Abstract
The gene for glutathione-S-transferase (GST) M1 (GSTM1), a member of the GST-superfamily, is widely studied in cancer risk with regard to the homozygous deletion of the gene (GSTM1 null), leading to a lack of corresponding enzymatic activity. Many of these studies have reported inconsistent findings regarding its association with cancer risk. Therefore, we employed in silico, in vitro, and in vivo approaches to investigate whether the absence of a functional GSTM1 enzyme in a null variant can be compensated for by other family members. Through the in silico approach, we identified maximum structural homology between GSTM1 and GSTM2. Total plasma GST enzymatic activity was similar in recruited individuals, irrespective of their GSTM1 genotype (positive/null). Furthermore, expression profiling using real-time PCR, western blotting, and GSTM2 overexpression following transient knockdown of GSTM1 in HeLa cells confirmed that the absence of GSTM1 activity can be compensated for by the overexpression of GSTM2.
Glutathione-S-transferases (GSTs) belong to a superfamily of ubiquitous, multifunctional dimeric cytosolic enzymes that play a very important role in the Phase II detoxification (or biotransformation) pathway in humans and confer protection against a wide array of toxic insults1,2. Several GST isoforms have been identified and characterised, forming seven distinct classes: α, μ, π, ο, τ, κ, and ζ3,4. Functionally, most GSTs catalyse the conjugation of the nucleophilic tripeptide glutathione to a wide range of electrophilic substrates for detoxification. However, the conjugation reaction can occasionally lead to the formation of compounds that are far more toxic than the initial substrate, thereby leading to disease outcomes1,5,6. Interestingly, a null variant is encountered for two members, GSTT1 and GSTM1, whereby the entire gene is homozygously deleted in a considerable proportion of different populations, resulting in the complete absence of the corresponding enzyme activity7,8. The GSTM1 gene is highly polymorphic and is located on chromosome 1p13.3. A wide range of variation in GSTM1 homozygous deletion polymorphism (approximately 20–67%) has been observed globally with regard to various ethnicities9,10,11,12. It is often hypothesised that, due to the lack of functional GSTT1 and/or GSTM1, the null phenotype is unable to efficiently perform the conjugation reaction (biotransformation) and the subsequent elimination of toxic products via urine and bile. The null variant of GSTΜ1 is of particular interest, as a plethora of studies have demonstrated the difference in susceptibility, exposure to environmental toxicants, resistance to chemotherapy treatment, variability in drug response, manifestation of several diseases, and, most importantly, cancerous outcomes.
The four other members of the GSTμ subfamily, i.e., GSTM2, GSTM3, GSTM4, and GSTM5, exhibit high levels of sequence homology and substrate specificity with GSTM113. Among these genes, GSTM1 has largely been studied due to its null genotype. Although a large number of studies have attempted to associate the GSTM1-null genotype with cancer risk, the results are inconclusive. Several studies have attempted to identify the association of GSTM1 null with cancer risk through meta-analysis using the existing literature; however, these analyses failed to show a significant association of GSTM1 with cancer14,15,16,17,18,19,20. These observations prompted us to search for the functional relevance of this “well known gene” with other family members that are relatively less studied. A possible explanation of the apparently inconsistent results could be that other members of the GST family compensate for the absence of a functional GSTM1 enzyme. In this study, we attempted to ascertain whether the other members of the GST family, particularly those belonging to the GSTM group, can compensate for the loss of the GSTM1 enzyme due to the absence of GSTM1 under normal physiological conditions.
Results
GSTM1 shares maximum homology with GSTM2
In our structural homology analysis, the members of the GST superfamily were found to share high sequence homology with each other when examined by ClustalW (http://www.genome.jp/tools/clustalw/) and a domain search using Pfam (http://pfam.sanger.ac.uk). Members of the same class (i.e., other GSTμ enzymes) share 75–99% sequence identity (maximum homology between GSTM1 and GSTM2), whereas the homology is approximately 25–30% with different classes (GSTθ and GSTπ). This finding prompted us to perform a 3D superimposition of the GSTM1 protein structure with other members of the GSTμ family, GSTT1, and GSTP1 through Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB-PDB). The results distinctly demonstrated that GSTM2 has the highest degree of identical 3D organisation with GSTM1 (root-mean-square deviation [RMSD] value of 0.7 Å). In addition, the enzyme expression pattern from the GeneCard database (http://www.genecards.org/) also suggests a similar pattern of expression among the family members, with maximum similarity in expression patterns in the case of GSTM1 and GSTM2 in different tissues.
