nucleotide polymorphisms (SNPs) in MLST [15], and has been used to demonstrate geographical and temporal differences amongst C. albicans isolates [16], the method examines only one genetic marker, has a low discriminatory power (i.e. discriminates isolates into one of three genotypes) and therefore should not be used as a definitive test for isolate relatedness [15]. Genotypes A and B predominate amongst C. albicans, whereas genotype C is quite rare [17].
The purpose of the present study was to characterise the population of C. albicans isolates recovered from histopathogically-defined CL and NCL lesions from a cohort of Irish oral leukoplakia patients using MLST and ABC genotyping in order to determine if these lesions could be associated with specific C. albicans lineages.
Materials and Methods
Ethics statement
Ethical approval for this study was obtained from the St. James's Hospital (SJH) and Adelaide and Meath Hospitals including the National Children's Hospital (AMNCH) Research Ethics Committee, Dublin, Ireland. Prior to enrollment in the study, all participants were provided with comprehensive patient information documentation and all participants included in the study provided written consent. All documentation provided to patients, including consent forms were pre-approved by the Research Ethics Committee. This paper does not include any identifying, or potentially identifying, patient information.
Study group
Oral rinse and swab samples were collected from 78 patients with oral leukoplakia attending the Oral Medicine and Dysplasia clinics at the Dublin Dental University Hospital (DDUH) between December 2006 and March 2009 (Table 1). Patients were examined and followed-up by two oral medicine consultants and an oral maxillofacial consultant. A full medical and dental history was recorded and a comprehensive oral examination undertaken. A clinical diagnosis of oral leukoplakia was made as described previously [2]. A histopathological diagnosis of CL was made based on the detection of hyphae and hyphal-associated polymorphonuclear leukocytes in CL biopsies. Similarly, the absence of hyphae in leukoplakia biopsies was used to define NCL lesions. Squamous cell carcinoma developed in two CL patients at the lesional site within the follow up two-year period. None of the NCL group developed malignant change to date. The mean age of the patient group was 57.8 years (range 29–87 years) and the sex distribution was 40 (51%) male and 38 (49%) female. Forty-nine patients were smokers (62.8%), 20 of whom were diagnosed with CL. Fifteen (48.3%) of the CL patients and 11 (23.4%) NCL patients wore upper dentures.
Table 1. Oral leukoplakia patient details and Candida species recovered.
| Patient | Sex | Age | Lesionsite | Denture wearer | Smoker | Degree of dysplasia | Candida cell density | Candida species isolated | |
| CL | Rinse cfu/ml | Swab cfu | |||||||
| 01 | M | 58 | PAL | Yes | Yes | Moderate | 40 | 420 | C. albicans |
| 02 | F | 52 | BM | Yes | Yes | No | 70 | 35 | C. albicans |
| 03 | M | 74 | TON | Yes | No | Severe | 0 | 415 | C. albicans |
| 04 | M | 38 | BM | No | Yes | Moderate | 157 | 42 | C. albicans |
| 05 | M | 60 | BM | Yes | Yes | Moderate | C | 380 | C. albicans |
| 06 | F | 77 | TON | No | No | Mild | 1450 | 120 | C. albicans |
| 07 | M | 73 | AR | Yes | Yes | No | 0 | 72 | C. albicans C. glabrata |
| 08 a | F | 58 | GIN | No | No | Severe (SCC) | 0 | 480 | C. albicans |
| 09 | F | 47 | BM | No | No | Severe | 0 | 1525 | C. albicans |
| 10 | M | 55 | BM | Yes | Yes | Mild | 2060 | 130 | C. albicans C. dubliniensis |
| 11 | F | 73 | BM | Yes | Yes | Moderate | 1201025 | 8447 | C. albicans C. parapsilosis C. guilliermondii |
| 12 | F | 44 | BM | No | Yes | Moderate | 2 | 15 | C. albicans |
| 13 | M | 29 | BM | Yes | No | Severe | 2256035 | 44191 | C. albicans C. tropicalis S. cerevisiae |
| 14 a | M | 63 | BM | No | Yes | Severe (SCC) | SC | 166 | C. albicans |
| 15 | F | 64 | BM | No | No | Severe | C | SC | C. albicans |
| 16 | F | 57 | BM | No | Yes | Severe | 450 | 400 | C. albicans |
| 17 | M | 41 | BM | No | Yes | Moderate | 160 | 10 | C. albicans |
| 18 | M | 69 | BM | No | Yes | Moderate | 27 | 0 | C. albicans |
| 19 | F | 76 | PAL | Yes | No | Severe | SC | 200 | C. albicans |
| 20 | M | 72 | PAL | No | No | Moderate | 290 | 0 | C. albicans |
| 21 | F | 63 | TON | No | No | Severe | C | SC | C. albicans |
| 22 | M | 77 | BM | Yes | Ex | Moderate | 0 | 0 | None |
| 23 | M | 45 | BM | Yes | Yes | Moderate | 10 | 2 | C. albicans |
| 24 | F | 60 | BM | No | No | Moderate | 240 | 125 | C. albicans |
| 25 | M | 47 | BM | No | Yes | Severe | C130 | 4406 | C. albicans C. glabrata |
| 26 | M | 63 | BM | No | Yes | Moderate | SC | SC | C. albicans |
| 27 | F | 60 | BM | Yes | Yes | Moderate | 280 | 92 | C. albicans |
| 28 | F | 46 | BM | Yes | Yes | Mild | 0 | 250 | C. albicans |
| 29 | M | 46 | TON | No | Yes | No | 293 | 220 | C. albicans |
| 30 | F | 40 | BM | No | Yes | Moderate | 215 | 185 | C. albicans |
| 31 | M | 56 | BM | Yes | Yes | Mild | 0 | 0 | None |
| NCL | |||||||||
| 32 | F | 79 | BM | Yes | Ex | Severe | 20 | C | C. albicans |
| 33 | M | 55 | BM | Yes | Ex | Severe | 50 | 0 | C. albicans |
| 34 | M | 62 | BM | No | Yes | Moderate | 0 | 2 | C. albicans |
| 35 | M | 46 | TON | No | Ex | Moderate | 150 | 0 | C. albicans |
| 36 | F | 82 | AR | Yes | Yes | Moderate | 3010 | 07 | C. albicans C. glabrata |
| 37 | F | 41 | BM | No | Yes | Severe | 48 | 18 | C. albicans |
| 38 | M | 83 | TON | Yes | Yes | Moderate | 172 | 150 | C. albicans |
| 39 | M | 68 | PAL | No | Yes | No | 4 | 0 | C. albicans |
| 40 | F | 39 | BM | No | No | Moderate | 10 | 0 | C. albicans |
| 41 | M | 59 | TON | No | Ex | Severe | 16 | 9 | C. albicans |
| 42 | F | 49 | TON | No | No | Severe | SC | 0 | C. albicans |
| 43 | F | 44 | BM | No | No | Severe | 83 | 32 | C. albicans |
| 44 | M | 51 | PAL | No | Yes | Moderate | 79 | 8 | C. albicans |
