Abstract
Germination ability is regulated by a combination of environmental and endogenous signals with both synergistic and antagonistic effects. Nitric oxide (NO) is a potent dormancy-releasing agent in many species, including Arabidopsis, and has been suggested to behave as an endogenous regulator of this physiological blockage. Distinct reports have also highlighted a positive impact of NO on seed germination under sub-optimal conditions. However, its molecular mode of action in the context of seed biology remains poorly documented. This review aims to focus on the implications of this radical in the control of seed dormancy and germination. The consequences of NO chemistry on the investigations on both its signaling and its targets in seeds are discussed. NO-dependent protein post-translational modifications are proposed as a key mechanism underlying NO signaling during early seed germination.
Keywords: abscisic acid, dormancy, germination, nitric oxide, seed, vigor
INTRODUCTION
Survival of plant species mainly relies on the sexual reproduction which gives rise to new populations. During this process, the adult angiosperm plants produce flowers which upon fertilization give rise to seeds, the main unit of dispersal of flowering plants. In the plant life cycle, the seed and seedling stages are key developmental stages conditioning the final yield of crops. Indeed, seed dormancy, viability, and germination vigor are among the main concerns for agricultural productivity. High vigor seed lots display a low dormancy and lead to seedlings able to withstand extreme stress conditions. If not completely released, dormancy will negatively influence seed germination, which is detrimental to crop yield. However, from an agronomical point of view, lack of dormancy is not a desirable trait as it may lead to pre-harvest sprouting (Bewley and Black, 1994). Therefore, the management of this trait is of fundamental concern for the seed industry and agriculture performance. Thus, investigation of seed quality, toward a better understanding of dormancy, germination and longevity, is of paramount agronomical importance. All these seed features are complex traits controlled by a large number of genes, which are affected by both developmental and environmental factors.
Numerous distinct nitrogen-containing compounds have been shown to positively influence seed germination especially by releasing seed dormancy and improving seed vigor in a wide range of species (Bethke et al., 2007b). These concentration-dependent effects could allow the sensing of the presence of these essential resources in the direct environment. The possibility that all these molecules could act in a similar way prompted plant biologists to look for a possible common nitrogen-containing intermediate and pinpointed nitric oxide (NO) as a possible candidate. Indeed, since its discovery, this radical has progressively emerged as an ubiquitous molecule in both animal and plant signaling networks (Baudouin, 2011). Increasing reports highlight its large implication in diverse signaling pathways regulating growth and developmental processes all along the plant life cycle. A key role for NO was further demonstrated in plant response to abiotic and biotic stresses. Instead of describing in details all these roles that have already been extensively discussed in recent reviews (Besson-Bard et al., 2008; Wilson et al., 2008; Moreau et al., 2010; Baudouin, 2011), we will focus on the implications of NO in the control of seed dormancy and germination with a particular emphasis on the experiments carried out on the model Angiosperm plant Arabidopsis thaliana. The present review also aims to provide outlooks for future investigation in this field.
DEFINITION AND GENERAL OVERVIEW ON SEED DORMANCY AND GERMINATION
SEED DORMANCY
Under natural conditions, an appropriate timing of seed germination is determinant to ensure optimal growth conditions for the young seedlings and guarantee the survival of the species (Bewley, 1997). Seed dormancy is one of the mechanisms contributing to this spatio-temporal adjustment and is defined as a block to the completion of germination of an intact viable seed placed under (temporary) favorable conditions in an otherwise unfavorable season (Bewley, 1997; Finch-Savage and Leubner-Metzger, 2006; Graeber et al., 2012). It may be due to certain properties of the seed coat, mobilization of reserve components, hormone levels, or the joint action of several of these factors (Koornneef et al., 2002). Thus, dormancy is determined by genetic factors but it can also be substantially modulated by environmental parameters (Graeber et al., 2012). Indeed, the alleviation of this blockage can be conditioned by several distinct environmental (temperature, humidity, light, nutrient concentration⋯) or physical (testa rupture⋯) factors. The exact conditions required for dormancy release and subsequent germination depend on the species and thus contribute to the adequacy of the plant to its environment by delaying germination until the seed meets appropriate conditions for its development. In addition, the depth of primary dormancy in mature seeds can depend on the conditions under which the mother plant was exposed such as temperature or availability of mineral elements (such as nitrate) in the soil (Alboresi et al., 2005; Kendall et al., 2011). Thus, seeds have developed a complex control of the depth of dormancy integrating diverse spatio-temporal parameters allowing a dynamic definition of the minimal requirements for germination. In addition, when a non-dormant seed encounters inappropriate conditions for germination, it can enter into a so-called secondary dormancy. Overall, these mechanisms contribute to the sensing of environmental conditions and can lead to dormancy cycling under natural conditions (Footitt et al., 2011).