Similar GST enzymatic activities in GSTM1 null and non-null groups
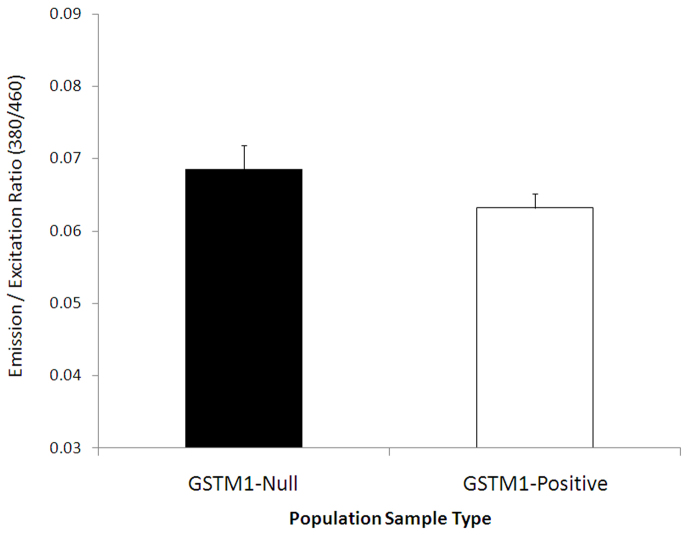
We recruited 275 healthy individuals for screening the GSTM1-null variant. Among the 275 individuals initially recruited, 68 (24.73%) were found to have a GSTM1-null (homozygous deletion for GSTM1) genotype; the remaining 207 (75.27%) individuals were positive for GSTM1 (had at least one functional GSTM1 allele). The null group was composed of 18 female and 50 male individuals; 36 female and 100 male participants were selected from the 207 GSTM1-positive individuals (matched in terms of age, gender, and tobacco usage to nullify possible confounding factors) for further studies. We measured the total plasma glutathione S-transferase enzymatic activity level in the GSTM1-null and -positive individuals. However, the detection of the actual GSTM1 concentration is difficult due to the limitations of antibody-based detection methods and high cross-reactivity among members of the GSTμ subfamily; thus, we measured the total plasma GST activity. For this purpose, a non-fluorescent dye, monochlorobimane (MCB), was used. No significant difference was observed in the overall plasma GST activity between the GSTM1-null and -positive individuals (Fig. 1). In other words, the GSTM1-null individuals exhibited the same catalytic efficiency for MCB as the GSTM1-positive individuals.
Figure 1. Total plasma GST activity in the GSTM1-positive and -null groups.
The total plasma enzyme activity (mean ± SEM, in mU.mg−1.min−1) was found to be similar in both groups, though GSTM1 isoenzyme activity should be undetectable in the GSTM1-null individuals. The similar level of total plasma GST activity indicates the presence of a counter-balance mechanism.
Overexpression of GSTM2 in GSTM1-null individuals
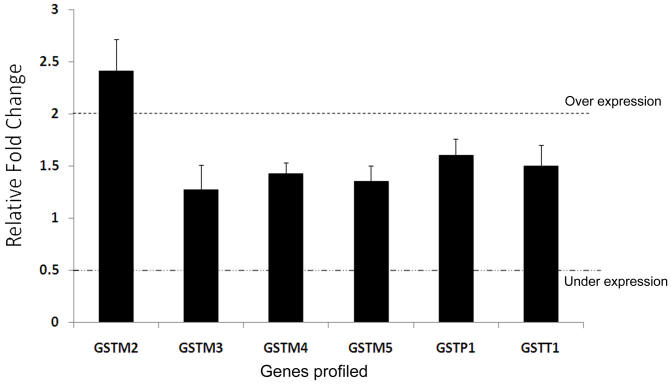
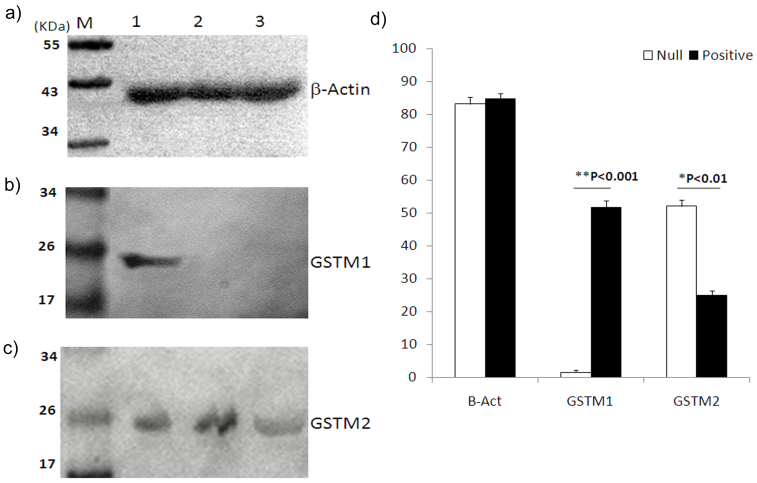