| 45 | F | 64 | BM | No | Yes | Severe | 39 | 10 | C. albicans |
| 46 | M | 76 | BM | No | Yes | Mild | 10 | 9 | C. albicans |
| 47 | F | 55 | AR | No | Yes | Mild | 50 | 11 | C. albicans |
| 48 | M | 40 | AR | No | Yes | Mild | 0 | 3 | C. albicans |
| 49 | F | 35 | FM | Yes | Yes | Moderate | 485113 | 01 | C. albicans C. krusei |
| 50 | F | 65 | BM | No | Yes | Moderate | 20 | 0 | C. albicans |
| 51 | M | 67 | BM | Yes | Yes | Moderate | 0 | 0 | None |
| 52 | F | 62 | FM | No | Yes | Moderate | 100 | 0 | C. albicans |
| 53 | M | 73 | GIN | No | Yes | Moderate | 0 | 0 | None |
| 54 | M | 58 | FM | No | Ex | Moderate | 110 | 13 | C. albicans |
| 55 | M | 78 | BM | No | Yes | Moderate | 40 | 12 | C. albicans |
| 56 | F | 51 | BM | No | Ex | Moderate | 0 | 203 | C. albicans |
| 57 | F | 55 | TON | Yes | No | Severe | 260 | 107 | C. albicans |
| 58 | F | 87 | AR | Yes | No | Mild | 108 | 0 | C. albicans |
| 59 | M | 66 | BM | No | Ex | No | 3040 | 0 | C. albicans |
| 60 | F | 66 | BM | No | Yes | Mild | 20 | 0 | C. albicans |
| 61 | M | 60 | PAL | No | Yes | Mild | 477 | 16 | C. albicans |
| 62 | F | 78 | BM | Yes | Yes | No | 150 | 41 | C. albicans |
| 63 | F | 60 | TON | Yes | Yes | Moderate | 40 | 0 | C. abicans |
| 64 | M | 38 | TON | No | Ex | Severe | 120 | 60 | C. albicans |
| 65 | M | 47 | BM | No | Yes | Moderate | 180 | 5 | C. albicans |
| 66 | F | 48 | PAL | No | Yes | Moderate | 43 | 2 | C. albicans |
| 67 | M | 81 | BM | Yes | Ex | Severe | CSC | 270120 | C. albicans C. tropicalis |
| 68 | F | 73 | BM | No | Yes | Severe | 0 | 0 | None |
| 69 | F | 56 | TON | No | Yes | Severe | 0 | 0 | None |
| 70 | F | 62 | BM | No | No | Severe | 0 | 0 | None |
| 71 | F | 56 | FM | No | No | Severe | 0 | 0 | None |
| 72 | M | 62 | AR | No | No | Severe | 0 | 0 | None |
| 73 | F | 61 | TON | No | No | Moderate | 0 | 0 | None |
| 74 | F | 61 | PAL | No | No | Severe | 0 | 0 | None |
| 75 | F | 40 | AR | No | No | No | 0 | 0 | None |
| 76 | M | 51 | GIN | No | No | No | 0 | 0 | None |
| 77 | M | 51 | BM | No | No | Severe | 0 | 0 | None |
| 78 | M | 54 | BM | No | No | Severe | 0 | 0 | None |
Both of these patients with CL went on to develop squamous cell carcinoma at the lesional site within a two-year follow up period.
Abbreviations; CL, Candida leukoplakia; NCL, Non-Candida leukoplakia; SCC, squamous cell carcinoma; SC, semi-confluent growth (approx. 1000 cfu per swab or per ml oral wash); C, confluent (approx. 5000 cfu per swab or per ml oral wash); BM, buccal mucosa; TON, tongue, PAL, palate, AR, alveolar ridge; FM, floor of mouth; GIN, gingivae.
Oral rinse samples were also taken from a control group of 110 healthy volunteers recruited from the DDUH Accident and Emergency Department for comparison. Of these, 56 (50.9%) were male and 54 (49.1%) were female, the mean age was 54.1 years (range 29–78 years) and none had oral leukoplakia. These samples were taken during the same time period as the leukoplakia samples (i.e. December 2006-March 2009). Twenty-one healthy volunteers were smokers (19.0%) and six (5.5%) wore upper dentures.
Patients with leukoplakia and healthy volunteers were excluded from the study if they met any of the following criteria: pregnancy or lactation, diabetes or asthma, steroid treatment during the last year, antibiotic or anti-fungal treatment in the previous six months.
Histopathological diagnosis of CL and NCL lesions
A DDUH oral surgeon performed incisional lesional biopsies. Biopsy samples were fixed in 10% (v/v) neutral buffered formalin (4% (v/v) formaldehyde). Histopathological investigation of biopsy samples was performed at the Central Pathology Laboratory at St. James's Hospital, Dublin. Fixed tissues were dehydrated with 70–100% (v/v) ethanol, cleared with xylene and embedded in paraffin wax. Tissue samples were then cut into 6–8 µm thick sections and routinely stained with haematoxylin and eosin. As candidal hyphae are poorly stained by hematoxylin/eosin, the periodic acid-Schiff (PAS) stain was also used, as hyphal structures stain strongly with PAS [18]. An oral and maxillofacial pathologist made diagnoses based on the histological features of each specimen.
Sampling of the oral cavity and leukoplakia lesions
Oral rinse samples were performed as described previously [19]. Candida isolates were recovered by culture following incubation at 37°C for 48 h on the chromogenic culture medium CHROMagar CandidaTM (CHROMagar Company, Paris, France). This medium permits the presumptive identification of several clinically important Candida species including C. albicans based on colony colour and morphology [20].
Swab samples were taken using sterile cotton transport swabs and then transferred to alginate gel transport medium (Venturi Transystem, Copan Italia s.p.a, Brescia, Italy). Swab samples were taken by rubbing the entire surfaces of the lesion for 30 s; the swabbed areas sampled depended on the size of the lesion. Swab samples were plated within 2 h on to CHROMagar CandidaTM medium and incubated as above.
Identification of Candida isolates
Following incubation, CHROMagar CandidaTM plates were examined, and the relative abundance of each colony type present was recorded. Selected examples of each colony type were chosen for detailed analysis. Isolates were initially presumptively identified on the basis of colony color and morphology [20], [21] and were definitively identified by substrate assimilation profiles using the API ID 32C yeast identification system (BioMérieux, Marcy l′Etoile, France) [22].
DNA extraction
Genomic DNA from C. albicans isolates was extracted using the Qiagen DNeasy blood and tissue kit (Qiagen Crawley, West Sussex, UK) according to the manufacturer's instructions.
ABC genotyping
Template DNAs extracted from isolates were assigned to genotypes A, B or C based on the differential PCR amplification of the 25S rRNA gene, as previously described [14].
