Abstract
Persistent infection of the mouse central nervous system (CNS) with mouse hepatitis virus (MHV) induces a demyelinating disease pathologically similar to multiple sclerosis and is therefore used as a model system. There is little information regarding the host factors that correlate with and contribute to MHV-induced demyelination. Here, we detail the genes and pathways associated with MHV-induced demyelinating disease in the spinal cord. High-throughput sequencing of the host transcriptome revealed that demyelination is accompanied by numerous transcriptional changes indicative of immune infiltration as well as changes in the cytokine milieu and lipid metabolism. We found evidence that a Th1-biased cytokine/chemokine response and eicosanoid-derived inflammation accompany persistent MHV infection and that antigen presentation is ongoing. Interestingly, increased expression of genes involved in lipid transport, processing, and catabolism, including some with known roles in neurodegenerative diseases, coincided with demyelination. Lastly, expression of several genes involved in osteoclast or bone-resident macrophage function, most notably TREM2 and DAP12, was upregulated in persistently infected mouse spinal cord. This study highlights the complexity of the host antiviral response, which accompany MHV-induced demyelination, and further supports previous findings that MHV-induced demyelination is immune-mediated. Interestingly, these data suggest a parallel between bone reabsorption by osteoclasts and myelin debris clearance by microglia in the bone and the CNS, respectively. To our knowledge, this is the first report of using an RNA-seq approach to study the host CNS response to persistent viral infection.
Introduction
Mouse hepatitis virus (MHV) is a positive-sense RNA virus of the coronaviridae family. MHV strain A59 is both hepatotropic and neurotropic, and intracranial inoculation of mice causes acute hepatitis and encephalitis and subsequent demyelination. MHV-,A59 replicates in the brain, spinal cord and liver, peaking at about five days post infection after which time infectious virus is cleared primarily by the CD8T cell response, also requiring CD4T cells. Although virus is cleared by 10-14 dpi from all organs, viral RNA persists in the central nervous system (CNS) and is accompanied by inflammatory demyelination, which peaks in the spinal cord at approximately one month post-infection [1-6]. Demyelinating strains of MHV (A59 and JHM) have been widely used as models for the study of the human demyelinating disease multiple sclerosis [1,7-10]. MHV-induced CNS disease involves recurring demyelination and remyelination in spinal cords of infected mice [5,6,11], suggesting that this model has pathological similarities to the remitting-relapsing forms of multiple sclerosis.
The mechanisms underlying MHV-induced demyelination remain unclear. Although MHV induces primary demyelination, axonal damage has also been observed in MHV-infected CNS [12]. Early pathological studies suggested that demyelination results from direct infection of and damage to oligodendrocytes, the myelin producing cells of the CNS [13,14]. More recent reports have implicated immunopathological mechanisms rather than oligodendrocyte damage in MHV-induced demyelination, although the specific pathways involved still remain to be determined. While RAG-/- mice, lacking mature B- and T-cells, do not undergo demyelination, neither CD4 nor CD8T-cells are absolutely required [2,8]. Virus-specific T cells persist in the CNS [15,16], and both macrophages and microglia are associated with demyelinating lesions [14]. Mice lacking B-cells exhibit more severe demyelination than immunocompetent mice, throughout the course of infection [2], but this is likely due at least in part to persistence of infectious virus. Additionally, several cytokines have been studied in the context of MHV-induced demyelination; only IP-10 is reported to affect the clinical course of disease [17-21]. While many demyelinating disorders are also presumed to involve dysregulation of lipid metabolism and transport [22-25], there are to date no reports of alterations of lipid metabolism during MHV infection.
To comprehensively characterize the gene expression changes and pathways affected by MHV-induced demyelination in the murine CNS, we employed high-throughput RNA sequencing (RNA-seq) using spinal cords from MHV-A59-infected mice at 33 days post-infection, around the time of maximum MHV-induced demyelination as quantified luxol fast blue staining of spinal cord sections [1,2,5,26]. At this time post infection, demyelination is robust and viral genome expression is detectable [1,2,5,16,26]. To our knowledge, this is the first report of using RNA-seq to detail the host response to virus-induced demyelinating disease. We found that the host response to MHV persistence was characterized by a Th1-biased cytokine response, eicosanoid-associated inflammation, ongoing antigen presentation, lymphocyte proliferation and activation, and lipid processing changes. In addition, the expression levels of several genes involved in osteoclast function, or the function of bone-resident macrophages, were augmented in the demyelinating CNS, most likely being expressed by brain resident microglia. Two of these genes, TREM2 and DAP12, were among the most highly induced during MHV-induced demyelinating disease. Interestingly, natural polymorphisms in these genes lead to a demyelinating disorder in humans concurrent with osteopetrosis, or bone cysts [27-30]. The increased expression of genes that directly affect bone resorption and osteoclasts during demyelination leads to the intriguing possibility that osteoclasts and microglia may have parallel roles in maintaining the specialized extracellular matrix of their respective compartments.
Materials and Methods
Mice, viruses, and cells
C57/Bl6 (B6) mice were obtained from the National Cancer Institute (Frederick, MD). Four-week-old mice were inoculated intracranially with 2000 plaque-forming units (pfu) of MHV-A59 [7] or an equal volume of uninfected cell lysate (mock infected) in 25 µL of PBS containing 0.75% BSA, and tissues were harvested at the indicated dpi. Tissues were flash frozen for later RNA isolation, fixed in phosphate buffered formalin for tissue embedding, or frozen in saline with 0.167% gelatin for titering of infectious virus. Standard plaque assays were performed on transformed mouse fibroblast L2 cells as described previously [31].
Ethics Statement
All mouse procedures were performed on protocols approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee (IACUC) (Reference Assurance # A3079-01). Intracranial inoculations were carried out under anesthesia with isoflurane and all efforts were made to minimize suffering. The University Of Pennsylvania School Of Medicine is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Isolation of primary murine cells
Bone marrow-derived macrophages (BMM) were generated from the hind limbs of B6 mice as described previously [32,33]. Briefly, bone marrow was differentiated into macrophages by growth in L929 fibroblast secreted M-CSF. Macrophages were seeded after 6-7 days in vitro.
Neurons were isolated from the hippocampi of E15.5-16.5 mouse embryos and plated on poly-L-lysine coated wells without astrocyte feeder layers as described previously [34,35]. Hippocampal neuron cultures were typically >95% pure as assessed by immunostaining with an antibody recognizing MAP2 (1:2; AP14, kindly provided by Virginia Lee, University of Pennsylvania).
Primary glial cultures were established as described previously [36], with the following modifications. For isolation of astrocytes, confluent mixed glial cultures were shaken on day 4-7 in vitro to dissociate the loose top layer of microglia, which were discarded. The remaining adherent cells, mostly astrocytes, were trypsinized and re-plated at a 1:2 dilution. This procedure was repeated on day 13-15. Prior to seeding, the astrocytes were passed through a series of three suspension cell culture plates for 20 minutes each at room temperature to allow adhesion of contaminating microglia and fibroblasts. For isolation of microglia, mixed glial cultures were shaken on d13-15 and the cells in the supernatant re-plated. The medium was replaced after 30 minutes to remove contaminating astrocytes, which adhere more slowly. Astrocyte and microglial cultures were each >95-99% pure as assessed by immunofluorescence staining with monoclonal antibodies directed against GFAP for astrocytes (Dako Z0334) and CD11b for microglia (Abcam ab6332) as described previously [35].
Mouse oligodendrocyte cultures were derived from a mixed population of cells isolated from the forebrains of 1- to 3-day-old neonatal C57/BL6 mice and cultured in serum-free growth medium containing PDGF, FGF2, and NT-3, as previously described [37]. When confluent, the oligodendrocyte-lineage cells were purified using a modified wash down procedure to remove astrocytes and other contaminating cells [37].
Persistence experiment
Brain lysates were cleared of cell debris by centrifugation at 3,000 rpm for fifteen minutes. A portion of the supernatant was removed and centrifuged at 13,000 rpm for three minutes. Twenty microliters of this supernatant was then injected into each of 3-5 naïve four-week old B6 mice. These mice were observed for clinical symptoms of disease, and their brains, spinal cords, and livers were harvested at the indicated dpi. Extracts from the second round of mice were titered by standard plaque assay and assayed for characteristic cytopathic effects of MHV on L2 mouse fibroblast cells. The L2 fibroblast lysates were then assayed for viral RNA using polymerase chain reaction (PCR) with primers specific to the viral subgenomic mRNA7.
Real time quantitative reverse transcriptase-PCR (qRT-PCR)
Total RNA was isolated from homogenized tissue or cultured cells using either RNeasy mini kit columns (Qiagen) or TRIzol (Gibco) according to the manufacturer’s instructions and depleted of DNA using Turbo DNase (Ambion). qRT-PCR was performed as described previously [38] on each biological replicate. Briefly, 350 ng of total RNA (from cells or tissue) was transcribed into cDNA using the Superscript III reverse transcript kit (Invitrogen). Then, 2 µl of cDNA was combined with 12.5 µl of iQ5 SYBR green mix (Bio-Rad, Hercules, CA), 6.5 µl of DEPC-treated water, and 4 µl of mixed primers (5 µM each), DNA was amplified using an iQ5 iCycler (Bio-Rad), and cycle threshold (C T) values were recorded. Basal mRNA levels were expressed as ΔC T, that is, relative to the level of β-actin mRNA (ΔC T= C T(gene of interest)-C T(ß-actin)). The expression levels in infected spinal cords relative to those in mock-infected spinal cords (fold changes) were expressed as 2-∆∆CT.
Illumina library preparation and sequencing
Tissues were solubilized in TRIzol, and total RNA was isolated and DNase-treated as described above. Poly-A-containing mRNA was selected from 20 µg of RNA pooled from 5 mice using the Dynabeads mRNA direct kit (Invitrogen). The mRNA was fragmented, a 5’ phosphate was added using T4 polynucleotide kinase (Invitrogen), and the library was size-selected (60-200 nucleotides) on a 15% TBE-urea gel. Illumina v1.5 RNA adapters were ligated to ends of the fragmented products using T4 RNA ligase 2 (New England Biolabs). A cDNA library was generated using Superscript II (Invitrogen) and PCR-amplified using Phusion HiFi DNA polymerase (New England Biolabs). The library was purified on a 6% TBE gel (Invitrogen). The quality of the library was verified prior to Illumina sequencing by cloning into Zero Blunt TOPO (Invitrogen) and analyzing 6-10 clones by standard sequencing to determine a) the presence of adapters, b) appropriate size selection, c) the abundance of ribosomal RNA, and d) the diversity of the library products. The verified size-selected poly-A stranded cDNA library was submitted to the Penn Genome Frontiers Institute (PGFI, University of Pennsylvania, Philadelphia, PA) for sequencing. The Illumina Solexa GAII platform was used to obtain 50-bp single-end sequencing reads, which were analyzed using the v1.5 pipeline.
Computational Analysis
The reads were aligned to the mm9 genome using Bow tie [39]. Differential expression was analyzed using DESeq [40] in the R statistical environment (www.r-project.org/). Transcripts with a fold induction ≥ 2 and Benjamini-Hochbery adjusted p-value ≤ 0.05 were considered significant and included in downstream analysis. Gene ontology (GO) analysis [41] was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) online resource. Pathway analysis was also performed using DAVID [42], and enriched categories and pathways were further examined by qRT-PCR analysis. Genes involved in myelination, osteoclast function, oligodendrocyte differentiation, lipid/cholesterol transport and metabolism, and cytokine activity were queried from the GO database (http://www.geneontology.org/), and the resulting Mus musculus gene information from these lists were used to assess the expression of these genes in the MHV datasets. The data were deposited in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology under accession number GSE44333.