Real-time PCR was performed to explore the contribution of other GST family members compensating for GSTM1 activity in null individuals in vivo. We evaluated the expression pattern of several members of the GST family (GSTM1-GSTM5, GSTT1, and GSTP1) in presence or absence of GSTM1. For this purpose, 15 age, sex, and tobacco usage-matched individuals were selected from both the GSTM1-positive and -null groups, and the gene expression levels of the seven aforementioned GST members were examined. A two-step quantitative real-time polymerase chain reaction (qRT-PCR) approach was implemented using SYBR-Green I. The preliminary results indicated that GSTM2 was expressed at an approximately 2.4-fold higher level in the lymphocytes of GSTM1-null individuals compared to the GSTM1-positive individuals; however, no significant difference was observed in the case of the other GST enzymes (Fig. 2). We also measured the expression pattern of GSTM1 and GSTM2 in the lymphocytes of the GSTM1-positive and -null individuals by western blotting and found that expression of the GSTM2 protein was considerably higher (~2-fold) in the GSTM1-null individuals compared to the GSTM1-positive individuals (Fig. 3).
Figure 2. Gene expression profiling of GST enzymes.
Normalised gene expression profile (mean ± SEM) for GST family members in 15 GSTM1-null individuals with respect to the 15 age and sex-matched GSTM1-positive individuals. The significantly high GSTM2 expression (2.4 fold) under normal physiological conditions indicates a compensatory mechanism in the individuals completely lacking the GSTM1 enzyme.
Figure 3. Western blot analysis reveals high GSTM2 expression in the GSTM1-null individuals.
Representative figure for (a) β-actin from one GSTM1 positive (Lane 1) and two null individuals (Lanes 2 and 3). (b) A GSTM1-positive individual showing a specific 26-kDa band for GSTM1, whereas no band is observed for the GSTM1-null individuals. (c) GSTM2 is present in all three individuals, though with various intensities. (d) A densitometric analysis (mean ± SEM; pixels/ng) of the target proteins in the western blot. A total of 17 GSTM1 null and 16 GSTM1 positive samples were analyzed. All the blots are representative cropped images and every set have been processed simultaenously, under similar conditions. Representative original blots with cropped demarcations (3b & 3c) are provided in supplementary figure 1.