MLST
Twenty-five C. albicans isolates from CL lesions and 19 isolates from NCL lesions, from separate patients in each case (with the exception of two isolates, CL12 and CL122 which were recovered from separate CL lesions in patient 12), were subjected to MLST as described previously [12]. For comparison, MLST was also performed on 34 oral carriage (OC) C. albicans isolates recovered from oral rinse samples of 34 separate control healthy subjects attending the DDUH Accident and Emergency Department. All DNA sequencing reactions were performed commercially by Source Bioscience LifeSciences (Dublin, Ireland) using ABI 3730xl DNA analysers and dye-labelled terminators (Applied Biosystems, Foster City, CA, USA). Sequences were analysed using BioNumerics version 5.0 software (Applied Maths NV, Saint-Martens-Latem, Belgium) and the online C. albicans MLST database (http://calbicans.mlst.net). All MLST data generated in the present study have been deposited in the C. albicans MLST database (http://calbicans.mlst.net).
The genetic relatedness of the isolates investigated in the present study to each other and to selected strains from the online MLST database was evaluated using the software program MEGA version 5 to generate dendrograms based on the unweighted-pair group method with arithmetic averages (UPGMA) [23]. The UPGMA clustering algorithm defines MLST clades that correlate well with those previously defined by DNA fingerprinting using the C. albicans-specific repetitive sequence-containing Ca3 probe [24].
The online Based Upon Related (eBURST) algorithm (http://eburst.mlst.net) was used to subdivide MLST allelic profile datasets into hypothetical non-overlapping groups of clonal complexes (CCs) composed of a single putative founder diploid sequence type (DST) and its closely related descendant DSTs [25], [26].
Statistical Analyses
Statistical analyses such as Fisher's exact tests and two-tailed Student's t-tests for independent sample proportions were carried out using Graphpad software (Graphpad Software Inc., CA, USA). A power analysis was undertaken based on the comparison of two independent samples. The parameters for this one sided test used a desired power value of 75% with a α value 0.05 (http://stat.ubc.ca/~rollin/stats/ssize/b2.html).
Results
Oral leukoplakia and healthy control subjects
Clinical details of the oral leukoplakia patients are shown in Table 1. Seventy-eight patients with oral leukoplakia were enrolled in the study, of whom 31/78 (39.7%) had histopathologically-defined CL. Biopsies from the remaining 47 patients (60.3%) were classified as NCL following histopathological investigation (Table 1). None of the healthy subjects included in the study had clinical signs of oral leukoplakia. The buccal mucosa was the most prevalent lesional site in the CL group (22/31; 71%), followed by the tongue (4/31; 12.9%), palate (3/31; 9.7%), gingivae (1/31; 3.2%) and alveolar ridge (1/31; 3.2%) (Table 1). The buccal mucosa was also the most prevalent lesional site in the NCL group (21/47; 44.7%), followed by the tongue (9/47; 19.1%), alveolar ridge (6/47; 12.8%), palate (5/47; 10.6%), the floor of the mouth (4/47; 8.5%) and gingivae (2/47; 4.3%) (Table 1).
Candida detection by oral rinse sampling
Candida isolates were recovered from oral rinse samples in 55/78 oral leukoplakia patients (70.5%). Candida albicans was the most prevalent Candida species recovered and was isolated from all 55 Candida-positive oral leukoplakia patients, either alone (48 patients) or in combination with other Candida species or Saccharomyces cerevisiae (seven patients) (Table 1). Oral rinse sampling of 24/31 (77.4%) patients with CL yielded Candida species, all of whom harboured C. albicans, either alone (20 patients) or in combination with other Candida species or S. cerevisiae (four patients) (Table 1). Oral rinse sampling of 31/47 (66%) NCL patients yielded Candida species, all of whom harboured C. albicans, either alone (28 patients) or in combination with other Candida species (3 patients) (Table 1).
The average Candida cell density recovered by oral rinse sampling of the 55 Candida-positive patients was 719 cfu/ml (range 2 cfu/ml to approximately 6000 cfu/ml) (Table 1). Oral rinse sampling of 34/110 (30.9%) healthy control subjects yielded C. albicans. The median and mean average Candida cell density values recovered from these 34 Candida-positive controls was 18 and 32 cfu/ml, respectively (range 1–150 cfu/ml). No other yeast species were recovered from the healthy control subjects.
Candida detection by lesional swab sampling
Candida species were recovered by swab sampling of oral leukoplakia lesions from 50/78 (64.1%) of the total oral leukoplakia patient cohort. Candida albicans was the predominant species recovered and was isolated from 48/50 (96%) Candida-positive patients either alone (42 patients) or in combination with other Candida species or S. cerevisiae (six patients). One patient (patient 49) yielded C. krusei only and another patient (patient 36) yielded C. glabrata only using the swab method. Both of these patients yielded C. albicans by oral rinse (Table 1).
Swab sampling of lesional tissues from 27/31 (87.1%) patients with CL yielded positive cultures for Candida species, all of which consisted of C. albicans either alone (23 patients) or in combination with other Candida species or S. cerevisiae (Table 1). Swab sampling of lesional tissues from 23/47 (48.9%) patients with NCL yielded positive cultures for Candida species, 21/23 (91.3%) of which consisted of C. albicans either alone (20 patients) or in combination with C. tropicalis (patient 67). One patient yielded C. glabrata only (patient 36) and one patient yielded C. krusei only (patient 49) (Table 1).
Interestingly, a comparison of oral rinse and lesional swab Candida culture data revealed that swab samples from oral lesions in eight patients [five CL patients (03, 07, 08, 09 and 28) and three NCL patients (34, 48 and 56)] were culture-positive for Candida, whereas the corresponding oral rinse samples were culture-negative (Table 1). The opposite was true for patient 40, from whom Candida was recovered by oral rinse, but not by lesional swabbing (Table 1).
MLST analysis of C. albicans
Isolates from CL (n = 25), NCL (n = 19) and healthy controls (n = 34), were analysed with the consensus C. albicans MLST scheme [12]. MLST resulted in a dataset of 2,883 bp for each isolate. Single nucleotide polymorphisms (SNPs) were identified in 84 of the 2,883 bp (2.9%) analysed. The AAT1a gene yielded 11 SNPs, (13.1%) the ACC1 gene yielded six (7.1%) SNPs, the ADP1 gene yielded 14 (16.7%) SNPs, the MPIb gene yielded 12 (14.2%) SNPs, the SYA1 gene also yielded 11 (13.1%) SNPs, the VPS13 gene yielded 15 (17.9%) SNPs and the ZWF1 gene yielded 15 (17.9%) SNPs.
New allelic profiles and DSTs were identified in 56 isolates (Table 2). Of the seven loci investigated for each isolate, five new alleles were identified including four new ZWF1b alleles, and one MPIb allele. Only two DSTs were identified in duplicate; the newly identified DST2108 was identified in an isolate (OL37) recovered from an NCL lesion and in an isolate (OC304) recovered from a healthy individual. Another newly identified DST, 2105, was identified in isolates CL12 and CL122, however, these isolates were recovered from separate CL lesions in the same patient, patient 12 (Table 2 and Figure 1).