Results
MHV genome and mRNA, but not infectious virus, persist in the central nervous system (CNS) of infected B6 mice
MHV persistence is a well-documented phenomenon, but the extent to which viral transcription and translation occur at late time points (one month or later) post infection are not clear [1,13,43]. Here, we have employed a highly sensitive assay for detection of infectious virus in conjunction with targeted amplification of genomic and messenger RNA to investigate the presence of viral components in the spinal cord during chronic disease. Four-week-old C57Bl/6 (B6) mice were inoculated intracranially with MHV-A59 and infectious viral titers in several organs quantified. During acute infection, that is, the first week, mice exhibited characteristic hunching, ataxia, depression, ruffled fur, and weight loss.
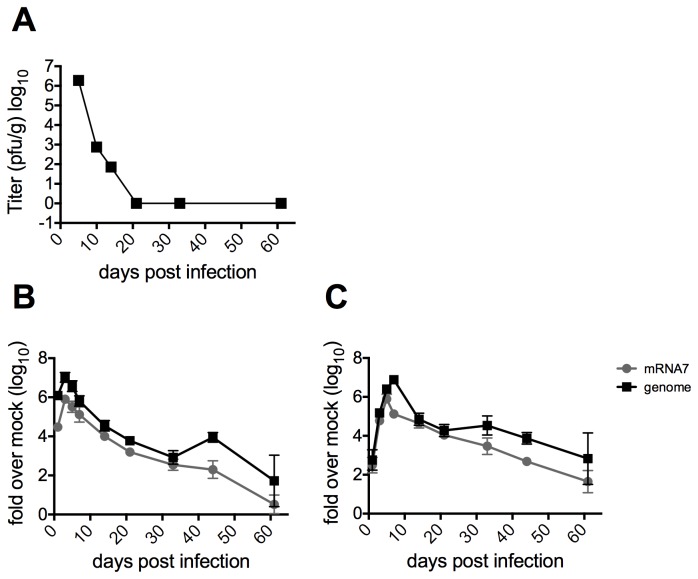
MHV titers peaked 5 days post infection (dpi), and by 10-14 dpi infectious virus was no longer detectable in any of the organs as assayed by standard plaque assay (Figure 1A , data not shown). By 10-14 dpi, the mice began regaining weight and resumed their normal grooming habits and activity levels. Although infectious virus and clinical signs were no longer detectable, viral genomic RNA and subgenomic mRNAs persisted in the brain (Figure 1B ) and, to higher levels, in the spinal cord (Figure 1C ). These viral mRNAs were detectable by reverse transcriptase-quantitative polymerase chain reaction (qRT-PCR) until at least 60 dpi, suggesting a low level of ongoing transcription. During the persistent phase of infection, some mice exhibited minor weight loss, ruffled fur and slight depression, while a few mice also experienced hindlimb paresis of several days’ duration. By 60 dpi, all mice had completely recovered to similar weights as uninfected animals and exhibited normal hindlimb movement. Luxol fast blue staining of spinal cord sections taken from mice at 33 dpi, approximately at the peak of demyelinating disease [1,2,5,26] showed characteristic demyelinating lesions (data not shown).
Figure 1. Replication of MHV in mice.
Four-week old B6 mice were infected intracranially with 2000 pfu of MHV-A59 and sacrificed at the indicated days post infection. A) Brains were harvested and placed in gelatin saline, homogenized and infectious virus titered on mouse L2 fibroblasts. Titers shown are averages from five mice per group. B) Brains and C) spinal cords were harvested and RNA isolated on Qiagen RNeasy columns. qRT-PCR was performed to quantify relative abundance of MHV-A59 genomic and subgenomic RNA mRNA7. Data are plotted as means with SEM of 5-8 mice.
To investigate whether very low levels of infectious virus (below the level of detection by plaque assay) are produced during the demyelinating phase of MHV infection, we used an in vivo bioassay to probe for infectious MHV in persistently infected tissues. Brain lysates from the above infected (donor) mice (taken on 5, 7, 14, 33, and 44 dpi) were injected into naïve four-week-old recipient mice, which were observed for clinical signs of infection (Table 1 ) and sacrificed 5 and 7 dpi, at the peak times of replication and encephalitis, respectively. The recipient mice that were inoculated with brain lysates of donor mice sacrificed 5 or 14 dpi, but not from donor mice sacrificed 33 or 44 dpi, exhibited clinical symptoms of MHV disease. Recipient mice that were inoculated with 5 dpi donor mouse extract experienced weight loss, ataxia, hunching, and ruffled fur, as well as visible necrosis in the liver on dpi 7. Mice inoculated with 14 dpi donor mouse extract experienced minimal weight loss and clinical signs but had detectable titers of infectious virus in both the brain and liver 5 and 7 dpi (Table 1 and data not shown).
Table 1. Virus persistence in mice.
| Day of isolation from donor mouse (brain)* | Day of isolation from recipient mouse (brain)** | Clinical symptoms*** | Cytopathic effect of brain extract on L2 cells (recipient mouse)**** |
|---|---|---|---|
| 5 | 5 | Weight loss, ataxia, hunching, ruffled fur | Yes |
| 7 | Weight loss, ataxia, hunching, ruffled fur, necrotic liver | Yes | |
| 14 | 5 | None, brain titer | Yes |
| 7 | Weight loss, brain titers | Yes | |
| 33 | 5 | No | No |
| 7 | No | No | |
| 44 | 5 | No | No |
| 7 | No | No |
* Four-week old mice were infected intracranially with 2000pfu of MHV-A59 and brains were harvested from donor mice on indicated day post infection.
** The cleared brain lysate from donor mice was injected into naïve four-week old recipient mice and brains were harvested from these mice on days 5 and 7 post infection.
*** Recipient mice were observed for clinical symptoms until they were sacrificed. The titers, if there were assayed, are indicated here.
**** Lysate from recipient mice was placed on L2 cells and cells were watched for cytopathic effects which include syncytia, cell death, and rounding.
Finally, the brains and livers harvested from recipient mice were assessed for infectious virus by treatment of L2 fibroblasts, which are sensitive to MHV infection, with these extracts. Cells were then assayed for viral nucleocapsid mRNA7 and genome by qRT-PCR as well as observed for cytopathic effects (Table 1 ). Again, only the extracts from recipient mice that had been inoculated with extract from 5 or 14 dpi donor mice produced any cytopathic effect or viral RNA in L2 cells. These data confirm that infectious virus is not present in the CNS, as it was not detectable even by a very sensitive bioassay, during MHV-induced demyelinating disease, while both genomic and subgenomic viral RNAs remained detectable.
MHV-induced demyelinating disease alters the transcriptome of host spinal cord tissue
To characterize the host response during the demyelinating phase of disease, total RNA was extracted from spinal cords of both mock- and MHV-infected B6 mice sacrificed 33 dpi. Poly-adenylated RNA was selected and the resulting library subjected to 50-bp Illumina Solexa GAII sequencing (Table S1 ). We used the DESeq package (http://www-huber.embl.de/users/anders/DESeq/) to analyze the changes in the host transcriptome during the demyelinating phase of MHV-A59 infection and found 909 transcripts for which expression was significantly altered (894 upregulated, 14 downregulated). Although 24,321 of the 27,731 annotated transcripts in the mouse genome (NCBI Build 37) were expressed at similar levels in mock-infected and MHV-infected animals (Figure S1 ), only 2,502 transcripts were not expressed at all in either mock-infected or MHV-infected spinal cord.
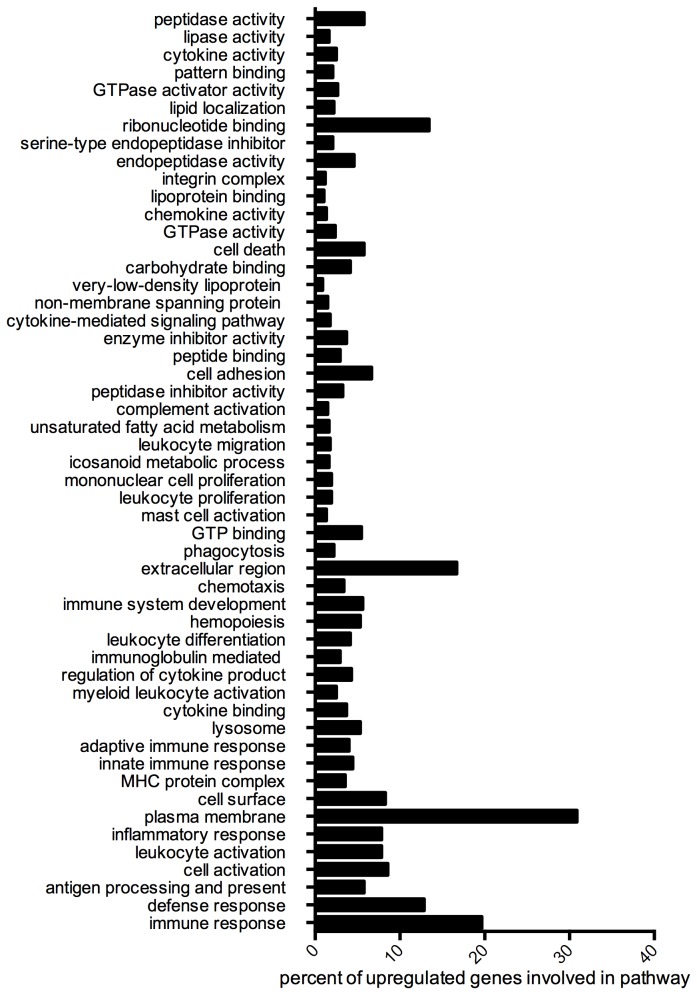
Next, we used the gene ontology (GO) database and the database for annotation, visualization and integrated discovery (DAVID) to characterize the 894 transcripts for which expression was upregulated in infected spinal cord (Table S2 ). Subsets of these genes were found to be involved in antigen presentation and processing, inflammation, defense, and immune responses, chemotaxis and cytokine signaling, complement, and lipid processing, transport, storage, and metabolism (Figure 2 and Table S3 ). A small number of these genes were expressed at undetectable levels in spinal cord from mock-infected mice but at significant levels in spinal cord from MHV-infected mice (65 genes, starred in Table S2 ). These genes encode proteins that perform immunological functions, including surface markers expressed exclusively on myeloid or lymphoid cells (24 genes), cytokines (5 genes), lysosomal components (3 genes), granzyme B, apolipoproteins C-II and C-IV, and a complement component (Cfb). Neither complement components nor active B/T-cells nor their products would be expected to be present in the spinal cord in naïve mice; therefore, these data illuminate the utility of this assay for detecting transcriptional changes reflective of infiltrating immune cells. The 50 most highly differentially expressed transcripts, which are representative of the functional categories in Figure 2 , are listed in Table 2 .
Figure 2. Gene Ontology Analysis.
The transcripts for which expression was significantly upregualted (894 transcripts) were analyzed using the functional annotation tool in DAVID and using only the molecular function, cellular component, and biological process terms in the gene ontology database. The most significant and non-redundant categories are represented here. The percentages of the 894 upregulated genes that are involved in each category are represented.