Restoration of cellular function by GSTM2 in GSTM1-null individuals
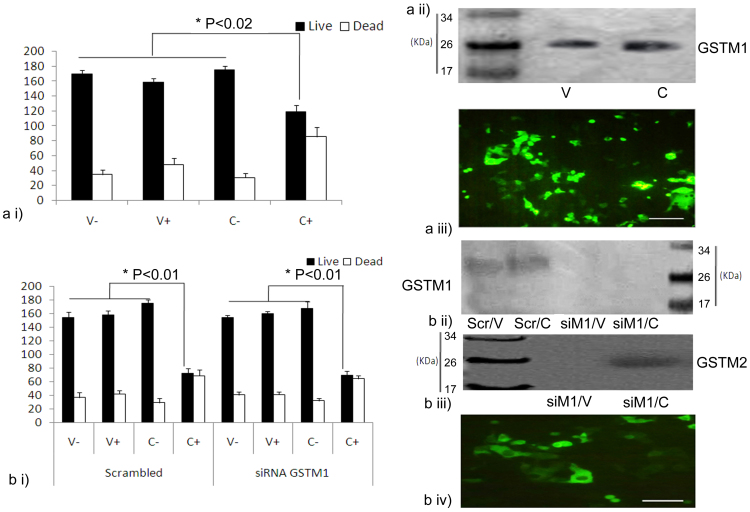
To verify the observed compensatory role of GSTM2 in the absence of GSTM1, a cell culture-based approach was employed. We used green fluorescent protein (GFP)-tagged plasmid constructs of GSTM1 and/or GSTM2 in HeLa cells and evaluated the capacity of the transfected cells to cleave the glutathione-sulphoraphane (GSH-SF) conjugate, an isothiocyanate intermediate that is naturally produced in the body during post-digestion, is normally broken down to free sulphoraphane (SF) by the catalytic action of GSTM121. Sulforaphane induces cell death, mainly through apoptosis, by acting as a growth inhibitor in such cancer cell lines as HeLa and HT2922,23,24. However, it is not known, whether the GSTM2 enzyme can perform a similar function. Indeed, if the GSTM2 enzyme catalyses the breakdown of this conjugate in a manner comparable to that of GSTM1, the functional similarity between these two isozymes would be established. It is known that HeLa cells express only basal levels of GSTM1, but not GSTM225. We over-expressed GSTM1 and GSTM2 in HeLa cells following the transient knockdown of GSTM1 by siRNA. The breakdown of the GSH-SF conjugate and subsequent release of free SF in each case was estimated from the percentage of cell death, as measured by a trypan blue exclusion assay26. The level of expression of GSTM1 and GSTM2 in each case was confirmed by western blotting of whole-cell lysates. The death rate of the cells in the presence of the GSH-SF conjugate either with GSTM1 or GSTM2 (over the background of GSTM1 knockdown) was similar (Fig. 4). This result clearly demonstrated that both GSTM1 and GSTM2 had the same functional efficiency. In other words, GSTM2 could effectively compensate for the loss of GSTM1 under physiological conditions.
Figure 4. Evidence for the functional similarity of the isozymes GSTM1 and GSTM2.
Group (a): i) Trypan blue cell viability assay for GSTM1 in HeLa cells. ii) Western blotting analysis of GSTM1 overexpression in transfected (C) and control cells (V). iii) GFP profile in transfected HeLa cells. Group (b): i) Trypan blue cell viability assay of HeLa cells transfected with GSTM2 and treated either with scrambled or GSTM1-targeted si-RNA (V-, empty vector without GSH-SF treatment, V+, empty vector with GSH-SF treatment, C-, plasmid construct with either GSTM1 (in figure a) or GSTM2 (in figure b) without GSH-SF treatment, C+, plasmid construct with GSH-SF treatment). ii) Western blotting analysis of GSTM1 in the siRNA-treated HeLa cells. iii) Western blot analysis of GSTM2 in siRNA-treated HeLa cells. iv) GFP profile of transfected HeLa cells after siRNA treatment. All the blots are representative cropped images and every set have been processed under similar conditions as detailed in the methods section.
Discussion
The majority of polymorphisms found to affect genes involved in carcinogenesis are single-nucleotide polymorphisms. In contrast, the complete absence of a function in the form of null allele is relatively rare; thus, the GSTM1 homozygous deletion genotype has attracted much attention of researchers worldwide. Extensive studies have been attempted to link the GSTM1-null genotype with disease, particularly cancer. However, the results of association studies correlating GSTM1 with disease risk have been inconclusive, and numerous studies, including a large number of meta-analysis reports, have failed to demonstrate a significant association14,16,17. A meta-analysis of 98 case-control studies was conducted to test the association of GSTM1 null with lung cancer risk, revealing a poor association in both random and fixed effect models; however, no increase risk was seen when only the five largest studies (>500 cases each) were considered27. Analysing 130 case-control studies on GSTM1 null with lung cancer risk revealed a similar observation15. GSTM1 is highly polymorphic, and the prevalence of the GSTM1-null genotype in different populations ranges from 64% to as high as 100% in Kiribati natives28. The frequency also suggests that this gene has not encountered strong environmental selection pressure during evolution and that there might be other enzymes involved in similar chemical detoxification. Based on the results of association studies, it can be clearly understood that GSTM1, a low-penetrant gene, is not a major determinant for cancer association. However, cancer risk can be modulated due to this polymorphism. Therefore, it is important to test the predictive value of the GSTM1-null variant before population-based association studies are conducted. Accordingly, in the present study, we attempted to highlight the role of other family members, particularly in the absence of a functional GSTM1 allele.