Table 2. MLST DSTs, allelic profiles and ABC genotypes of C. albicans isolates recovered from OL patients and healthy oral carriers.
| Isolate a | Source | DST | MLST loci | MLST cladeb | CC | Htz | ABC Genotype c | ||||||
| AAT | ACC | ADP1 | MPI | SYA | VPS13 | ZWF | |||||||
| CL10 | CL | 1235 | 2 | 2 | 2 | 31 | 2 | 24 | 5 | 1 | 1 | 14 | A |
| CL09 | CL | 1234 | 2 | 2 | 2 | 27 | 2 | 24 | 5 | 1 | 1 | 14 | A |
| CL29 | CL | 1659 | 2 | 2 | 2 | 2 | 2 | 24 | 5 | 1 | 1 | 15 | A |
| CL01 | CL | 408 | 2 | 5 | 5 | 2 | 2 | 20 | 5 | 1 | 1 | 17 | A |
| CL20 | CL | 1400 | 2 | 5 | 5 | 31 | 2 | 6 | 5 | 1 | 1 | 22 | A |
| CL08 d | CL | 1239 | 2 | 23 | 5 | 2 | 2 | 63 | 5 | 1 | S | 18 | A |
| CL15 | CL | 1237 | 73 | 5 | 5 | 9 | 2 | 127 | 20 | 1 | S | 7 | A |
| CL23 | CL | 228 | 8 | 5 | 20 | 6 | 43 | 69 | 22 | 8 | 23 | 14 | A |
| CL25 | CL | 538 | 62 | 12 | 21 | 1 | 6 | 30 | 4 | 11 | 5 | 23 | A |
| CL12 | CL | 2105 | 6 | 3 | 37 | 2 | 38 | 46 | 237 | 7 | 16 | 27 | A |
| CL122 | CL | 2105 | 6 | 3 | 37 | 2 | 38 | 46 | 237 | 7 | 16 | 27 | A |
| CL04 | CL | 149 | 2 | 5 | 5 | 2 | 2 | 21 | 20 | 1 | 1 | 18 | A |
| CL17 | CL | 1240 | 13 | 10 | 15 | 24 | 50 | 37 | 15 | 3 | 4 | 21 | B |
| CL21 | CL | 1294 | 21 | 26 | 14 | 18 | 72 | 86 | 84 | 6 | 8 | 21 | B |
| CL22 | CL | 1682 | 4 | 31 | 6 | 4 | 61 | 15 | 201 | 15 | 58 | 11 | B |
| CL16 | CL | 2106 | 13 | 10 | 15 | 4 | 7 | 32 | 15 | 3 | 4 | 21 | B |
| CL24 | CL | 79 | 2 | 5 | 5 | 9 | 2 | 6 | 5 | 1 | 1 | 17 | C |
| CL13 | CL | 1236 | 13 | 32 | 15 | 6 | 7 | 140 | 15 | 3 | 4 | 28 | C |
| CL14 d | CL | 1681 | 13 | 10 | 83 | 6 | 84 | 32 | 15 | 3 | S | 19 | C |
| CL05 | CL | 1232 | 14 | 3 | 6 | 4 | 56 | 3 | 8 | 4 | S | 7 | C |
| CL02 | CL | 1432 | 4 | 26 | 61 | 4 | 34 | 60 | 202 | 4 | S | 8 | C |
| CL06 | CL | 1233 | 33 | 7 | 38 | 31 | 78 | 122 | 15 | 8 | S | 19 | C |
| CL07 | CL | 1680 | 28 | 14 | 38 | 102 | 31 | 47 | 15 | 8 | S | 17 | C |
| CL03 | CL | 1431 | 4 | 35 | 6 | 4 | 58 | 15 | 200 | 15 | S | 6 | C |
| CL11 | CL | 2107 | 4 | 13 | 6 | 4 | 61 | 29 | 201 | 15 | 30 | 12 | C |
| OL62 | NCL | 1648 | 8 | 3 | 2 | 4 | 2 | 6 | 5 | 1 | 1 | 9 | A |
| OL65 | NCL | 1047 | 8 | 2 | 5 | 2 | 2 | 6 | 5 | 1 | 1 | 16 | A |
| OL66 | NCL | 1651 | 2 | 2 | 5 | 2 | 2 | 63 | 20 | 1 | 1 | 8 | A |
| OL32 | NCL | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 | 1 | 1 | 23 | A |
| OL64 | NCL | 1650 | 3 | 3 | 5 | 3 | 57 | 3 | 6 | 1 | 10 | 10 | A |
| OL47 | NCL | 1238 | 35 | 2 | 4 | 4 | 49 | 4 | 4 | 2 | 2 | 3 | A |
| OL61 | NCL | 1496 | 35 | 2 | 4 | 4 | 49 | 26 | 4 | 2 | 2 | 7 | A |
| OL34 | NCL | 481 | 8 | 14 | 8 | 4 | 2 | 3 | 8 | 4 | 3 | 4 | A |
| OL57 | NCL | 1683 | 62 | 3 | 3 | 3 | 26 | 16 | 95 | 9 | 6 | 19 | A |
| OL59 | NCL | 1685 | 67 | 3 | 10 | 1 | 6 | 8 | 20 | 11 | S | 8 | A |
| OL37 | NCL | 2108 | 35 | 4 | 4 | 4 | 4 | 41 | 4 | 2 | 3 | 19 | A |
| OL38 | NCL | 37 | 2 | 5 | 5 | 2 | 2 | 21 | 5 | 1 | 1 | 22 | A |
| OL55 | NCL | 1224 | 2 | 2 | 2 | 2 | 2 | 20 | 5 | 1 | 1 | 16 | A |
| OL58 | NCL | 1684 | 28 | 14 | 38 | 2 | 106 | 122 | 15 | 8 | S | 22 | B |
| OL39 | NCL | 1429 | 3 | 3 | 3 | 3 | 67 | 16 | 95 | 9 | S | 13 | B |
| OL43 | NCL | 2109 | 4 | 82 | 6 | 4 | 61 | 32 | 201 | 15 | S | 11 | B |
| OL63 | NCL | 1649 | 8 | 3 | 2 | 2 | 2 | 6 | 5 | 1 | 1 | 16 | C |
| OL33 | NCL | 1686 | 11 | 8 | 4 | 3 | 7 | 19 | 4 | 5 | S | 23 | C |
| OL35 | NCL | 1652 | 32 | 3 | 43 | 3 | 3 | 32 | 94 | 9 | S | 15 | C |
| OC102 | OC | 1660 | 3 | 5 | 5 | 2 | 2 | 76 | 5 | 1 | 1 | 18 | A |