Table 2. Top 50 Differentially Expressed Genes**.
| NCBI Accession*** | Gene symbol | Fold Change (log2) | P-value |
|---|---|---|---|
| Antigen presentation | |||
| NM_010378 | H2-Aa | 8.091196867 | 5.24E-38 |
| NM_010545 | Cd74 | 7.559236329 | 1.23E-37 |
| NM_207105 | H2-Ab1 | 7.164195919 | 9.97E-33 |
| NM_010382 | H2-Eb1 | 6.078669952 | 7.39E-26 |
| NM_013532 | Lilrb4* | 7.153079999 | 5.53E-20 |
| NM_009857 | Cd8a* | 7.247316069 | 5.82E-18 |
| NM_010724 | Psmb8 | 4.256179268 | 6.92E-15 |
| Complement | |||
| NM_021334 | Itgax | 7.520522753 | 3.12E-27 |
| NM_009779 | C3ar1 | 5.279252321 | 2.59E-19 |
| NM_007572 | C1qa | 4.42367613 | 3.59E-17 |
| NM_009777 | C1qb | 4.147839729 | 1.56E-15 |
| NM_007574 | C1qc | 4.195323372 | 1.72E-15 |
| Associated with lymphocytes (T, B,NK) | |||
| NM_152839 | Igj | 7.28755371 | 1.55E-31 |
| NM_007654 | Cd72 | 6.36344867 | 5.95E-21 |
| NM_008534 | Ly9 | 6.437381518 | 5.53E-20 |
| NM_144539 | Slamf7 | 6.979726259 | 1.10E-19 |
| NM_013706 | Cd52 | 5.404143569 | 1.87E-19 |
| NM_009850 | Cd3g | 6.89889651 | 4.92E-18 |
| NM_009857 | Cd8a* | 7.247316069 | 5.82E-18 |
| NM_013489 | Cd84 | 4.961200073 | 2.36E-17 |
| NM_008147 | Gp49a* | 8.808431827 | 6.92E-15 |
| NM_009845 | Cd22 | 5.403189838 | 6.92E-15 |
| Cytokines and receptors | |||
| NM_008599 | Cxcl9 | 8.457703361 | 7.22E-26 |
| NM_013653 | Ccl5 | 8.138965713 | 3.68E-23 |
| Lipid transport and metabolism | |||
| NM_009853 | Cd68* | 5.132714539 | 1.32E-18 |
| NM_009695 | Apoc2 | 5.960434921 | 1.48E-17 |
| NM_007385 | Apoc4 | 6.306136984 | 3.17E-16 |
| NM_031195 | Msr1* | 5.445227457 | 1.24E-15 |
| Associated with myeloid cells (macrophages, DCs): | |||
| NM_053110 | Gpnmb* | 8.23606446 | 6.56E-42 |
| NM_009977 | Cst7* | 7.995478697 | 5.88E-36 |
| NM_027836 | Ms4a7 | 7.415825374 | 2.26E-26 |
| NM_010705 | Lgals3* | 5.361533229 | 7.20E-22 |
| NM_010821 | Mpeg1 | 4.794347075 | 2.26E-19 |
| NM_011662 | Tyrobp* | 4.812736142 | 4.20E-19 |
| NM_009853 | Cd68* | 5.132714539 | 1.32E-18 |
| NM_031254 | Trem2* | 4.734450912 | 1.35E-18 |
| NM_145634 | Cd300lf* | 5.577795412 | 3.07E-17 |
| NM_009690 | Cd5l | 7.137739453 | 3.80E-17 |
| NM_031195 | Msr1* | 5.445227457 | 1.24E-15 |
| NM_011426 | Siglec1 | 6.552776952 | 2.08E-15 |
| NM_008147 | Gp49a* | 8.808431827 | 6.92E-15 |
| Pathogen destruction | |||
| NM_017372 | Lyz2 | 7.261341918 | 4.11E-35 |
| NM_013590 | Lyz1 | 7.022574167 | 1.51E-32 |
| NM_020008 | Clec7a | 6.380384706 | 4.54E-26 |
| NM_007807 | Cybb | 6.264961456 | 1.38E-25 |
| NM_010705 | Lgals3* | 5.361533229 | 7.20E-22 |
| NM_021792 | Iigp1 | 5.022735652 | 2.36E-17 |
| Osteoclast function | |||
| NM_053110 | Gpnmb* | 8.23606446 | 6.56E-42 |
| NM_175406 | Atp6v0d2 | 8.109994462 | 4.53E-25 |
| NM_016873 | Wisp2 | 6.415273428 | 2.94E-22 |
| NM_013532 | Lilrb4* | 7.153079999 | 5.53E-20 |
| NM_011662 | Tyrobp* | 4.812736142 | 4.20E-19 |
| NM_031254 | Trem2* | 4.734450912 | 1.35E-18 |
| NM_007388 | Acp5(TRAP) | 6.54687635 | 4.82E-18 |
| NM_145634 | Cd300lf* | 5.577795412 | 3.07E-17 |
| Other | |||
| NM_009977 | Cst7 | 7.995478697 | 5.88E-36 |
| NM_023044 | Slc15a3 | 5.351815911 | 2.18E-19 |
| NM_019984 | Tgm1 | 6.482855955 | 3.05E-18 |
| Unknown function | |||
| NM_028595 | Ms4a6c | 5.706251433 | 2.36E-17 |
| NM_001033767 | Gm4951 | 5.673905156 | 3.80E-17 |
| NM_001081957 | Gm11428 | 6.094571593 | 6.44E-16 |
* These genes are represented in more than one category
** Alternatively spliced isoforms were collapsed into one gene category. The top 50 differentially expressed genes as determined by DESeq are represented. Fold change values are log2 transformed.
*** The common gene symbol and the associated NCBI accession number are both listed.
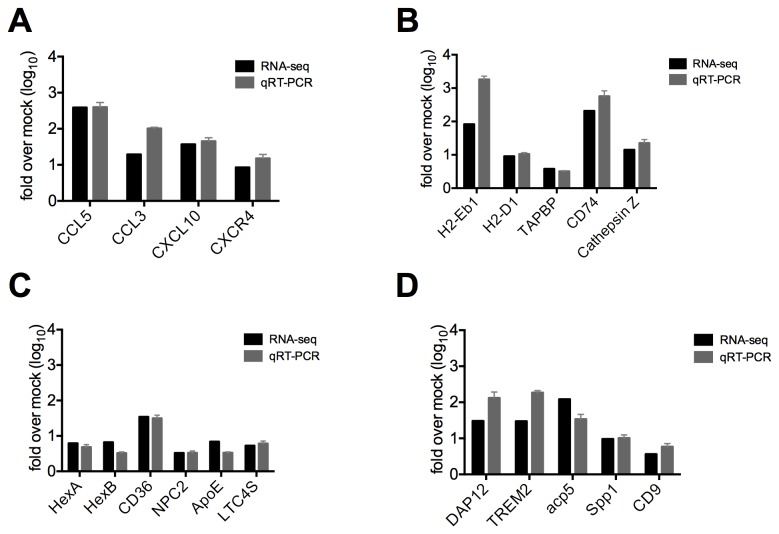
The RNA-seq results were validated by performing qRT-PCR on several transcripts for which expression was significantly increased in infected tissue and all transcripts assayed exhibited similar fold changes in expression between the two techniques (Figure S2 ). Additionally, total RNA was extracted from the spinal cords of a second set of infected mice and the relative expression of several genes in four categories of genes that were upregulated during demyelination (described below) were validated by qRT-PCR. Thus the RNA-seq data were validated technically by qRT-PCR and biologically by comparison with RNA from an independent set of infected mice (Figure 3 ).
Figure 3. Biological validation of genes of interest.
The expression of genes in several categories that were upregulated during MHV induced chronic disease A) Th1 chemokines; B) antigen presentation; C) lipid metabolism and neurodegenerative disease and D) osteoclast differentiation, maturation, and function was quantified by qRT-PCR analysis of spinal cord derived RNA from an independent set of infected mice. Oligonucleotide primers and qRT-PCR were used to detect expression of 4-5 genes in each category. Fold changes of mRNA transcripts in infected mice were determined relative to mock infected and compared to mRNA fold change levels determined in the RNA-seq experiment. All genes analyzed had significantly increased expression in infected mice as compared to mock infected, similar to observations in the RNA-seq analysis. qRT-PCR data plotted are means with SEM of 5 mice.
Th1 cytokine and chemokine gene expression is upregulated during MHV-chronic CNS disease
The expression levels of genes encoding several cytokines/chemokines, as well as their receptors, were upregulated in the spinal cords of infected mice harvested 33 dpi (Table S4 ). Twenty-six of these genes encode cytokine or chemokine activities, 29 are involved in cytokine production or regulation, and many others encode proteins involved in cytokine binding or signaling pathways (Figure 2 and Table S3 ). The genes encoding two Th1 chemokines, CXCL9 and CCL5, were among the top fifty most highly upregulated genes (Table 2 ). Increased expression levels of four Th1 chemokines were validated by qRT-PCR in biological replicates from mice persistently infected with A59 (Figure 3A ).
Genes involved in antigen presentation pathways have increased expression during MHV-chronic CNS disease
GO analysis also indicated that expression of 34 genes involved in antigen processing and presentation was upregulated in spinal cord during the persistent phase of MHV infection (Figure 2 and Table S3 ). Twenty of these genes encode major histocompatibility complex alleles, including eight MHC II and 12 MHC I alleles, 14 of which are non-classical (Table 2 , Tables S3 and S5 ). In addition to MHC alleles, CD74, the gamma chain of the MHC II protein, and CD8α, the MHC I receptor [44], are also highly overrepresented in the spinal cords of persistently infected animals (Table 2 , Figure 3B and Table S5 ). The increased expression levels of MHC alleles H2-Eb1 and H2-D1, CD74 (a transporter associated with antigen processing binding protein (TAPBP) [45,46]), and cathepsin Z (a lysosomal cysteine protease potentially involved in the degradation of viral antigens [47]), were confirmed by qRT-PCR using the spinal cord of MHV-infected animals in a second experiment (Figure 3B ).
Genes associated with lipid metabolism and transport have increased expression during MHV chronic CNS disease
We found that MHV infection induced major changes in the transcription of genes involved in many facets of lipid metabolism. In fact, expression levels of 22 genes whose protein products are components of chylomicrons, low-density lipoprotein, or very-low-density lipoprotein particles were increased in the demyelinating spinal cord. Interestingly, expression levels of 12 genes involved in lipid transport, including apolipoproteins E, C1, C2, and C4, were also upregulated (Table 2 , Tables S3 and S6 ). Eleven other genes for which expression was enhanced in MHV-infected spinal cord encode proteins with lipase or phospholipase activity (Figure 2 , Table 2 , Tables S3 and S6 ). For example, CD36 [48], CD68 [49], MSR1, and four other genes (Table S6 ) encode macrophage scavenger receptors responsible for phagocytosing macromolecules, cellular debris, and/or oxidized lipid-protein particles.
Eicosanoids, lipid-derived mediators of inflammation that are classified as leukotrienes or prostanoids, are synthesized in response to a variety of stimuli. Several eicosanoid genes in either the prostaglandin or leukotriene pathways showed increased expression in infected CNS tissue (Table S6 ). The expression levels of ten other genes either upstream of or involved in eicosanoid metabolism were upregulated in the MHV infected CNS (Figure 2 , Tables S3 and S6 ). Additionally, a subset of lipid processing genes that have increased expression in infected tissue include several genes known to be involved in human neurodegenerative diseases (Figure 3C ). Mutations in hexaminidase A, hexaminidase B, Niemann-Pick type C2, and ATP-binding cassette transporter A1 (ABCA1) cause Tay-Sachs [50], Sandhoff disease [51], Niemann-Pick type C2 (NPC2) [52], and Tangier disease [53], respectively.