Abscisic acid (ABA) is considered as the pivotal hormone responsible for the induction and maintenance of seed dormancy (Nambara et al., 2010). ABA is accumulated during seed maturation reaching high levels in dry seeds. Dry dormant seeds were found to contain higher amounts of ABA than dry after-ripened non-dormant seeds (Ali-Rachedi et al., 2004). Upon imbibition, a significant decrease in ABA content was observed in both dormant and non-dormant seeds (Ali-Rachedi et al., 2004). However, after 3 days of imbibition a significant up-accumulation of ABA was detected in dormant seeds only. Exposition of dormant seeds to common dormancy-releasing treatments such as cold-stratification or exogenous nitrate supply leaded to ABA levels similar to non-dormant seeds and prevented the increase in ABA observed when dormancy is maintained (Ali-Rachedi et al., 2004). Reactive oxygen species (ROS) and NO counteract the positive effect of ABA on seed dormancy maintenance. Exogenous application of fluridone (an inhibitor of ABA synthesis) also efficiently released seed dormancy by reducing ABA levels highlighting the requirement for de novo ABA synthesis for the maintenance of this blockage and the existence of a dynamic equilibrium between ABA synthesis and catabolism during seed imbibition (Ali-Rachedi et al., 2004). In addition, recent experiments demonstrated that two independent dormancy-releasing treatments led to similar proteome adjustments supporting the occurrence of shared molecular mechanisms underpinning seed dormancy release (Arc et al., 2012). Furthermore, recent data emphasize the importance of redox control of seed proteome in dormancy release (Marx et al., 2003; Bykova et al., 2011a,b). Thus, ROS and NO appear as good candidates, acting synergistically to release dormancy, putatively acting upstream of ABA.
SEED GERMINATION
Seed germination is temporally defined as the sequence of molecular and physiological events initiated upon imbibition of non-dormant seed and leading to the radicle protrusion through the seed external envelopes (testa and endosperm) that marks the end of germination sensu stricto (Bewley, 1997). Seed germination constitutes a pivotal physiological transition and is associated with a strong modification of the transcriptome (~one-third of the genome) and metabolism over a short time period (around 36–48 h for non-dormant Arabidopsis seeds) relatively to the plant life cycle. During this process, the initially quiescent dry seed successively go through three major steps of water uptake (Bewley, 1997; Weitbrecht et al., 2011). The first step consists in a rapid imbibition of the initially quiescent seeds that lead to the progressive resumption of metabolic activity, gene expression (transcription), protein synthesis and processing and DNA repair (Weitbrecht et al., 2011). The recapitulation of the metabolic activity mainly depends on the stored proteins and metabolites. The importance of the compounds accumulated in the seeds during the maturation was further highlighted by the finding that stored mRNAs and proteins are sufficient for germination sensu stricto (Rajjou et al., 2004; Sano et al., 2012). De novo protein synthesis from the stored mRNAs occurs during the very early step of germination. During this period, the proteins translated are similar to those accumulated during the late maturation and already abundant in seeds reflecting an early recapitulation of the corresponding gene expression program during early germination (Rajjou et al., 2006, 2012). During the second step of water uptake, the water content only slightly increases while important metabolic changes take place inside the seeds. A significant shift is observed during this step from maturation to germination program of development that includes the preparation for seedling establishment (Lopez-Molina et al., 2002; Nonogaki et al., 2007). This two steps time course is consistent with a model proposing that recapitulation of the late maturation program occurs during early germination up to an ABA-dependent developmental checkpoint after which the seed can either activate its germination program or maintain a dormant state notably depending on the sensing of environmental conditions during early imbibition (Lopez-Molina et al., 2002; Rajjou et al., 2012). During this period, seeds maintain their desiccation tolerance. At the end of this second step, if the “decision” to pursue toward germination is taken, the growth potential of the embryo progressively overcome the mechanical constraints imposed by the surrounding layers leading to the successive rupture of the testa and the endosperm (Nonogaki, 2006; Bentsink and Koornneef, 2008). The protrusion of the radicle through the seed coat is thus achieved as a result of important cell elongation without any cell division (Sliwinska et al., 2009) and occurs concomitantly with an important resumption of water uptake. The ABA/gibberellins (GAs) balance coordinate this last step with a decrease in ABA leading to the progressive release its inhibitory effect on endosperm rupture while an important increase in bioactive GAs levels both enhanced the growth potential of the embryo and induced hydrolytic enzymes that weaken the barrier tissues (Bewley and Black, 1994; Muller et al., 2006; Finkelstein et al., 2008).