| OC100 | OC | 1428 | 2 | 23 | 5 | 102 | 2 | 20 | 20 | 1 | S | 10 | A |
| OC107 | OC | 1664 | 4 | 5 | 6 | 2 | 2 | 106 | 5 | 1 | S | 16 | A |
| OC116 | OC | 1673 | 2 | 3 | 10 | 2 | 2 | 94 | 2 | 1 | S | 16 | A |
| OC106 | OC | 1663 | 4 | 5 | 4 | 4 | 139 | 26 | 4 | 2 | 2 | 11 | A |
| OC119 | OC | 275 | 35 | 2 | 4 | 4 | 4 | 4 | 4 | 2 | 2 | 8 | A |
| OC103 | OC | 1661 | 40 | 24 | 41 | 21 | 4 | 76 | 27 | 2 | S | 13 | A |
| OC109 | OC | 1666 | 4 | 2 | 14 | 4 | 139 | 41 | 67 | 2 | S | 7 | A |
| OC112 | OC | 1669 | 36 | 2 | 6 | 4 | 49 | 41 | 4 | 2 | S | 4 | A |
| OC110 | OC | 1667 | 8 | 14 | 8 | 4 | 56 | 10 | 8 | 4 | 3 | 9 | A |
| OC115 | OC | 1672 | 4 | 60 | 6 | 4 | 4 | 41 | 4 | 2 | S | 11 | A |
| OC117 | OC | 1674 | 13 | 3 | 6 | 34 | 62 | 8 | 47 | 5 | S | 16 | A |
| OC108 | OC | 1665 | 5 | 27 | 37 | 4 | 34 | 105 | 12 | 11 | S | 20 | A |
| OC114 | OC | 1671 | 4 | 19 | 6 | 4 | 61 | 15 | 201 | 15 | 58 | 8 | A |
| OC301 | OC | 2110 | 35 | 4 | 4 | 4 | 49 | 4 | 4 | 2 | 3 | 11 | A |
| OC303 | OC | 2111 | 2 | 3 | 6 | 2 | 2 | 5 | 5 | 1 | 1 | 19 | A |
| OC304 | OC | 2108 | 35 | 4 | 4 | 4 | 4 | 41 | 4 | 2 | 3 | 19 | A |
| OC305 | OC | 2112 | 2 | 5 | 5 | 2 | 2 | 5 | 12 | 1 | 1 | 17 | A |
| OC306 | OC | 492 | 6 | 3 | 37 | 2 | 38 | 32 | 12 | 7 | 16 | 20 | A |
| OC307 | OC | 1118 | 55 | 3 | 4 | 54 | 6 | 45 | 15 | 8 | 12 | 11 | A |
| OC310 | OC | 2113 | 60 | 7 | 21 | 1 | 50 | 11 | 15 | 11 | 11 | 15 | A |
| OC311 | OC | 1133 | 2 | 5 | 5 | 2 | 2 | 5 | 5 | 1 | 1 | 22 | A |
| OC312 | OC | 24 | 2 | 5 | 5 | 2 | 2 | 24 | 5 | 1 | 1 | 18 | A |
| OC313 | OC | 2114 | 35 | 4 | 4 | 4 | 4 | 4 | 26 | 2 | 3 | 15 | A |
| OC315 | OC | 171 | 8 | 3 | 6 | 2 | 2 | 6 | 5 | 1 | 1 | 16 | A |
| OC316 | OC | 444 | 2 | 5 | 5 | 4 | 2 | 6 | 5 | 1 | 1 | 17 | A |
| OC105 | OC | 659 | 11 | 26 | 6 | 4 | 34 | 60 | 119 | 4 | 9 | 10 | B |
| OC104 | OC | 1662 | 3 | 26 | 6 | 4 | 34 | 60 | 55 | 4 | S | 4 | B |
| OC111 | OC | 1668 | 14 | 14 | 30 | 4 | 56 | 3 | 8 | 4 | S | 8 | B |
| OC302 | OC | 2115 | 4 | 35 | 6 | 4 | 61 | 15 | 5 | 15 | 62 | 12 | B |
| OC309 | OC | 299 | 4 | 17 | 21 | 19 | 27 | 83 | 22 | 12 | 7 | 15 | B |
| OC113 | OC | 1670 | 8 | 7 | 6 | 4 | 56 | 3 | 118 | 4 | S | 5 | C |
| OC308 | OC | 924 | 8 | 14 | 6 | 4 | 7 | 3 | 8 | 4 | 2 | 9 | C |
| OC314 | OC | 2116 | 60 | 7 | 21 | 1 | 7 | 55 | 15 | 11 | 11 | 26 | C |
With regard to isolates recovered from CL and NCL patients, the numeral part of the isolate name refers to the corresponding patient numbers shown in Table 1, column one (e.g. isolate CL10 was recovered from patient 10). Isolates CL12 and CL122 were recovered from two separate CL lesions in patient 12. CL and NCL isolates were recovered from lesional swabs, whereas OC isolates were recovered from oral rinse samples.
MLST clades defined according to Odds et al. [17].
ABC genotypes were assigned based on the presence or absence of an intron in the 25S rRNA gene [14].
Two CL patients included in this study (patients 08 and 14, Table 1) developed squamous cell carcinoma at the lesional site within a two-year follow up period. Isolate CL14 (patient 14) and isolate CL08 (patient 8) belonged to ABC genotypes C and A, respectively.
Abbreviations: DST, diploid sequence type; CC, clonal complex; Htz, number of heterozygous sites; CL, Candida leukoplakia; NCL, non-Candida leukoplakia; OC, oral carriage; S, singleton.
Figure 1. UPGMA dendrogram depicting the genetic relatedness of all C. albicans isolates subjected to MLST and ABC genotyping analysis in the present study.