Differential expression of oligodendrocyte-associated genes is not associated with MHV- chronic CNS disease
Myelin- and oligodendrocyte-associated genes were not among those expressed at higher levels in MHV-A59 infected versus mock-infected spinal cord (Figure S3 and Table S7 ). More specifically, genes encoding proteo-lipid protein 1 (PLP1), MBP, myelin oligodendrocyte protein (MOG), myelin-associated protein (MAG), and other structural protein components of the myelin sheath were expressed at similar levels in mock and infected tissue. Similarly, Olig1, 2, and 3 and Sox 8, 9, and 10, which are transcription factors associated with oligodendrocyte differentiation [54-60] (Figure S3 and Table S7 ), were not more highly expressed in spinal cords of infected mice.
Increased expression of genes with roles in osteoclast development and function, including TREM2 and DAP12 is associated with MHV chronic CNS disease
Microglia are CNS-resident macrophages and have specialized functions for maintaining homeostasis in their specific environment. Unsurprisingly, 36 genes involved in the differentiation, maturation, and function of myeloid-lineage cells showed increased expression in the 33-dpi MHV-infected spinal cord (Table 2 , Figure 2 , Tables S2 and S3 ). These genes are involved in scavenging (CD68, MSR1), signaling (Ms4a7, Tyrobp, TREM2, Gp49a, and AIM [61-66]), pathogen destruction (Lgals3 [67]), and monocyte differentiation (Tyrobp, TREM2, Mpeg1, and gpnmb [68-73]). Two of the genes for which expression was most highly induced during MHV persistence were TREM2 (Triggering receptor expressed on myeloid cells 2) and DAP12 (DNAX-activating protein of 12kDa), both of which are involved in myeloid differentiation and function (Table 2 ). In humans, congenital natural polymorphisms that cause loss of function mutations in either TREM2 or DAP12 result in Nasu-Hakola disease, which is characterized by late-onset demyelination in the CNS and osteopetrosis [27-30]. Interestingly, several other myeloid-specific genes in addition to TREM2 and DAP12 that exhibited increased expression levels in demyelinating spinal cord are also important for osteoclast differentiation and function (Figure 3D and Table S8 ). Indeed increased expression of tartrate-resistant acid phosphatase (Acp5), bone sialoprotein (spp1), osteopontin and oncostatin M receptor was associated with chronic MHV induced CNS disease (Table S4 ).
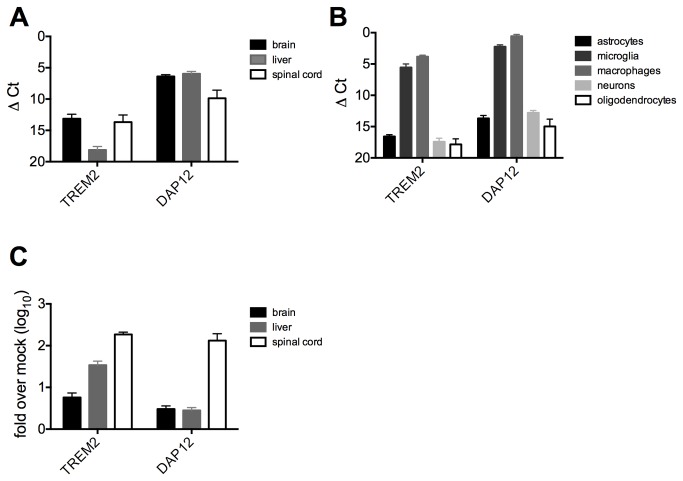
Because natural mutations in TREM2 and DAP12 lead to demyelinating disease in humans and these genes are associated with demyelination in this model, we chose to study the expression of TREM2 and DAP12 mRNA expression levels in the brain, spinal cord, and liver in 33-dpi infected and mock-infected animals (Figure 4A ). In the uninfected mouse, TREM2 is expressed at high levels in the brain and spinal cord, but at minimal levels in the liver, while DAP12 shows higher and more ubiquitous expression in all organs assayed. We also assessed the basal expression levels of TREM2 and DAP12 in CNS cell types cultured in vitro (Figure 4B ). DAP12 was expressed in all cell types, with much higher expression in myeloid cells, whereas TREM2 was expressed solely in myeloid cells, with astrocytes, oligodendrocytes, and neurons showing minimal to no detectable expression of this gene (Figure 4B ). In the 33-dpi MHV-infected animals, TREM2 expression was upregulated strongly in the spinal cord and to lesser extents in the brain and liver (Figure 4C ). Infection increased DAP12 expression significantly only in the infected spinal cord and not in the brain or liver (Figure 4C ).
Figure 4. TREM2/DAP12 mRNA expression.
A) Expression of TREM2/DAP12 mRNA was determined by qRT-PCR in RNA from spinal cord, brain, and liver of mock infected mice (n=3-5 mice). B) Expression of basal levels of TREM2/DAP12 mRNA was determined by qRT-PCR in RNA from primary cell cultures (oligodendrocytes, n= 2; macrophages, n= 4; neurons, n= 6; astrocytes and microglia, n=5). C) Virus induced levels of TREM2/DAP12 mRNA expression were determined by qRT-PCR in RNA from spinal cord, brain, and liver of infected mice, 33 dpi. Data plotted are means with SEM.
Discussion and Conclusions
Other and we have reported that MHV genome RNA persists in the CNS for the lifetime of the mouse despite the inability to detect infectious virus during MHV chronic disease [2,74,75]. A previous study comparing persisting and non-persisting strains of MHV indicates that virus-specific CD8T-cells persist only when viral RNA is present and that viral persistence also correlates with demyelination [76]. Therefore, although infectious virus does not appear to persist in the CNS of MHV-infected mice, the viral products that do persist may be the viral mediators of the demyelinating pathology. Here, we report that even by using an extremely sensitive in vivo bioassay, persistently infected CNS tissue is unable to transfer infectious virus to naïve hosts. We furthermore demonstrated that viral mRNA as well as genomic RNA persists suggesting viral transcription is occurring and perhaps mRNA is translated into protein that is responsible for maintenance of T cells during persistence as well as the demyelinating pathology. The nature of persistent MHV mRNA is not yet known and requires additional studies, now underway, to characterize the viral sequences, including their mutation rate(s), the presence of six subgenomic mRNAs, leader-junction diversity in subgenomic species, and relative abundance of negative-sense RNA species.
Many different biological activities are hypothesized to lead to demyelination and neurodegeneration in the CNS. Activated lymphocytes and inflammation-mediated responses may directly or indirectly induce death in oligodendrocytes, thus interfering with maintenance of the myelin sheath. The role of oligodendrocytes, the cells that specialize in myelination, in supporting MHV persistence and demyelinating disease has been disputed. Previous studies have shown that MHV persists selectively in oligodendrocytes [11,13,14,77-82]. An early study reported that MHV antigens were present in the oligodendrocytes of demyelinating lesions four weeks post infection [4]. Some studies show that oligodendrocytes are directly damaged during MHV infection but it has also been shown that oligodendrocytes in the MHV infected brain are not apoptotic, and MHV-induced demyelination correlates with inflammation rather than direct damage to oligodendrocytes [14,83,84]. In contrast to MHV infection, immunization against myelin basic protein (MBP) results in direct damage to oligodendrocytes and EAE [85-87] and leads to reduced expression of many oligodendrocyte or myelin-specific genes in the CNS [88,89]. Here we report no changes in expression of oligodendrocyte specific, or myelination-relevant genes during MHV-induced demyelinating disease supporting the hypothesis that oligodendrocytes are not specifically damaged during MHV persistence. It is important to note here that remyelination also occurs during MHV induced chronic disease and that while not maximal until ten weeks post infection, remyelination-associated genes may be upregulated at 33 dpi. However, our data suggests remyelinating processes at this time post infection do not appear to involve increased expression of oligodendrocyte specific genes.
Activated lymphocytes and inflammatory responses can also be damaging to neurons and other cells of the CNS and may contribute to the immune-mediated pathology observed during demyelination. We report here that Th1 chemokines (CCL5, CCL3, CXCL10, CXCR3, CXCR4) have augmented expression during MHV-induced demyelinating disease. CCL5 and CCL3 attract T-cells and other leukocytes to sites of inflammation [90-93], while CXCR4 is a T-cell receptor that promotes chemotaxis to sites of inflammation [94]. CXCL10 (IP-10) and its receptor CXCR3 recruit many immune cells to inflammatory sites and have been previously shown to influence MHV-induced demyelination [17]. Additionally, CXCL10 has been extensively studied in two models of multiple sclerosis, MHV, and experimentally induced autoimmune encephalitis (EAE). Additionally, antibody-mediated blockade of this cytokine has ameliorated disease in both models [17,95]. The combination of increased expression of several canonical Th1 cytokines/chemokines and absent expression of the canonical Th2 cytokines/chemokines [96-101] supports the hypothesis that the host is continuing to mount a cell-mediated immune response to the virus, as previously suggested [102].
Antigen presenting cells activate and recruit lymphocytes to sites of infection and this pathway remains upregulated during MHV-induced demyelinating disease. Several non-classical MHC alleles have augmented expression as well and the roles of these alleles in persistent infection, CNS infection, and demyelination are completely unknown. The MHC II locus is a confirmed multiple sclerosis susceptibility locus in humans [103], and it is possible that certain MHC alleles may also be important for the establishment of MHV-induced demyelination. Although infectious virus does not persist in the CNS, the CD8T-cell response to viral antigen is known to continue in the CNS through this time period [16,76,104,105], and CD8T-cell exhaustion has been proposed to contribute to MHV persistence [106]. Indeed, the sustained activation of lymphocytes, which is associated with sustained antigen presentation, is established not just for MHV-induced demyelination [1,43,107-112], but also for EAE [87,113,114] and multiple sclerosis [115-117], lending evidence to the theory that demyelination is an immunopathological process possibly triggered by diverse biological activities.
The importance of lipid metabolism in general in myelination and demyelination is undisputed. As lipid is a major component of the myelin sheath, changes in lipid metabolism may influence the composition, degradation, or regeneration of myelin in the CNS. Thus, the set of genes related to lipid transport, differentially expressed during MHV-induced chronic disease may be important not only for the formation of the myelin sheath but also for remyelination during MHV infection or in diseases such as MS. Indeed by 33 dpi in the MHV model, while demyelination is the predominant pathology; the remyelination process has likely begun [6]. Thus, increased expression of many lipid processing genes during MHV-induced chronic CNS disease likely represents both processes.
Several genes associated with neurodegenerative disease in humans have highly increased expression during MHV-induced demyelinating disease (HexA, HexB, NPC2, ABCA1, apoE, CD36) [50]. Patients with genetic disorders affecting these genes have greatly reduced amounts of myelination. Furthermore, a variant of apolipoprotein E, apoEε4, is a genetic risk factor for Alzheimer’s disease [118,119], and focal demyelination has been observed near amyloid-beta plaques in both humans and mice [120]. CD36 has been shown to be involved in peripheral nerve remyelination after injury [121]. The expression levels of both CD36 and ABCA1 are also upregulated in demyelinating spinal cord tissue from mice with EAE [88]. In addition, CD36 [122] and apoE [123,124] have recently been associated with the active phase of relapsing-remitting MS.