Although no enzyme activity is expected in individuals with a null genotype. There are some interesting observations in which GSTM1 activity was identified in GSTM1-null individuals, though the authors failed to present any supportive evidence29,30. These findings support our observation of the total GST activity being similar in individuals, irrespective of the presence or absence of GSTM1. This situation is possible only if another member of the GST family compensates for the loss of GSTM1 in the null individuals. In our study, the enhanced expression of GSTM2, both at the mRNA and protein level, confirmed the role of a compensatory mechanism by a family member in the absence of GSTM1. Moreover, our in vitro functional assays clearly demonstrated a rescue of catalytic activity towards Glutathione-sulforaphane breakdown by GSTM2 over expression in cells where GSTM1 was knocked down. Taken together, these observations, i.e., structural and functional, strengthen our hypothesis of the compensatory role of GSTM2 in the absence of a functional GSTM1 gene. Therefore, a new assessment of the association studies connecting the GSTM1-null phenotype with disease incidence is required, and such studies must be supplemented with functional proof to substantiate their findings. In addition, we might also extrapolate the results of this study to hypothesise that, while studying a disease involving a gene family with high sequence homology and overlapping substrate specificity, the examination of only one gene will not provide the proper insight of the disease in question, as multiple numbers of gene families can function simultaneously.
Methods
Study samples
Healthy study participants were selected from the East Midnapore district, West Bengal, India. We collected blood samples (approx. 5 ml each) from 275 individuals (ages between 15 and 70 years). All the participants were recruited after a thorough screening by physicians, and each provided informed consent before they were included in the study. The non-physician interviewer examined the participants on the basis of a structured questionnaire that elicited information about their lifetime residential history, occupation, diet, and smoking habit. This study was conducted in accord with the Helsinki II Declaration and approved by the ethics committee of CSIR-Indian Institute of Chemical Biology.
Isolation of plasma, DNA, RNA, and protein
Blood samples were centrifuged at 1000 × g for 10 minutes at 4°C to isolate plasma. Nucleic acids and proteins were isolated from blood using the Qiagen DNA, RNA, protein isolation kit following the manufacturer's protocol (Qiagen, Hilden, Germany). The concentration and quality of the nucleic acids were measured using a NanoDrop instrument (NanoDrop Technologies, Wilmington, DE, USA) and agarose gel electrophoresis. The protein concentration was determined using the standard protocol of Bradford (Amresco, OH, USA) assay using bovine serum albumin (HiMedia, India) as a standard; the specific enzyme activity was expressed in mUmg−1min−1.
Screening of GSTM1-null samples
For identifying and confirming the GSTM1-null variant, two exons (exon 2 and exon 7) were amplified separately31. To ensure that the absence of PCR products for any template was due to the presence of a null mutation and not the result of amplification failure, GSTM2 exon 1 (FP, 5′-CTGTCTGCAGAATCCACAGC-3′, and RP, 5′-CTGCAGCTGCTCCACACTT-3′) was amplified as a positive control. Cycling was performed using an Eppendorf Mastercycler (Hamburg, Germany), as follows: a pre-PCR step of a 5-min denaturation at 94°C, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing for 30 sec, and extension at 72°C for 30 sec, and a final 5-min incubation at 72°C; the annealing temperature was 69°C (for M1_exon 2) or 58°C (for both M1_exon 7 and M2_exon 1). All PCR products were separated by polyacrylamide gel (6%) electrophoresis, stained with ethidium bromide, and photographed under UV light.
Total plasma GST enzyme activity assay
The total plasma GST enzyme activity was measured using the Bio-Vision Fluorometric activity assay kit (CA, USA) following the manufacturer's protocol. Monochlorobimane (MCB), a non-fluorescent substrate, was used; MCB fluoresces blue upon reaction with glutathione, and the level of fluorescence is directly proportional to the enzyme activity. The fluorescence was quantified using a micro-plate fluoro-spectrometer (LS 55, Perkin Elmer) at Ex/Em of 380/460 nm32. The plasma protein concentration was estimated using the Bradford assay, as described above, prior to the enzyme activity analysis.