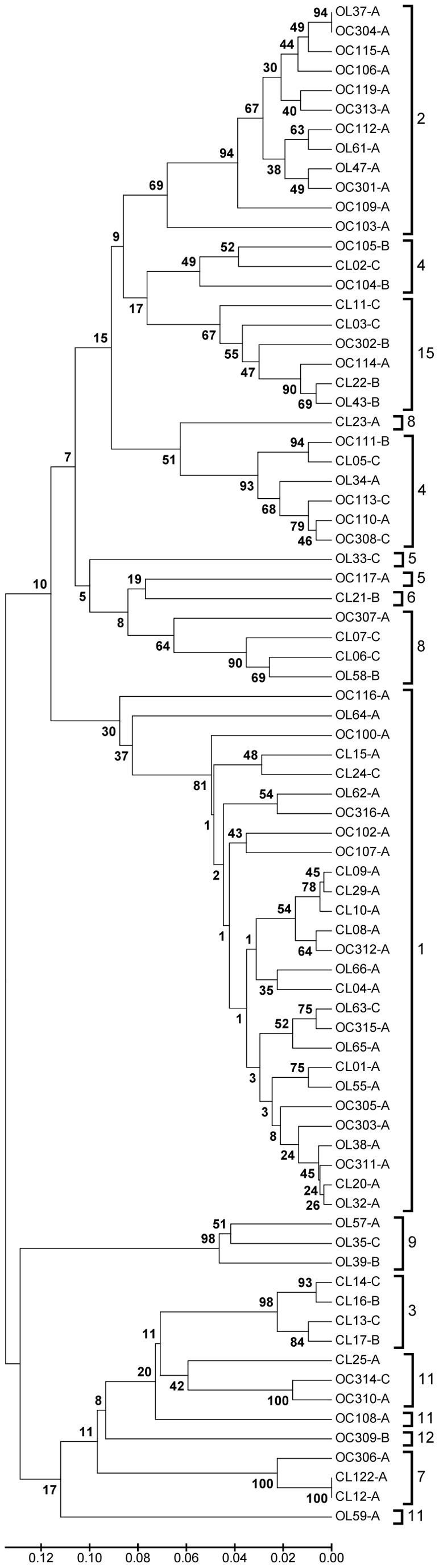
Individual isolates recovered from CL (n = 25) and NCL (n = 19) patients are indicated with the letters CL and OL in the isolate names, respectively. The numeral part of the isolate name refers to the corresponding patient numbers as shown in Table 1, column 1. Isolates recovered from age and sex-matched healthy control patients (n = 34) by oral rinse are indicated using the letters OC in the isolate name. Hyphenated letters A, B or C following each isolate name indicate ABC genotypes. The scale bar indicates p- distance. Distinct MLST clades are indicated by separated square parenthesis to the right of the isolate names and labelled with previously designated clade numbers [17]. Numbers at clade branches indicate bootstrap support levels, based on 1000 replications. Overall, the population analysis based on MLST suggests no clonal enrichment of C. albicans isolates recovered from CL or NCL lesions. Isolates recovered from CL patients are distributed among eight clades, isolates from NCL patients are distributed among eight clades and isolates from healthy carriers are distributed among nine clades, with clade 1 predominating in all three groups.
A UPGMA dendrogram was constructed using the MLST allelic profile data resulting from 44 isolates recovered from oral leukoplakia lesions, 34 OC isolates and 42 isolates representative of previously documented MLST clades previously included in the C. albicans MLST database (data not shown), to determine the genetic relatedness of isolates in the present study to those previously included in the MLST database, and to assign newly identified DSTs to clades. This tree identified the presence of distinct MLST clades as previously described [17]. A smaller UPGMA dendrogram was constructed based on the concatenated MLST SNPs of only the 78 isolates included in the current study (Figure 1). This dendrogram shows that isolates recovered from both CL and NCL patient groups are distributed amongst 11 distinct MLST clades, and OC isolates are distributed amongst nine distinct MLST clades, with clade 1 predominant in all three groups of patients (Figure 1 and Table 2). Of the 78 isolates investigated by MLST, 27/78 (34.6%) belonged to C. albicans MLST clade 1, consisting of nine CL isolates, eight NCL isolates and 10 OC isolates. A further 12/78 (15.4%) isolates belonged to MLST clade 2, of which nine were OC isolates and three were NCL isolates. There was an absence of CL isolates in clade 2. The third most predominant clade was clade 4, which contained 9/78 (11.5%) isolates including six OC isolates, two CL isolates and one NCL isolate. Interestingly, all four isolates belonging to clade 3 in the present study were recovered from CL lesions, and all three isolates belonging to clade 9 in the present study were recovered from NCL lesions (Figure 1 and Table 2).
The level of heterozygosity amongst the 84 polymorphic nucleotides were determined for the C. albicans isolates investigated in the present study, as significant losses of heterozygosity can be associated with minor genetic switches or microvariation [15], [27]. The average number of heterozygous sites in isolates recovered from CL lesions was 16.9±6.2 sites compared to 13.9±14.1 sites in isolates recovered from NCL lesions and 13.4±5.4 sites in the OC isolates recovered from healthy patients (Table 2). These differences are most likely the result of the minor differences in clade prevalence amongst isolates from each patient group.
According to eBURST analysis, an alternative method of predicting evolutionary patterns and founding genotypes, the most predominant clonal complex (CC) identified amongst 21/78 (26.9%) isolates from the present study was CC1. CC1 isolates were recovered from seven CL patients, seven NCL patients and seven OC subjects (Table 2). Five isolates belonged to CC2 and six to CC3 (Table 2). Singletons that were not assigned to any CCs were identified in isolates recovered from 8/25 (32%) CL patients, 6/19 (31.6%) NCL patients and 12/34 (35.3%) OC isolates (Table 2).
ABC genotyping of C. albicans isolates
The same 78 isolates that were investigated by MLST were subjected to ABC genotyping, including isolates from CL (n = 25), NCL (n = 19), and OC (n = 34). Genotype A was the most common genotype and was identified in 51/78 (65.4%) isolates. Twelve isolates (15.4%) were identified as genotype B and 15 (19.2%) were identified as genotype C (Table 2). Of the 34 OC isolates, 26 (76.5%) were identified as genotype A, five were identified as genotype B (14.7%), and the remaining three were genotype C (8.8%). In contrast, only 25/44 (56.8%) oral leukoplakia isolates (12 CL and 13 NCL) belonged to genotype A (Table 3). Genotype B was identified in seven (15.9%) oral leukoplakia isolates (four CL and three NCL). Interestingly, 12/44 (27.3%) oral leukoplakia isolates were identified as genotype C, (nine CL and three NCL). The proportions of genotypes A–C among CL and OC isolates were significantly different (P<0.02 with Fisher's exact test). Isolates from CL lesions were significantly enriched with genotype C 9/25 (36%) when compared to OC isolates (3/34, 8.8%) from healthy carriers (P<0.05 with two sample Student's t-test). Due to the small number of CL isolates available for investigation in the present study, a power analysis was undertaken to determine if the sample size was sufficient to reliably detect this increased prevalence of genotype C. Using a desired power value of 75% with an α value 0.05, a sample size of 25 was indicated for each sample group. As 25 CL isolates and 34 OC isolates were included in the study, the results of the power analysis indicated that sufficient isolates were investigated to support the significance of the observed increased prevalence of genotype C isolates amongst CL isolates relative to OC isolates.
Table 3. Distribution of ABC genotypes in isolates recovered from oral leukoplakia patients and healthy carriers.
| Patient Group | Number of isolates | Genotypes identified a n (%) |
| CL | 25 | A = 12 (48)B = 4 (16)C = 9 (36) |
| NCL | 19 | A = 13 (68.4)B = 3 (15.8)C = 3 (15.8) |
| OC | 34 | A = 26 (76.5)B = 5 (14.7)C = 3 (8.8) |
The prevalence of genotype A and genotype C isolates differed significantly between the CL and OC groups (P<0.02).
Abbreviations: CL, Candida leukoplakia; NCL, non-Candida leukoplakia; OC, oral carriage.