Additionally, genes involved in the eicosanoid production pathway are also upregulated in the demyelinating CNS. Our data indicate that upregulation of expression of eicosanoid-derived inflammatory mediators may be involved in demyelinating disease, although whether these eicosanoids contribute to or limit the pathology remains undetermined. Regardless, the modulation of eicosanoid-mediated inflammation in demyelinating CNS tissue is under active investigation [125].
Macrophages and microglia promote viral clearance and clear cellular debris; therefore, changes in these cells, as well as in oligodendrocytes, may promote myelin loss and/or alter the host ability to remyelinate during acute infection. Macrophages/microglia have been clearly associated with the pathology of MHV induced demyelinating disease [14,126,127]. A group of myeloid-specific genes that have proven importance for osteoclast function or bone maintenance was also upregulated during MHV-induced demyelination (Table S4 ). Two of the most highly regulated genes were DAP12 and TREM2. DAP12 is a signaling adapter that associates with multiple receptors in different cell types [128] and signals through an ITAM motif [62]. TREM2 signals exclusively through DAP12 and is expressed on myeloid cells in response to unknown ligands [129,130]. Polymorphisms in either of these genes found in individuals with, Nasu-Hakola disease cause demyelination and osteopetrosis, presumably resulting from defects in microglia and osteoclasts respectively [27-30]. DAP-12 deficient mice also exhibit a similar phenotype [131]. DAP12 expression is also associated with NKG2D T-cells in the CNS, which have been demonstrated to contribute to MHV-induced demyelination [132]. Other myeloid-specific genes that were upregulated during MHV induced chronic disease are also important for osteoclast differentiation and function, for example Acp5, Spp1, osteopontin, oncostatinM. Mice deficient in Acp5 exhibit osteopetrosis [133,134]. Bone sialoprotein (Spp1) anchors osteoclasts to bone and is a major component of the bone extracellular matrix [135], while osteopontin has been examined as a modulator of demyelination due to its increased expression in EAE tissue and cerebrospinal fluid from multiple sclerosis patients [136,137]. Lastly, the oncostatin M receptor is also highly expressed in MHV-infected and EAE- and MS-affected tissues [88,138]. Interestingly, infusion of oncostatin M into mice with chemically induced demyelination also regulated the expression of several genes involved in the differentiation of oligodendrocyte precursor cells into mature myelinating cells [139].
Previous studies have confirmed the expression of TREM2 in human microglia, with conflicting results concerning the expression of TREM2 in neurons or oligodendrocytes [140]. Here we show that TREM2 expression is most highly expressed in microglia and macrophages, while DAP12 expression was detected in neurons and astrocytes as well. Interestingly, these genes are more highly induced at 33 dpi in the spinal cord as compared to the brain. This is consistent with studies suggesting that MHV persistence ( [141] and Figure 1) as well as demyelination [142] is to a greater is extent in the spinal cord than in the brain.
Taken together, our findings suggest that osteoclasts and microglia may have several parallel functions. It is known that osteoclasts reabsorb bone (which is created by osteoblasts) using superoxides to destroy the bone matrix and specialized forms of phagocytosis [143,144]. Regulation of osteoclast and osteoblast function is critical for bone homeostasis, and a lack of osteoclast activity caused by mutations in any of several genes leads to osteopetrosis or bone cysts [144,145]. Similarly, microglia, along with inflammatory macrophages, may reabsorb damaged myelin and thus allow oligodendrocytes to remyelinate. Osteoclasts have been reported to present antigen, activate T-cells, and secrete cytokines in vitro [146], as microglia have been shown to do in the CNS [147]. Osteoclasts also respond to multiple cytokines that regulate their differentiation and bone-resorptive activity [148], while microglia and inflammatory macrophages are activated by cytokines to scavenge myelin debris [126]. The parallels between osteoclastic and microglial functions in bone remodeling of skeletal tissue and myelin remodeling of the CNS may currently be underappreciated. In support of this model, our findings here indicate a unique role for a subset of bone remodeling proteins in the remyelinating CNS during persistent MHV infection, perhaps by specific expression in activated microglia.
Finally, this study provides the first transcriptome-wide description of gene expression in the spinal cord of mice with MHV-induced demyelination. The data described here provide a basis on which to investigate these pathways in more detail. This includes understanding the pathways contributing to demyelination and/or perhaps remyelination, defining the cell types in which the expression of each of these classes of genes is induced as well as the times post-infection at which each group of genes is upregulated. Further studies will also focus on assessing the influences of translational and post-translational mechanisms that likely constitute another level of the host response to chronic MHV infection.
Supporting Information
Expression of RefSeq transcripts. Expression of each transcript was determined using DESeq in the R statistical background. The current RefSeq database (26,823 transcripts) for Mus musculus was divided into four categories: significantly increased expression (log2 fold change over mock ≥ and p < 0.05), significantly decreased expression (log2 fold change over mock ≤ 1 and p < 0.05), similar expression (-1< log2 fold change over mock < 1), and no expression (neither mock nor MHV-A59 infected had any reads to the transcript).
(TIFF)
RNA-seq technical validation. The expre≤ssion of nine transcripts was analyzed by RNA-seq and qRT-PCR using the same spinal cord RNAs (experiment 1) and by qRT-PCR of spinal cord RNAs from an independent mouse infection experiment (experiment 2). In each experiment, the samples were obtained from spinal cords of both infected and mock infected mice (3-5 mice), sacrificed at 33dpi. All nine transcripts were significantly induced using both techniques of measurement and in RNA samples from both experiments. T-tests were performed to assess significant induction in the qRT-PCR analysis. Data plotted are means with SEM.
(TIFF)
Oligodendrocyte-specific gene expression. As described in the legend to Figure 3, qRT-PCR primers were used to detect the level of expression of oligodendrocyte specific gene transcripts in spinal cords recovered from mock or MHV-A59 infected mice (33dpi). None of the genes had significantly altered expression in infected mice compared to mock. Data shown are means with SEM.
(TIFF)
RNA-seq library statistics.
(XLSX)
Genes whose expression was increased in the MHV-infected spinal cord.
(XLSX)
Gene ontology analysis of 894 upregulated genes.
(XLSX)
Cytokines and cytokine receptors.
(XLSX)
Antigen Presentation.
(XLSX)
Lipid and cholesterol transport and metabolism.
(XLSX)
Oligodendrocyte markers, differentiation and function.
(XLSX)
Osteoclast and ossification.
(XLSX)
Acknowledgments
We thank Dr. Dennis Kolson for discussion and reading the manuscript.
Funding Statement
This study was supported by grant RG 3843A6/1 from the National Multiple Sclerosis Society and National Institutes of Health (NIH) grant RO1-NS-054695. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Matthews AE, Weiss SR, Paterson Y (2002) Murine hepatitis virus--a model for virus-induced CNS demyelination. J Neurovirol 8: 76-85. doi:10.1080/13550280290049534. PubMed: 11935460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matthews AE, Lavi E, Weiss SR, Paterson Y (2002) Neither B cells nor T cells are required for CNS demyelination in mice persistently infected with MHV-A59. J Neurovirol 8: 257-264. doi:10.1080/13550280290049697. PubMed: 12053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lavi E, Gilden DH, Highkin MK, Weiss SR (1984) MHV-A59 pathogenesis in mice. Adv Exp Med Biol 173: 237-245. doi:10.1007/978-1-4615-9373-7_24. PubMed: 6331114. [DOI] [PubMed] [Google Scholar]
- 4. Woyciechowska JL, Trapp BD, Patrick DH, Shekarchi IC, Leinikki PO et al. (1984) Acute and subacute demyelination induced by mouse hepatitis virus strain A59 in C3H mice. J Exp Pathol 1: 295-306. PubMed: 6100984. [PubMed] [Google Scholar]
- 5. Lavi E, Gilden DH, Wroblewska Z, Rorke LB, Weiss SR (1984) Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology 34: 597-603. doi:10.1212/WNL.34.5.597. PubMed: 6324031. [DOI] [PubMed] [Google Scholar]
- 6. Kristensson K, Holmes KV, Duchala CS, Zeller NK, Lazzarini RA et al. (1986) Increased levels of myelin basic protein transcripts in virus-induced demyelination. Nature 322: 544-547. doi:10.1038/322544a0. PubMed: 2426599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Das Sarma J, Iacono K, Gard L, Marek R, Kenyon LC et al. (2008) Demyelinating and nondemyelinating strains of mouse hepatitis virus differ in their neural cell tropism. J Virol 82: 5519-5526. doi:10.1128/JVI.01488-07. PubMed: 18385249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu GF, Dandekar AA, Pewe L, Perlman S (2000) CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J Immunol 165: 2278-2286. PubMed: 10925317. [DOI] [PubMed] [Google Scholar]
- 9. Wu GF, Dandekar AA, Pewe L, Perlman S (2001) The role of CD4 and CD8 T cells in MHV-JHM-induced demyelination. Adv Exp Med Biol 494: 341-347. doi:10.1007/978-1-4615-1325-4_51. PubMed: 11774490. [DOI] [PubMed] [Google Scholar]
- 10. Bergmann CC, Lane TE, Stohlman SA (2006) Coronavirus infection of the central nervous system: host-virus stand-off. Nat Rev Microbiol 4: 121-132. doi:10.1038/nrmicro1343. PubMed: 16415928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knobler RL, Lampert PW, Oldstone MB (1982) Virus persistence and recurring demyelination produced by a temperature-sensitive mutant of MHV-4. Nature 298: 279-280. doi:10.1038/298279a0. PubMed: 6283382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dandekar AA, Wu GF, Pewe L, Perlman S (2001) Axonal damage is T cell mediated and occurs concomitantly with demyelination in mice infected with a neurotropic coronavirus. J Virol 75: 6115-6120. doi:10.1128/JVI.75.13.6115-6120.2001. PubMed: 11390613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lampert PW, Sims JK, Kniazeff AJ (1973) Mechanism of demyelination in JHM virus encephalomyelitis. Acta Neurophathol 24: 76-85. doi:10.1007/BF00691421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu GF, Perlman S (1999) Macrophage infiltration, but not apoptosis, is correlated with immune-mediated demyelination following murine infection with a neurotropic coronavirus. J Virol 73: 8771-8780. PubMed: 10482631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bergmann CC, Dimacali E, Stohl S, Marten N, Lai MM et al. (1998) Viral evolution and CTL epitope stability during JHMV infection in the central nervous system. Adv Exp Med Biol 440: 751-757. PubMed: 9782354. [DOI] [PubMed] [Google Scholar]