Expression profile of the GSTM group using real-time PCR
A two-step qRT-PCR approach was considered using SYBR-Green I (Brilliant SYBR Green QPCR master Mix, Agilent Technologies, CA, USA). Total RNA (1 μg) isolated from each sample was treated with DNase I (Applied Biosystems, Foster City, CA) prior to cDNA synthesis using the MMuLV-based reverse transcriptase enzyme (RevertAid H Minus First strand cDNA synthesis kit, Fermentas Life Sciences, USA). Primers were designed for the exon-exon boundary using Primer 3 software, and the β-actin gene was used as an internal control (Table 1). Each sample, in duplicate, was amplified as follows: one cycle of 95°C for 1 minute for pre-incubation, followed by 40 cycles of 95°C for 30 seconds, 60°C for 1 min, and 72°C for 1 minute, with a subsequent melting curve analysis using Mx3000p (Stratagene, Agilent Technologies, CA, USA). In addition, after qRT, the amplified product was further analysed by PAGE. An efficiency correction was performed using Agilent software. The fold change in the target gene expression in the GSTM1-positive versus the GSTM1-null samples was calculated using the formula 2−ΔΔCT following the general guidelines discussed by Schmittgen and Livak33. The fold difference was calculated after the data were normalised with the internal control. A less than 0.5 fold change was considered to be under-expression, whereas a > 2.0-fold increase was considered to be overexpression.
Table 1. Primers used in gene expression profiling.
| Primer | Sequence (5′ → 3′) |
|---|---|
| M1FP | AGCGGCCATGGTTTGCAGGAA |
| M1RP | TTCTCCAAGCCCTCAAAGCGG |
| M2FP | CCAGAGCAACGCCATCCT |
| M2RP | GATTCCCCGCACAGGTTGT |
| M3FP | TCACCATGTCGTGCGAGTCGT |
| M3RP | TCATAGTCAGGAGCTTCCCCGCA |
| M4FP | TGGAGAACCAGGCTATGGACGT |
| M4RP | CCAGGAACTGTGAGAAGTGCTG |
| M5FP | AAGCACAACCTGTGTGGGGAGA |
| M5RP | AGCACAGTCTGACCAGCTCCAT |
| P1FP | TATTTCCCAGTTCGAGGCCGCT |
| P1RP | AACTTGGGGAGCTGCCCGTATA |
| T1FP | CCTGCCGCGCTGTTTACATCT |
| T1RP | GCCACACTCTCCGTCAAGGTGA |
| β-actin F | CGAGCACAGAGCCTCGCCTT |
| β-actin R | TCATCATCCATGGTGAGCTGGCG |
Western blotting
Protein lysates were prepared using 1% sodium dodecyl sulphate (SDS) lysis buffer and resolved by 12.5% SDS-PAGE for 2 hours at 150 mA, followed by dry transfer (i-Blot protein transfer apparatus, Invitrogen, USA) and incubation with primary antibodies. Rabbit anti-human GSTM1 antibody (Upstate Biotechnology, Lake Placid, NY, USA), rabbit anti-human GSTM2 antibody (Lifespan Biosciences, Inc, Seattle, WA) and rabbit anti-human beta-actin antibody (Santa Cruz Biotechnologies, CA, USA) were used in 1:1000 dilutions, followed by goat anti-rabbit IgG (GE Healthcare) as the secondary antibody (1:2000 dilutions).
Cell culture and transfection
HeLa cells were obtained from the national cell repository of the National Center for Cell Science (Pune, India). GFP-tagged plasmid constructs for GSTM1 and GSTM2 were purchased from OriGene Technologies Inc. (Rockville, MD, USA). The plasmids were propagated in E. coli DH5alpha and purified using Qiagen plasmid purification kits (Qiagen, Valencia, CA, USA). A 20-mM stock solution of the GSH-SF conjugate (USBio, Swampscott, MA, USA) was prepared in molecular-grade water (Invitrogen, Carlsbad, CA, USA) and stored at −20°C until use. Water was used as vehicle/control for the GSH-SF treatment. Empty GFP vector and scrambled siRNA were used as the control for the overexpression and knockdown experiments, respectively. For GSTM1 knockdown, HeLa cells were transfected with scrambled or GSTM1 siRNA (Dharmacon Inc., USA) using the Dharmafect-1 transfection reagent according to manufacturer's instructions. At 72 hours post-RNAi GSTM2 transfection (over the background of GSTM1 knockdown) was carried out with GSTM2 or control plasmids using Lipofectamine LTX (Invitrogen, Carlsbad, CA, USA) reagent according to the manufacturer's protocols34. At 48 hours post-GSTM2 transfection, the expression efficiency was estimated to be ~ 80%, as based on GFP reporter expression. The cells were serum starved overnight and were treated with 15 μM GSH-SF or vehicle for additional 48 hours22. Post-treatment, cell viability was then assessed by trypan blue exclusion assay using a Neubauer haemocytometer under an inverted bright- field microscope (Leica Microsystems, GmbH, Germany). The data represent the total counts of 200 cells, expressed as percentage, from each group across three independent experiments.