Discussion
The present study observed that Candida isolates were more frequently recovered by oral rinse sampling of patients with oral leukoplakia (70.5% positive) than of healthy controls (30.9% positive). Unsurprisingly, C. albicans was the most frequently identified Candida species, and was recovered from all Candida-positive oral leukoplakia patients either alone, or in combination with other species. No Candida species other than C. albicans were recovered from the healthy control population. Oral leukoplakia patients were histopathologically-diagnosed as having CL or NCL in the present study. Candida species were recovered from swab sampling of 64.1% of the oral leukoplakia patients, and in 87.1% of patients with CL. Interestingly, comparative oral rinse and lesional swab sampling methods revealed that in eight oral leukoplakia patients, lesional swab yields of Candida species did not correlate with corresponding oral rinse sampling Candida yields (Table 1). It is possible that the oral rinse sampling method, and to a lesser extent swab sampling, was not vigorous enough to dislodge strongly adherent and/or invasive Candida blastospores or hyphae from these CL lesions.
MLST is a very effective tool for C. albicans population analyses [11]. Overall, the population analysis based on MLST suggests that no clonal enrichment exists in isolates recovered from CL or NCL lesions. The isolates recovered from both CL and NCL patient groups are distributed amongst 11 distinct MLST clades, and OC isolates are distributed amongst nine distinct MLST clades. Clade 1 predominated in all three groups of patients with 27 isolates (34.6%) belonged to this clade. Twelve isolates (15.4%) belonged to MLST clade 2, and nine (11.5%) isolates belonged to MLST clade 4. This study therefore supports previous findings [17], [28], that clades 1, 2 and 4 comprise more than the 2/3 of isolates in the MLST database.
Interestingly, there was an absence of CL isolates in clade 2, whereas 9/12 (75%) clade 2 isolates consisted of OC isolates representing 26.5% (9/34) of all OC isolates included in the study. All four isolates belonging to clade 3 in the present study were recovered from CL lesions. Furthermore, all three isolates belonging to clade 9 in the present study were recovered from NCL lesions. It is likely that increasing the numbers of CL, NCL and OC isolates in larger future studies would result in the diversification of these clades (Fiure 1).
Currently, there are 2,218 C. albicans isolate profiles in the MLST database (http://calbicans.mlst.net/) comprising 2,086 unique allelic profiles and their corresponding DSTs (date accessed: 10th April 2013), of which 373 (17.9%) isolates are described as oral isolates comprising 290 distinct DSTs. According to the eBURST algorithm, these oral isolate DSTs can be divided into 24 different CCs, with CC1 predominating. The remaining 131/290 (45.2%) DSTs are classified as singletons. In the present study, analysis of 78 C. albicans MLST profiles using the eBURST algorithm showed that 50 isolates were distributed among 17 CCs, with CC1 predominating. Twenty-six (33.3%) isolates were assigned as singletons. This suggests that, like the UPGMA clade-based method, eBURST identifies no significant clonal enrichment in isolates recovered from oral leukoplakia patients. Overall, our MLST data correlates with previous biotyping studies [10] which failed to find clonal distinction among C. albicans strains recovered from CL lesions.
ABC genotyping of C. albicans isolates was included in the present study to provide further isolate discriminatory data. Similarly to previous studies [13], [16], [17], [29] genotype A predominated in all groups examined, the majority (92.6%) of isolates belonging to MLST clade 1 were identified as genotype A. Surprisingly, genotype C was identified more frequently in the current study than in the aforementioned studies, particularly in isolates from oral leukoplakia lesions (Table 3), being identified in 9/25 (36%) and 3/19 (15.8%) of isolates recovered from CL and NCL lesions, respectively (Table 3). In contrast, only three genotype C isolates were identified among the 34 (8.8%) OC isolates investigated. However, the MLST data clearly did not support any suggestion of clonal enrichment (Figure 1 and Table 2). It is important to emphasise that MLST has been shown to be an exquisitely informative tool for inferring clonal relationships between C. albicans isolates [17] in contrast to ABC genotyping, which examines only one locus and can only separate isolates into three groups. ABC genotypes A and B are determined by the absence or presence of an intron in the 25S rRNA gene, respectively. Genotype C is a hybrid of the other two genotypes, containing the intron in only one allele. On balance, it is unlikely that the observed genotype C enrichment among isolates from leukoplakia lesions reflects a specific clonal enrichment. This suggestion is supported by the findings that ABC genotype C was identified in Candida leukoplakia isolates belonging to MLST clades 1, 3, 4, 8 & 15, and in non-Candida leukoplakia isolates belonging to MLST clades 1, 5 & 9.
The oral leukoplakia isolates investigated in the present study might represent a geographical clonal population of C. albicans from Ireland. However, this seems unlikely as the OC control isolates were also from the same geographical location but exhibited a different ABC genotype distribution (Table 3). Further investigations based on a larger patient numbers are required and should include individuals from disparate geographical locations and ethnic backgrounds in order to decipher this phenomenon further. Additional investigations might involve comparative SNP microarray or whole genome sequence analysis of several CL and NCL isolates from each ABC genotype.
Very few previous studies actually correlated C. albicans ABC genotypes with specific medical conditions. Most previous studies that used ABC genotyping did so as a secondary typing method in combination with another, more discriminatory typing method such as microsatellite typing or MLST. Candida albicans ABC genotypes tend to be more often associated with particular MLST clades rather than with particular medical conditions [17]. For example, one previous study identified an enrichment of MLST clade 1 C. albicans isolates recovered from sub-gingival sites in patients with untreated periodontal disease; 21/31 isolates investigated belonged to MLST clade 1 and all were found to belong to ABC genotype A [19]. This finding was not unusual, as many previous studies have shown that MLST clade 1 consists predominantly of ABC genotype A isolates [17]. No ABC genotype C isolates were identified in the periodontal study. In a separate study on 14 oral C. albicans-positive patients with Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy (APECED) syndrome, a rare genetic disease characterised by autoimmunity to endocrine organs, ectodermal disorders and chronic mucocutaneous candidiasis, 9/14 patients yielded genotype A isolates, six of which belonged to MLST clade 1 [30]. One patient yielded genotype B isolates (MLST clade 8) and four patients yielded genotype C isolates, two of which belonged to MLST clade 4, one to clade 15, whereas the remaining isolate was a singleton.
Based mainly on case reports, the potential for malignant transformation of CL is well recognised [31], [32], and Candida has previously been implicated as having an aetiological role in initiation of carcinoma in leukoplakia lesions associated with acetylaldehyde production [33], [34]. A recent study reported that C. albicans adhesion, tissue invasion and damage of epithelial cells is influenced by a combination of fungal morphology and activity and epithelial cell type and stage of differentiation, suggesting that epithelial cells differ in their susceptibility to C. albians [35]. This study also showed that C. albicans can invade oral epithelial cells by induced endocytosis and by active penetration.
In the present study two cases of malignant transformation occurred in diagnosed CL lesions, one infected with an MLST clade 1, ABC genotype A isolate and the other infected with an MLST clade 3, ABC genotype C isolate, during the follow up period of two years (Table 1). Overall, the results of the present study do not show enrichment of specific C. albicans clonal lineages among isolates recovered from CL lesions although the apparent increased association of ABC genotype C isolates with CL lesions is intriguing. Our findings suggest that C. albicans invades leukoplakia lesions giving rise to Candida leukoplakia and that lesional factors are probably more significant than the C. albicans lineage. Larger studies are required to further investigate the role of Candida in CL.