- 16. MacNamara KC, Chua MM, Nelson PT, Shen H, Weiss SR (2005) Increased epitope-specific CD8+ T cells prevent murine coronavirus spread to the spinal cord and subsequent demyelination. J Virol 79: 3370-3381. doi:10.1128/JVI.79.6.3370-3381.2005. PubMed: 15731231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsunoda I, Lane TE, Blackett J, Fujinami RS (2004) Distinct roles for IP-10/CXCL10 in three animal models, Theiler’s virus infection, EAE, and MHV infection, for multiple sclerosis: implication of differing roles for IP-10. Mult Scler 10: 26-34. doi:10.1191/1352458504ms982oa. PubMed: 14760949. [DOI] [PubMed] [Google Scholar]
- 18. Lane TE, Liu MT, Chen BP, Asensio VC, Samawi RM et al. (2000) A central role for CD4(+) T cells and RANTES in virus-induced central nervous system inflammation and demyelination. J Virol 74: 1415-1424. doi:10.1128/JVI.74.3.1415-1424.2000. PubMed: 10627552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stohlman SA, Yao Q, Bergmann CC, Tahara SM, Kyuwa S et al. (1995) Transcription and translation of proinflammatory cytokines following JHMV infection. Adv Exp Med Biol 380: 173-178. doi:10.1007/978-1-4615-1899-0_28. PubMed: 8830475. [DOI] [PubMed] [Google Scholar]
- 20. Parra B, Bergmann CC, Hinton DR, Atkinson R, Stohlman SA (2001) IFN-gamma secreted by virus-specific CD8+ T cells contribute to CNS viral clearance. Adv Exp Med Biol 494: 335-340. doi:10.1007/978-1-4615-1325-4_50. PubMed: 11774489. [DOI] [PubMed] [Google Scholar]
- 21. Scott EP, Branigan PJ, Del Vecchio, Weiss SR (2008) Chemokine expression during mouse hepatitis virus-induced encephalitis: Contributions of the spike and background gene. J Neurovirol 14(1):5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adibhatla RM, Hatcher JF (2008) Altered Lipid Metabolism in Brain Injury and Disorders. Subcell Biochem 49: 241-268. doi:10.1007/978-1-4020-8831-5_9. PubMed: 18751914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ledeen RW (1984) Lipid-metabolizing enzymes of myelin and their relation to the axon. J Lipid Res 25: 1548-1554. PubMed: 6397559. [PubMed] [Google Scholar]
- 24. Chrast R, Saher G, Nave KA, Verheijen MH (2011) Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res 52: 419-434. doi:10.1194/jlr.R009761. PubMed: 21062955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adibhatla RM, Hatcher JF (2007) Role of Lipids in Brain Injury and Diseases. Future Lipidol 2: 403-422. doi:10.2217/17460875.2.4.403. PubMed: 18176634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sutherland RM, Chua MM, Lavi E, Weiss SR, Paterson Y (1997) CD4+ and CD8+ T cells are not major effectors of mouse hepatitis virus A59-induced demyelinating disease. J Neurovirol 3: 225-228. doi:10.3109/13550289709018297. PubMed: 9200070. [DOI] [PubMed] [Google Scholar]
- 27. Paloneva J, Kestilä M, Wu J, Salminen A, Böhling T et al. (2000) Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 25: 357-361. doi:10.1038/77153. PubMed: 10888890. [DOI] [PubMed] [Google Scholar]
- 28. Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J et al. (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71: 656-662. doi:10.1086/342259. PubMed: 12080485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cella M, Buonsanti C, Strader C, Kondo T, Salmaggi A et al. (2003) Impaired differentiation of osteoclasts in TREM-2-deficient individuals. J Exp Med 198: 645-651. doi:10.1084/jem.20022220. PubMed: 12913093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nataf S, Anginot A, Vuaillat C, Malaval L, Fodil N et al. (2005) Brain and bone damage in KARAP/DAP12 loss-of-function mice correlate with alterations in microglia and osteoclast lineages. Am J Pathol 166: 275-286. doi:10.1016/S0002-9440(10)62251-1. PubMed: 15632019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gombold JL, Hingley ST, Weiss SR (1993) Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J Virol 67: 4504-4512. PubMed: 8392595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caamaño J, Alexander J, Craig L, Bravo R, Hunter CA (1999) The NF-kappa B family member RelB is required for innate and adaptive immunity to Toxoplasma gondii. J Immunol 163: 4453-4461. PubMed: 10510387. [PubMed] [Google Scholar]
- 33. Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR (2011) Cell type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rall GF, Mucke L, Oldstone MB (1995) Consequences of cytotoxic T lymphocyte interaction with major histocompatibility complex class I-expressing neurons in vivo. J Exp Med 182: 1201-1212. doi:10.1084/jem.182.5.1201. PubMed: 7595191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bender SJ, Phillips JM, Scott EP, Weiss SR (2010) Murine coronavirus receptors are differentially expressed in the central nervous system and play virus strain-dependent roles in neuronal spread. J Virol 84: 11030-11044. doi:10.1128/JVI.02688-09. PubMed: 20739537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roth-Cross JK, Martínez-Sobrido L, Scott EP, García-Sastre A, Weiss SR (2007) Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81: 7189-7199. doi:10.1128/JVI.00013-07. PubMed: 17459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feigenson K, Reid M, See J, Crenshaw EB 3rd, Grinspan JB (2009) Wnt signaling is sufficient to perturb oligodendrocyte maturation. Mol Cell Neurosci 42: 255-265. doi:10.1016/j.mcn.2009.07.010. PubMed: 19619658. [DOI] [PubMed] [Google Scholar]
- 38. Rose KM, Elliott R, Martínez-Sobrido L, García-Sastre A, Weiss SR (2010) Murine Coronavirus Delays Expression of a Subset of Interferon-Stimulated Genes. J Virol 84: 5656-5669. doi:10.1128/JVI.00211-10. PubMed: 20357099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. doi:10.1186/gb-2009-10-3-r25. PubMed: 19261174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. doi:10.1186/gb-2010-11-10-r106. PubMed: 20979621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44-57. PubMed: 19131956. [DOI] [PubMed] [Google Scholar]
- 43. Bender SJ, Weiss SR (2010) Pathogenesis of murine coronavirus in the central nervous system. J Neuroimmune Pharmacol 5: 336-354. doi:10.1007/s11481-010-9202-2. PubMed: 20369302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swain SL (1983) T cell subsets and the recognition of MHC class. Immunol Rev 74: 129-142. doi:10.1111/j.1600-065X.1983.tb01087.x. PubMed: 6226585. [DOI] [PubMed] [Google Scholar]
- 45. Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P (1996) Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5: 103-114. doi:10.1016/S1074-7613(00)80487-2. PubMed: 8769474. [DOI] [PubMed] [Google Scholar]
- 46. Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A et al. (1992) Presentation of viral antigen by MHC class I molecules is dependent on a putative peptide transporter heterodimer. Nature 355: 644-646. doi:10.1038/355644a0. PubMed: 1538752. [DOI] [PubMed] [Google Scholar]
- 47. Santamaría I, Velasco G, Pendás AM, Fueyo A, López-Otín C (1998) Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J Biol Chem 273: 16816-16823. doi:10.1074/jbc.273.27.16816. PubMed: 9642240. [DOI] [PubMed] [Google Scholar]
- 48. Endemann G, Stanton LW, Madden KS, Bryant CM, White RT et al. (1993) CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem 268: 11811-11816. PubMed: 7685021. [PubMed] [Google Scholar]
- 49. Ramprasad MP, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D (1996) Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc Natl Acad Sci U S A 93: 14833-14838. doi:10.1073/pnas.93.25.14833. PubMed: 8962141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okada S, O’Brien JS (1969) Tay-Sachs Disease: Generalized Absence of a Beta-D-N-Acetylhexosaminidase Component. Science 165: 698-700. doi:10.1126/science.165.3894.698. PubMed: 5793973. [DOI] [PubMed] [Google Scholar]
- 51. Cantz M, Kresse H (1974) Sandhoff disease: defective glycosaminoglycan catabolism in cultured fibroblasts and its correction by beta-N-acetylhexosaminidase. Eur J Biochem 47: 581-590. doi:10.1111/j.1432-1033.1974.tb03729.x. PubMed: 4279821. [DOI] [PubMed] [Google Scholar]
- 52. Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT et al. (2000) Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290: 2298-2301. doi:10.1126/science.290.5500.2298. PubMed: 11125141. [DOI] [PubMed] [Google Scholar]
- 53. Schmitz G, Kaminski WE, Porsch-Ozcürümez M, Klucken J, Orsó E et al. (1999) ATP-binding cassette transporter A1 (ABCA1) in macrophages: a dual function in inflammation and lipid metabolism? Pathobiology 67: 236-240. doi:10.1159/000028100. PubMed: 10725792. [DOI] [PubMed] [Google Scholar]
- 54. Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I et al. (2000) Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25: 317-329. doi:10.1016/S0896-6273(00)80897-1. PubMed: 10719888. [DOI] [PubMed] [Google Scholar]
- 55. Zhou Q, Wang S, Anderson DJ (2000) Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25: 331-343. doi:10.1016/S0896-6273(00)80898-3. PubMed: 10719889. [DOI] [PubMed] [Google Scholar]
- 56. Takebayashi H, Ohtsuki T, Uchida T, Kawamoto S, Okubo K et al. (2002) Non-overlapping expression of Olig3 and Olig2 in the embryonic neural tube. Mech Dev 113: 169-174. doi:10.1016/S0925-4773(02)00021-7. PubMed: 11960707. [DOI] [PubMed] [Google Scholar]
- 57. Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D et al. (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165-170. doi:10.1101/gad.215802. PubMed: 11799060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stolt CC, Lommes P, Friedrich RP, Wegner M (2004) Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 131: 2349-2358. doi:10.1242/dev.01114. PubMed: 15102707. [DOI] [PubMed] [Google Scholar]
- 59. Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A et al. (2003) The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17: 1677-1689. doi:10.1101/gad.259003. PubMed: 12842915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M (1998) Sox10, a novel transcriptional modulator in glial cells. J Neurosci 18: 237-250. PubMed: 9412504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gingras MC, Lapillonne H, Margolin JF (2001) CFFM4: a new member of the CD20/FcepsilonRIbeta family. Immunogenetics 53: 468-476. doi:10.1007/s002510100345. PubMed: 11685457. [DOI] [PubMed] [Google Scholar]
- 62. Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH (1998) Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391: 703-707. doi:10.1038/35642. PubMed: 9490415. [DOI] [PubMed] [Google Scholar]
- 63. Daws MR, Lanier LL, Seaman WE, Ryan JC (2001) Cloning and characterization of a novel mouse myeloid DAP12-associated receptor family. Eur J Immunol 31: 783-791. doi:10.1002/1521-4141(200103)31:3. PubMed: 11241283. [DOI] [PubMed] [Google Scholar]
- 64. Arm JP, Gurish MF, Reynolds DS, Scott HC, Gartner CS et al. (1991) Molecular cloning of gp49, a cell-surface antigen that is preferentially expressed by mouse mast cell progenitors and is a new member of the immunoglobulin superfamily. J Biol Chem 266: 15966-15973. PubMed: 1714901. [PubMed] [Google Scholar]
- 65. DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW et al. (1997) Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15: 453-457. doi:10.1038/sj.onc.1201206. PubMed: 9242382. [DOI] [PubMed] [Google Scholar]