Statistical analyses
All the data are expressed as the mean ± S.E. The statistical analyses were performed with the Mann–Whitney test or Student's t-test, as applicable. GraphPad was used for the analyses.
Author Contributions
P.B., S.P. and A.K.G. conceived and designed the experiments. P.B., S.P., D.P. and P.B. conducted the experiments. P.B., N.G., A.B. and A.K.G. analysed and interpreted the results. P.B., M.B. and A.K.G. wrote the manuscript. A.K.G. supervised the project. All the authors contributed to the scientific planning and discussions.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We sincerely thank Council of Scientific and Industrial Research (CSIR), India, for providing the Senior Research Associateship (to P.B.) and Senior Research fellowship (to P. Banerjee). This work was supported by the CSIR Network Project NWP-0052 and CSIR-Emeritus Scientist Grants No 1(53)/2012/AKG to A.K.G. and CSIR Network Project BSC 0206 to A.B. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Strange R. C., Spiteri M. A., Ramachandran S. & Fryer A. A. Glutathione-S-transferase family of enzymes. Mutat. Res. 482, 21–26 (1997). [DOI] [PubMed] [Google Scholar]
- Rossini A. et al. Frequencies of GSTM1, GSTT1, and GSTP1 polymorphisms in a Brazilian population. Genet. Mol. Res. 1, 233–240 (2002). [PubMed] [Google Scholar]
- McLellan R. A. et al. Characterization of a human glutathione S-transferase mu cluster containing a duplicated GSTM1 gene that causes ultrarapid enzyme activity. Mol. Pharmacol. 52, 958–965 (1997). [DOI] [PubMed] [Google Scholar]
- White D. L., Li D., Nurgalieva Z. & El-Serag H. B. Genetic variants of glutathione S transferase as possible risk factors for hepatocellular carcinoma a HuGE systematic review and meta-analysis. Am. J. Epidemiol. 167, 377–389 (2008). [DOI] [PubMed] [Google Scholar]
- Cotton S. C., Sharp L., Little J. & Brockton N. Glutathione S-transferase polymorphisms and colorectal cancer: a HuGE review. Am. J. Epidemiol. 151, 7–32 (2000). [DOI] [PubMed] [Google Scholar]
- Strange R. C. & Fryer A. A. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci. Publ. 148, 231–249 (1999). [PubMed] [Google Scholar]
- Board P. et al. Genetic heterogeneity of the human glutathione transferases: a complex of gene families. Pharmacol Ther 48, 357–369 (1990). [DOI] [PubMed] [Google Scholar]
- Pemble S. et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem. J. 300, 271–276 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L. R. et al. Breast cancer and CYPIA1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res. 58, 65–70 (1998). [PubMed] [Google Scholar]
- Bell D. A. et al. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. J Natl. Cancer Inst. 85, 1159–1164 (1993). [DOI] [PubMed] [Google Scholar]
- Roth M. J. et al. Association between GSTM1*0 and squamous dysplasia of the esophagus in the high risk region of Linxian, China. Cancer Lett 156, 73–81 (2000). [DOI] [PubMed] [Google Scholar]
- Seidegård J. et al. Isoenzyme(s) of glutathione transferase (class Mu) as a marker for the susceptibility to lung cancer: a follow up study. Carcinogenesis 11, 33–36 (1990). [DOI] [PubMed] [Google Scholar]
- Pearson W. R. et al. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am. J. Hum. Genet. 53, 220–233 (1993). [PMC free article] [PubMed] [Google Scholar]
- Hosgood 3rd H. D., Berndt S. I. & Lan Q. GST genotypes and lung cancer susceptibility in Asian populations with indoor air pollution exposures: a meta-analysis. Mutat. Res. 636, 134–143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Song H., Higgins J. P., Pharoah P. & Danesh J. Five glutathione s-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med. 3, e91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sull J. W., Ohrr H., Kang D. R. & Nam C. M. Glutathione S-transferase M1 status and breast cancer risk: a meta-analysis. Yonsei Med. J. 45, 683–689 (2004). [DOI] [PubMed] [Google Scholar]