Acknowledgments
We acknowledge Imperial College London for hosting the C. albicans MLST website and the Wellcome Trust for funding the database and website. We thank Dr. M.E. Bougnoux at the Institut Pasteur, Paris, France for her advice and assistance, and for the continuous curation and management of the C. albicans MLST database.
Funding Statement
This study was supported by the Dublin Dental University Hospital Microbiology Research Unit. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Odds FC (1988) Ecology of Candida and epidemiology of candidosis. In: Odds FC editor. Candida and Candidosis. London: Bailière Tindall. 68–92.
- 2. Warnakulasuriya S, Johnson NW, van der Waal I (2007) Nomenclature and classification of potentially malignant disorders of the oral mucosa. J Oral Pathol Med 36: 575–580. [DOI] [PubMed] [Google Scholar]
- 3. Lehner T (1964) Chronic Candidiasis. Trans St Johns Hosp Dermatol Soc 50: 8–21. [PubMed] [Google Scholar]
- 4. Cawson RA, Lehner T (1968) Chronic hyperplastic candidiasis–Candidal leukoplakia. Br J Dermatol 80: 9–16. [DOI] [PubMed] [Google Scholar]
- 5. Krogh P, Holmstrup P, Thorn JJ, Vedtofte P, Pindborg JJ (1987) Yeast species and biotypes associated with oral leukoplakia and lichen planus. Oral Surg Oral Med Oral Pathol 63: 48–54. [DOI] [PubMed] [Google Scholar]
- 6. Sitheeque MA, Samaranayake LP (2003) Chronic hyperplastic candidosis/candidiasis (Candidal leukoplakia). Crit Rev Oral Biol Med 14: 253–267. [DOI] [PubMed] [Google Scholar]
- 7. Jepsen A, Winther JE (1965) Mycotic infection in oral leukoplakia. Acta Odontol Scand 23: 239–256. [DOI] [PubMed] [Google Scholar]
- 8. Holmstrup P, Bessermann M (1983) Clinical, therapeutic, and pathogenic aspects of chronic oral multifocal candidiasis. Oral Surg Oral Med Oral Pathol 56: 388–395. [DOI] [PubMed] [Google Scholar]
- 9. McCullough M, Jaber M, Barrett AW, Bain L, Speight PM, et al. (2002) Oral yeast carriage correlates with presence of oral epithelial dysplasia. Oral Oncol 38: 391–393. [DOI] [PubMed] [Google Scholar]
- 10. Bartie KL, Williams DW, Wilson MJ, Potts AJ, Lewis MA (2001) PCR fingerprinting of Candida albicans associated with chronic hyperplastic candidosis and other oral conditions. J Clin Microbiol 39: 4066–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bougnoux ME, Morand S, d'Enfert C (2002) Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans . J Clin Microbiol 40: 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, et al. (2003) Collaborative consensus for optimized multilocus sequence typing of Candida albicans . J Clin Microbiol 41: 5265–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tavanti A, Gow NA, Senesi S, Maiden MC, Odds FC (2003) Optimization and validation of multilocus sequence typing for Candida albicans . J Clin Microbiol 41: 3765–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCullough MJ, Clemons KV, Stevens DA (1999) Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea . J Clin Microbiol 37: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte JA, et al. (2006) Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol 44: 3647–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCullough MJ, Clemons KV, Stevens DA (1999) Molecular epidemiology of the global and temporal diversity of Candida albicans . Clin Infect Dis 29: 1220–1225. [DOI] [PubMed] [Google Scholar]
- 17. Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, et al. (2007) Molecular phylogenetics of Candida albicans . Eukaryot Cell 6: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker DM, Arendorf TM (1990) Candidal leucoplakia, chronic multifocal candidosis and median rhomboid glossitis. In: Samaranayake LP, MacFarlane TW editors. Oral Candidosis. Guilford, UK: John Wright, 184–199.
- 19. McManus BA, Maguire R, Cashin PJ, Claffey N, Flint S, et al. (2012) Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J Clin Microbiol 50: 3335–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Odds FC, Bernaerts R (1994) CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol 32: 1923–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coleman DC, Sullivan DJ, Bennett DE, Moran GP, Barry HJ, et al. (1997) Candidiasis: the emergence of a novel species, Candida dubliniensis . AIDS 11: 557–567. [DOI] [PubMed] [Google Scholar]
- 22. Pincus DH, Coleman DC, Pruitt WR, Padhye AA, Salkin IF, et al. (1999) Rapid identification of Candida dubliniensis with commercial yeast identification systems. J Clin Microbiol 37: 3533–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 24. Odds FC, Jacobsen MD (2008) Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell 7: 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spratt BG, Hanage WP, Li B, Aanensen DM, Feil EJ (2004) Displaying the relatedness among isolates of bacterial species – the eBURST approach. FEMS Microbiol Lett 241: 129–134. [DOI] [PubMed] [Google Scholar]
- 27. Bougnoux ME, Diogo D, François N, Sendid B, Veirmeire S, et al. (2006) Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J Clin Microbiol 44: 1810–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen KW, Chen YC, Lo HJ, Odds FC, Wang TH, et al. (2006) Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J Clin Microbiol 44: 2172–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wrobel L, Whittington JK, Pujol C, Oh SH, Ruiz MO, et al. (2008) Molecular phylogenetic analysis of a geographically and temporally matched set of Candida albicans isolates from humans and nonmigratory wildlife in central Illinois. Eukaryot Cell 7: 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McManus BA, McGovern E, Moran GP, Healy CM, Nunn J, et al. (2011) Microbiological screening of Irish patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy reveals persistence of Candida albicans strains, gradual reduction in susceptibility to azoles, and incidences of clinical signs of oral candidiasis without culture evidence. J Clin Microbiol 49: 1879–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cawson RA (1966) Chronic oral candidiasis and leukoplakia. Oral Surg Oral Med Oral Pathol 22: 582–591. [DOI] [PubMed] [Google Scholar]
- 32. Williamson DM (1969) Chronic hyperplastic candidiasis and squamous carcinoma. Br J Dermatol 81: 125–127. [DOI] [PubMed] [Google Scholar]
- 33. Krogh P (1990) The role of yeasts in oral cancer by means of endogenous nitrosation. Acta Odontol Scand 48: 85–88. [DOI] [PubMed] [Google Scholar]
- 34. Gainza-Cirauqui ML, Nieminen MT, Novak Frazer L, Aguirre-Urizar JM, Moragues MD, et al. (2013) Production of carcinogenic acetaldehyde by Candida albicans from patients with potentially malignant oral mucosal disorders. J Oral Pathol Med 42: 243–249. [DOI] [PubMed] [Google Scholar]
- 35. Dalle F, Wächtler B, L'Ollivier C, Holland G, Bannert N, et al. (2010) Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol 12: 248–271. [DOI] [PubMed] [Google Scholar]