- 66. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G et al. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514-518. doi:10.1038/nature07725. PubMed: 19158675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cherayil BJ, Weiner SJ, Pillai S (1989) The Mac-2 antigen is a galactose-specific lectin that binds IgE. J Exp Med 170: 1959-1972. doi:10.1084/jem.170.6.1959. PubMed: 2584931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Owen TA, Smock SL, Prakash S, Pinder L, Brees D et al. (2003) Identification and characterization of the genes encoding human and mouse osteoactivin. Crit Rev Eukaryot Gene Expr 13: 205-220. doi:10.1615/CritRevEukaryotGeneExpr.v13.i24.120. PubMed: 14696968. [DOI] [PubMed] [Google Scholar]
- 69. Safadi FF, Xu J, Smock SL, Rico MC, Owen TA et al. (2001) Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biochem 84: 12-26. PubMed: 11746512. [DOI] [PubMed] [Google Scholar]
- 70. Spilsbury K, O’Mara MA, Wu WM, Rowe PB, Symonds G et al. (1995) Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood 85: 1620-1629. PubMed: 7888681. [PubMed] [Google Scholar]
- 71. Bakker AB, Baker E, Sutherland GR, Phillips JH, Lanier LL (1999) Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proc Natl Acad Sci U S A 96: 9792-9796. doi:10.1073/pnas.96.17.9792. PubMed: 10449773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aoki N, Kimura S, Takiyama Y, Atsuta Y, Abe A et al. (2000) The role of the DAP12 signal in mouse myeloid differentiation. J Immunol 165: 3790-3796. PubMed: 11034384. [DOI] [PubMed] [Google Scholar]
- 73. Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M et al. (2006) Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 177: 3520-3524. PubMed: 16951310. [DOI] [PubMed] [Google Scholar]
- 74. Lavi E, Gilden DH, Highkin MK, Weiss SR (1984) Persistence of mouse hepatitis virus A59 RNA in a slow virus demyelinating infection in mice as detected by in situ hybridization. J Virol 51: 563-566. PubMed: 6086966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Das Sarma J, Fu L, Tsai JC, Weiss SR, Lavi E (2000) Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J Virol 74: 9206-9213. doi:10.1128/JVI.74.19.9206-9213.2000. PubMed: 10982367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Marten NW, Stohlman SA, Bergmann CC (2000) Role of viral persistence in retaining CD8(+) T cells within the central nervous system. J Virol 74: 7903-7710. PubMed: 10933698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ireland DD, Stohlman SA, Hinton DR, Kapil P, Silverman RH et al. (2009) RNase L mediated protection from virus induced demyelination. PLOS Pathog 5: e1000602 PubMed: 19798426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Powell HC, Lampert PW (1975) Oligodendrocytes and their myelin-plasma membrane connections in JHM mouse hepatitis virus encephalomyelitis. Lab Invest 33: 440-445. PubMed: 171479. [PubMed] [Google Scholar]
- 79. Knobler RL, Dubois-Dalcq M, Haspel MV, Claysmith AP, Lampert PW et al. (1981) Selective localization of wild type and mutant mouse hepatitis virus (JHM strain) antigens in CNS tissue by fluorescence, light and electron microscopy. J Neuroimmunol 1: 81-92. doi:10.1016/0165-5728(81)90010-2. PubMed: 6276437. [DOI] [PubMed] [Google Scholar]
- 80. Knobler RL, Rodriguez M, Lampert PW, Oldstone MB (1983) Virologic models of chronic relapsing demyelinating disease. Acta Neuropathol Suppl 9: 31-37. doi:10.1007/978-3-642-69094-5_4. PubMed: 6312726. [DOI] [PubMed] [Google Scholar]
- 81. Fleury HJA, Sheppard RD, Bornstein MB, Raine CS (1980). Neuropathol App Neurobiol 8: 165-179. [DOI] [PubMed] [Google Scholar]
- 82. Dubois-Dalcq ME, Doller EW, Haspel MV, Holmes KV (1982) Cell tropism and expression of mouse hepatitis viruses (MHV) in mouse spinal cord cultures. Virology 119: 317-331. doi:10.1016/0042-6822(82)90092-7. PubMed: 6281976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. González JM, Bergmann CC, Ramakrishna C, Hinton DR, Atkinson R et al. (2006) Inhibition of interferon-gamma signaling in oligodendroglia delays coronavirus clearance without altering demyelination. Am J Pathol 168: 796-804. doi:10.2353/ajpath.2006.050496. PubMed: 16507895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lane TE, Hosking MP (2010) The pathogenesis of murine coronavirus infection of the central nervous system. Crit Rev Immunol 30: 119-130. doi:10.1615/CritRevImmunol.v30.i2.20. PubMed: 20370625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Swanborg RH, Amesse LS (1971) Experimental allergic encephalomyelitis: species variability of the encephalitogenic determinant. J Immunol 107: 281–3. PubMed: 4103918. [PubMed] [Google Scholar]
- 86. Martenson RE, Deibler GE, Kies MW, Levine S, Alvord EC (1972) Myelin basic proteins of mammalian and submammalian vertebrates: encephalitogenic activities in guinea pigs and rats. J Immunol 109: 262-270. PubMed: 4114825. [PubMed] [Google Scholar]
- 87. Sedgwick JD, Mason DW (1986) The mechanism of inhibition of experimental allergic encephalomyelitis in the rat by monoclonal antibody against CD4. J Neuroimmunol 13: 217-232. doi:10.1016/0165-5728(86)90066-4. PubMed: 3097071. [DOI] [PubMed] [Google Scholar]
- 88. Spach KM, Pedersen LB, Nashold FE, Kayo T, Yandell BS et al. (2004) Gene expression analysis suggests that 1,25-dihydroxyvitamin D3 reverses experimental autoimmune encephalomyelitis by stimulating inflammatory cell apoptosis. Physiol Genomics 18: 141-151. doi:10.1152/physiolgenomics.00003.2004. PubMed: 15138306. [DOI] [PubMed] [Google Scholar]
- 89. Ibrahim SM, Mix E, Böttcher T, Koczan D, Gold R et al. (2001) Gene expression profiling of the nervous system in murine experimental autoimmune encephalomyelitis. Brain 124: 1927-1938. doi:10.1093/brain/124.10.1927. PubMed: 11571211. [DOI] [PubMed] [Google Scholar]
- 90. Schall TJ, Bacon K, Toy KJ, Goeddel DV (1990) Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature 347: 669-671. doi:10.1038/347669a0. PubMed: 1699135. [DOI] [PubMed] [Google Scholar]
- 91. Matsushima K, Larsen CG, DuBois GC, Oppenheim JJ (1989) Purification and characterization of a novel monocyte chemotactic and activating factor produced by a human myelomonocytic cell line. J Exp Med 169: 1485-1490. doi:10.1084/jem.169.4.1485. PubMed: 2926331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schall TJ, Bacon K, Camp RD, Kaspari JW, Goeddel DV (1993) Human macrophage inflammatory protein alpha (MIP-1 alpha) and MIP-1 beta chemokines attract distinct populations of lymphocytes. J Exp Med 177: 1821-1826. doi:10.1084/jem.177.6.1821. PubMed: 7684437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rot A, Krieger M, Brunner T, Bischoff SC, Schall TJ et al. (1992) RANTES and macrophage inflammatory protein 1 alpha induce the migration and activation of normal human eosinophil granulocytes. J Exp Med 176: 1489-1495. doi:10.1084/jem.176.6.1489. PubMed: 1281207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I et al. (1996) The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382: 829-833. doi:10.1038/382829a0. PubMed: 8752280. [DOI] [PubMed] [Google Scholar]
- 95. Liu MT, Keirstead HS, Lane TE (2001) Neutralization of the chemokine CXCL10 reduces inflammatory cell invasion and demyelination and improves neurological function in a viral model of multiple sclerosis. J Immunol 167: 4091-4097. PubMed: 11564831. [DOI] [PubMed] [Google Scholar]
- 96. Sallusto F, Mackay CR, Lanzavecchia A (1997) Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 277: 2005-2007. doi:10.1126/science.277.5334.2005. PubMed: 9302298. [DOI] [PubMed] [Google Scholar]
- 97. Sallusto F, Lenig D, Mackay CR, Lanzavecchia A (1998) Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med 187: 875-883. doi:10.1084/jem.187.6.875. PubMed: 9500790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Loetscher P, Uguccioni M, Bordoli L, Baggiolini M, Moser B et al. (1998) CCR5 is characteristic of Th1 lymphocytes. Nature 391: 344-345. doi:10.1038/34812. PubMed: 9450746. [DOI] [PubMed] [Google Scholar]
- 99. Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R et al. (1998) Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med 187: 129-134. doi:10.1084/jem.187.1.129. PubMed: 9419219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Taub DD, Lloyd AR, Conlon K, Wang JM, Ortaldo JR et al. (1993) Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med 177: 1809-1814. doi:10.1084/jem.177.6.1809. PubMed: 8496693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mosmann TR, Coffman RL (1989) TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol 7: 145-173. doi:10.1146/annurev.iy.07.040189.001045. PubMed: 2523712. [DOI] [PubMed] [Google Scholar]
- 102. Chen BP, Kuziel WA, Lane TE (2001) Lack of CCR2 results in increased mortality and impaired leukocyte activation and trafficking following infection of the central nervous system with a neurotropic coronavirus. J Immunol 167: 4585-4592. PubMed: 11591787. [DOI] [PubMed] [Google Scholar]
- 103. Gourraud PA, Harbo HF, Hauser SL, Baranzini SE (2012) The genetics of multiple sclerosis: an up-to-date review. Immunol Rev 248: 87-103. doi:10.1111/j.1600-065X.2012.01134.x. PubMed: 22725956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Marten NW, Stohlman SA, Smith-Begolka W, Miller SD, Dimacali E et al. (1999) Selection of CD8+ T cells with highly focused specificity during viral persistence in the central nervous system. J Immunol 162: 3905-3914. PubMed: 10201909. [PubMed] [Google Scholar]
- 105. Pewe L, Wu GF, Barnett EM, Castro RF, Perlman S (1996) Cytotoxic T cell-resistant variants are selected in a virus-induced demyelinating disease. Immunity 5: 253-262. doi:10.1016/S1074-7613(00)80320-9. PubMed: 8808680. [DOI] [PubMed] [Google Scholar]
- 106. Phares TW, Ramakrishna C, Parra GI, Epstein A, Chen L et al. (2009) Target-dependent B7-H1 regulation contributes to clearance of central nervous system infection and dampens morbidity. J Immunol 182: 5430-5438. doi:10.4049/jimmunol.0803557. PubMed: 19380790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhao J, Zhao J, Perlman S (2009) De novo recruitment of antigen-experienced and naive T cells contributes to the long-term maintenance of antiviral T cell populations in the persistently infected central nervous system. J Immunol 183: 5163-5170. doi:10.4049/jimmunol.0902164. PubMed: 19786545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bergmann CC, Altman JD, Hinton D, Stohlman SA (1999) Inverted immunodominance and impaired cytolytic function of CD8+ T cells during viral persistence in the central nervous system. J Immunol 163: 3379-3387. PubMed: 10477608. [PubMed] [Google Scholar]