- Vogl F. D. et al. Glutathione S-transferases M1, T1, and P1 and breast cancer: a pooled analysis. Cancer Epidemiol. Biomarkers Prev. 13, 1473–1479 (2004). [PubMed] [Google Scholar]
- Varela-Lema L. et al. Meta-analysis and pooled analysis of GSTM1 and CYP1A1 polymorphisms and oral and pharyngeal cancers: a HuGE-GSEC review. Genet Med 10, 369–384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras E. & Kitsios G. D. Synopsis and synthesis of candidate-gene association studies in chronic lymphocytic leukemia: the CUMAGAS-CLL information system. Am. J. Epidemiol. 170, 671–678 (2009). [DOI] [PubMed] [Google Scholar]
- Zhuo W. L. et al. Association studies of CYP1A1 and GSTM1 polymorphisms with esophageal cancer risk: evidence-based meta-analyses. Arch. Med. Res. 40, 169–179 (2009). [DOI] [PubMed] [Google Scholar]
- Traka M. et al. Broccoli consumption interacts with GSTM1 to perturb oncogenic signalling pathways in the prostate. PLoS One 3, e2568 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamet-Payrastre L. et al. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 60, 1426–1433 (2000). [PubMed] [Google Scholar]
- Frydoonfar H. R., McGrath D. R. & Spigelman A. D. Sulforaphane inhibits growth of a colon cancer cell line. Colorectal Dis. 6, 28–31 (2004). [DOI] [PubMed] [Google Scholar]
- Park S. Y. et al. Induction of apoptosis in HT-29 colon cancer cells by phloretin. J. Med. Food 10, 581–586 (2007). [DOI] [PubMed] [Google Scholar]
- Kenine E. C., Kara J. J., Dean R. & Henner W. D. Isolation and analysis of the gene and cDNA for a human Mu Class Glutathione S-Transferase GSTM4. J. Biol. Chem. 268, 16958–16965 (1993). [PubMed] [Google Scholar]
- Bhosle S. M., Huilgol N. G. & Mishra K. P. Enhancement of radiation-induced oxidative stress and cytotoxicity in tumor cells by ellagic acid. Clin. Chim. Acta 359, 89–100 (2005). [DOI] [PubMed] [Google Scholar]
- Carlsten C., Sagoo G. S., Frodsham A. J., Burke W. & Higgins J. P. Glutathione S-transferase M1 (GSTM1) polymorphisms and lung cancer: a literature-based systematic HuGE review and meta-analysis. Am. J. Epidemiol. 167, 759–774 (2008). [DOI] [PubMed] [Google Scholar]
- Syed R., Deeba F. & Jamil K. Role of GSTM1 gene polymorphism and its association with coronary artery disease. Journal of Clin. Med. Res. 2, 22–25 (2010). [Google Scholar]
- Gronau S., Koenig-Greger D., Jerg M. & Riechelmann H. Gene polymorphisms in detoxification enzymes as susceptibility factor for head and neck cancer? Otolaryngol. Head Neck Surg. 128, 674–680 (2003). [DOI] [PubMed] [Google Scholar]
- Konig-Greger D., Riechelmann H., Wittich U. & Gronau S. Genotype and phenotype of glutathione-S-transferase in patients with head and neck carcinoma. Otolaryngol. Head Neck Surg. 130, 718–725 (2004). [DOI] [PubMed] [Google Scholar]
- Ghosh P. et al. Cytogenetic damage and genetic variants in the individuals susceptible to arsenic-induced cancer through drinking water. Int. J. Cancer 118, 2470–7248 (2006). [DOI] [PubMed] [Google Scholar]
- Mak J. C. et al. Relationship between glutathione S-transferase gene polymorphisms and enzyme activity in Hong Kong Chinese asthmatics. Clin. Exp. Allergy 37, 1150–1157 (2007). [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D. & Livak K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- Mishra S. et al. Interaction of annexin A6 with alpha actinin in cardiomyocytes. BMC Cell Biol 12, 7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1