- 109. Castro RF, Evans GD, Jaszewski A, Perlman S (1994) Coronavirus-induced demyelination occurs in the presence of virus-specific cytotoxic T cells. Virology 200: 733-743. doi:10.1006/viro.1994.1237. PubMed: 8178457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zimprich F, Winter J, Wege H, Lassmann H (1991) Coronavirus induced primary demyelination: indications for the involvement of a humoral immune response. J Virol 65: 1789-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Perlman S, Dandekar AA (2005) Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol 5: 917-927. doi:10.1038/nri1732. PubMed: 16322745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gil-Cruz C, Perez-Shibayama C, Firner S, Waisman A, Bechmann I et al. (2012) T helper cell- and CD40-dependent germline IgM prevents chronic virus-induced demyelinating disease. Proc Natl Acad Sci U S A 109: 1233-1238. doi:10.1073/pnas.1115154109. PubMed: 22232667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sagar D, Lamontagne A, Foss CA, Khan ZK, Pomper MG et al. (2012) Dendritic cell CNS recruitment correlates with disease severity in EAE via CCL2 chemotaxis at the blood-brain barrier through paracellular transmigration and ERK activation. J Neuroinflammation 9: 245. doi:10.1186/1742-2094-9-245. PubMed: 23102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Johnson AB, Wiśniewski HM, Raine CS, Eylar EH, Terry RD (1971) Specific binding of peroxidase-labeled myelin basic protein in allergic encephalomyelitis. Proc Natl Acad Sci U S A 68: 2694-2698. doi:10.1073/pnas.68.11.2694. PubMed: 5288245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wilson HL (2012) B cells contribute to MS pathogenesis through antibody-dependent and antibody-independent mechanisms. Biologics 6: 117-123. PubMed: 22690126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sandberg-Wollheim M, Turesson I (1975) Lymphocyte subpopulations in the cerebrospinal fluid and peripheral blood in patients with multiple sclerosis. Scand J Immunol 4: 831-836. doi:10.1111/j.1365-3083.1975.tb02693.x. PubMed: 1082635. [DOI] [PubMed] [Google Scholar]
- 117. Chastain EM, Duncan DS, Rodgers JM, Miller SD (2011) The role of antigen presenting cells in multiple sclerosis. Biochim Biophys Acta 1812: 265-274. doi:10.1016/j.bbadis.2010.07.008. PubMed: 20637861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC et al. (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261: 921-923. doi:10.1126/science.8346443. PubMed: 8346443. [DOI] [PubMed] [Google Scholar]
- 119. Pericak-Vance MA, Bebout JL, Gaskell PC, Yamaoka LH, Hung WY et al. (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet 48: 1034-1050. PubMed: 2035524. [PMC free article] [PubMed] [Google Scholar]
- 120. Mitew S, Kirkcaldie MT, Halliday GM, Shepherd CE, Vickers JC et al. (2010) Focal demyelination in Alzheimer’s disease and transgenic mouse models. Acta Neuropathol 119: 567-577. doi:10.1007/s00401-010-0657-2. PubMed: 20198482. [DOI] [PubMed] [Google Scholar]
- 121. Eto M, Sumi H, Fujimura H, Yoshikawa H, Sakoda S (2008) Pioglitazone promotes peripheral nerve remyelination after crush injury through CD36 upregulation. J Peripher Nerv Syst 13: 242-248. doi:10.1111/j.1529-8027.2008.00183.x. PubMed: 18844791. [DOI] [PubMed] [Google Scholar]
- 122. Ferrandi C, Richard F, Tavano P, Hauben E, Barbié V et al. (2011) Characterization of immune cell subsets during the active phase of multiple sclerosis reveals disease and c-Jun N-terminal kinase pathway biomarkers. Mult Scler 17: 43-56. doi:10.1177/1352458510381258. PubMed: 20855355. [DOI] [PubMed] [Google Scholar]
- 123. Shi J, Tu JL, Gale SD, Baxter L, Vollmer TL et al. (2011) APOE ε4 is associated with exacerbation of cognitive decline in patients with multiple sclerosis. Cogn Behav Neurol 24: 128-133. doi:10.1097/WNN.0b013e31823380b5. PubMed: 21945984. [DOI] [PubMed] [Google Scholar]
- 124. Tamam Y, Tasdemir N, Yalman M, Tamam B (2011) Association of apolipoprotein E genotypes with prognosis in multiple sclerosis. Eur Rev Med Pharmacol Sci 15: 1122-1130. PubMed: 22165672. [PubMed] [Google Scholar]
- 125. Mirshafiey A, Jadidi-Niaragh F (2010) Prostaglandins in pathogenesis and treatment of multiple sclerosis. Immunopharmacol Immunotoxicol 32: 543-554. doi:10.3109/08923971003667627. PubMed: 20233088. [DOI] [PubMed] [Google Scholar]
- 126. Templeton SP, Kim TS, O’Malley K, Perlman S (2008) Maturation and localization of macrophages and microglia during infection with a neurotropic murine coronavirus. Brain Pathol 18: 40-51. doi:10.1111/j.1750-3639.2007.00098.x. PubMed: 17935605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Xue S, Sun N, Van Rooijen N, Perlman S (1999) Depletion of blood-borne macrophages does not reduce demyelination in mice infected with a neurotropic coronavirus. J Virol 73: 6327-6634. PubMed: 10400724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lanier LL (2009) DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev 227: 150-160. doi:10.1111/j.1600-065X.2008.00720.x. PubMed: 19120482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bouchon A, Dietrich J, Colonna M (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol 164: 4991-4995. PubMed: 10799849. [DOI] [PubMed] [Google Scholar]
- 130. Bouchon A, Hernández-Munain C, Cella M, Colonna M (2001) A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J Exp Med 194: 1111-1122. doi:10.1084/jem.194.8.1111. PubMed: 11602640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Bayless KJ, Meininger GA, Scholtz JM, Davis GE (1998) Osteopontin is a ligand for the alpha4beta1 integrin. J Cell Sci 111 (Pt 9): 1165-1174. PubMed: 9547293. [DOI] [PubMed] [Google Scholar]
- 132. Dandekar AA, O’Malley K, Perlman S (2005) Important roles for gamma interferon and NKG2D in gammadelta T-cell-induced demyelination in T-cell receptor beta-deficient mice infected with a coronavirus. J Virol 79: 9388-9396. doi:10.1128/JVI.79.15.9388-9396.2005. PubMed: 16014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H et al. (2011) Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet 43: 127-131. doi:10.1038/ng.748. PubMed: 21217755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lausch E, Janecke A, Bros M, Trojandt S, Alanay Y, De Laet C et al. (2011) Genetic deficiency of tartrate-resistant acid phosphatase associated with skeletal dysplasia, cerebral calcifications and autoimmunity. Nat Genet 43: 132-137. doi:10.1038/ng.749. PubMed: 21217752. [DOI] [PubMed] [Google Scholar]
- 135. Reinholt FP, Hultenby K, Oldberg A, Heinegård D (1990) Osteopontin--a possible anchor of osteoclasts to bone. Proc Natl Acad Sci U S A 87: 4473-4475. doi:10.1073/pnas.87.12.4473. PubMed: 1693772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Börnsen L, Khademi M, Olsson T, Sørensen PS, Sellebjerg F (2011) Osteopontin concentrations are increased in cerebrospinal fluid during attacks of multiple sclerosis. Mult Scler 17: 32-42. doi:10.1177/1352458510382247. PubMed: 20921238. [DOI] [PubMed] [Google Scholar]
- 137. Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR et al. (2001) The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 294: 1731-1735. doi:10.1126/science.1062960. PubMed: 11721059. [DOI] [PubMed] [Google Scholar]
- 138. Ruprecht K, Kuhlmann T, Seif F, Hummel V, Kruse N et al. (2001) Effects of oncostatin M on human cerebral endothelial cells and expression in inflammatory brain lesions. J Neuropathol Exp Neurol 60: 1087-1098. PubMed: 11706938. [DOI] [PubMed] [Google Scholar]
- 139. Glezer I, Rivest S (2010) Oncostatin M is a novel glucocorticoid-dependent neuroinflammatory factor that enhances oligodendrocyte precursor cell activity in demyelinated sites. Brain Behav Immun 24: 695-704. doi:10.1016/j.bbi.2010.01.005. PubMed: 20083191. [DOI] [PubMed] [Google Scholar]
- 140. Neumann H, Takahashi K (2007) Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol 184: 92-99. doi:10.1016/j.jneuroim.2006.11.032. PubMed: 17239445. [DOI] [PubMed] [Google Scholar]
- 141. Marten NW, Stohlman SA, Bergmann CC (2000) Role of viral persistence in retaining CD8(+) T cells within the central nervous system. J Virol 74: 7903-7710. doi:10.1128/JVI.74.17.7903-7910.2000. PubMed: 10933698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wang FI, Hinton DR, Gilmore W, Trousdale MD, Fleming JO (1992) Sequential infection of glial cells by the murine hepatitis virus JHM strain (MHV-4) leads to a characteristic distribution of demyelination. Lab Invest 66: 744-754. PubMed: 1318460. [PubMed] [Google Scholar]
- 143. Garrett IR, Boyce BF, Oreffo RO, Bonewald L, Poser J et al. (1990) Oxygen-derived free radicals stimulate osteoclastic bone resorption in rodent bone in vitro and in vivo. J Clin Invest 85: 632-639. doi:10.1172/JCI114485. PubMed: 2312718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Marks SC (1989) Osteoclast biology: lessons from mammalian mutations. Am J Med Genet 34: 43-54. doi:10.1002/ajmg.1320340110. PubMed: 2683780. [DOI] [PubMed] [Google Scholar]
- 145. Stark Z, Savarirayan R (2009) Osteopetrosis. Orphanet J Rare Dis 4: 5-. PubMed: 19232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Li H, Hong S, Qian J, Zheng Y, Yang J et al. (2010) Cross talk between the bone and immune systems: osteoclasts function as antigen-presenting cells and activate CD4+ and CD8+ T cells. Blood 116: 210-217. PubMed: 20304810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wucherpfennig KW (1994) Autoimmunity in the central nervous system: mechanisms of antigen presentation and recognition. Clin Immunol Immunopathol 72: 293-306. doi:10.1006/clin.1994.1145. PubMed: 8062444. [DOI] [PubMed] [Google Scholar]
- 148. Lee SK, Lorenzo J (2006) Cytokines regulating osteoclast formation and function. Curr Opin Rheumatol 18: 411-418. doi:10.1097/01.bor.0000231911.42666.78. PubMed: 16763463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of RefSeq transcripts. Expression of each transcript was determined using DESeq in the R statistical background. The current RefSeq database (26,823 transcripts) for Mus musculus was divided into four categories: significantly increased expression (log2 fold change over mock ≥ and p < 0.05), significantly decreased expression (log2 fold change over mock ≤ 1 and p < 0.05), similar expression (-1< log2 fold change over mock < 1), and no expression (neither mock nor MHV-A59 infected had any reads to the transcript).
(TIFF)
RNA-seq technical validation. The expre≤ssion of nine transcripts was analyzed by RNA-seq and qRT-PCR using the same spinal cord RNAs (experiment 1) and by qRT-PCR of spinal cord RNAs from an independent mouse infection experiment (experiment 2). In each experiment, the samples were obtained from spinal cords of both infected and mock infected mice (3-5 mice), sacrificed at 33dpi. All nine transcripts were significantly induced using both techniques of measurement and in RNA samples from both experiments. T-tests were performed to assess significant induction in the qRT-PCR analysis. Data plotted are means with SEM.
(TIFF)
Oligodendrocyte-specific gene expression. As described in the legend to Figure 3, qRT-PCR primers were used to detect the level of expression of oligodendrocyte specific gene transcripts in spinal cords recovered from mock or MHV-A59 infected mice (33dpi). None of the genes had significantly altered expression in infected mice compared to mock. Data shown are means with SEM.
(TIFF)
RNA-seq library statistics.
(XLSX)
Genes whose expression was increased in the MHV-infected spinal cord.
(XLSX)
Gene ontology analysis of 894 upregulated genes.
(XLSX)
Cytokines and cytokine receptors.
(XLSX)
Antigen Presentation.
(XLSX)
Lipid and cholesterol transport and metabolism.
(XLSX)
Oligodendrocyte markers, differentiation and function.
(XLSX)
Osteoclast and ossification.
(XLSX)