GERMINATION VIGOR
If the seed encounters suitable conditions for germination during its life, it may, if still viable, allow the young seedling establishment. But as a consequence of aging, the seed germination vigor can be severely affected. In other words, the capacity of a seed lot to germinate rapidly, uniformly and in a wide range of environmental conditions can be impaired or destroyed. As the seed germination process mainly relies on stored mRNA and proteins (Rajjou et al., 2004), damages at the DNA level can result in an aborted development of the seedling. Thus, cellular repair mechanisms especially at the DNA level but also for certain protein post-translational modifications (PTMs) play an essential role in seed vigor (Rajjou et al., 2012). Due to seed high vulnerability to injury, abiotic, and biotic stresses during imbibition, germination is considered as the most critical phase of the plant life cycle. The level of reactive oxygen and nitrogen species (respectively ROS and RNS), influenced by the storage and environmental conditions will determine a balance between the required signaling events and the detrimental oxidative damages (Bailly et al., 2008; Rajjou et al., 2008, 2012; Arc et al., 2011).
NITROGEN OXIDES IMPLICATION IN THE CONTROL OF SEED DORMANCY AND GERMINATION
NITRATE AND NITRITE AVAILABILITY: DETERMINANT FACTORS FOR SEED DORMANCY RELEASE AND SUBSEQUENT GERMINATION
Nitrate () is considered as a major nitrogen source for most plant species. Nitrate reduction into nitrite () is catalyzed by nitrate reductase (NR) that produces nitrogen-containing metabolites, such as amino acids and NO. Apart from being an essential nutrient, nitrate is also considered as a signaling molecule involved in both plant metabolism regulation and developmental processes (Krouk et al., 2010). In particular, nitrate has been shown to promote seed dormancy release and subsequent germination in numerous plant species (Bewley and Black, 1994). Most of the first experiments mainly investigated the effect of nitrate on these physiological processes although the principal product of its assimilation, nitrite can also alleviate seed dormancy (Bethke et al., 2006a).
Exogenous treatments with nitrates were shown to promote seed germination in Arabidopsis by reducing the light requirement (Hilhorst and Karssen, 1988; Batak et al., 2002). The enhancement of germination mediated by light absorbed by phytochrome-A operates via the very-low-fluence response (VLFR; Botto et al., 1996). Thus, nitrate could stimulate the accumulation of cGMP, which then promotes some phytochrome responses (Ludidi and Gehring, 2003). Moreover, a positive correlation between endogenous or applied nitrate levels and germination response to ethylene or GAs was reported for Chenopodium album seeds (Saini et al., 1985). In Arabidopsis, high nitrate feeding of mother plants is associated with higher nitrate content and lower dormancy of the seed progeny (Alboresi et al., 2005). This result suggests a negative correlation between nitrate levels in dry mature seeds and the depth of dormancy. In addition, mutation in the nitrate transporter NRT1.1/CHL1 resulted in lower sensitivity to exogenous nitrate indicating that this protein may be required for nitrate uptake by the seed (Alboresi et al., 2005). Moreover, mutants in the seed specific nitrate transporter AtNRT2.7, involved in nitrate loading into the vacuole during seed maturation, displayed reduced nitrate content and slightly increased dormancy (Chopin et al., 2007). Overall, nitrate availability in seeds appears as an important determinant of seed dormancy.
The reduced dormancy of NR deficient seeds, impaired in nitrate assimilation, along with the finding that glutamine, another nitrogen source did not affect seed germination suggest that the effect of nitrate is unrelated to plant nutrition (Alboresi et al., 2005). As stated in the previous part, exogenous nitrate application was proved to negatively affect ABA content during Arabidopsis seed imbibition (Ali-Rachedi et al., 2004). In addition, controlled nitrate supply to the mother plants led to ABA contents negatively correlated to the endogenous nitrate concentration in dry mature seeds (Matakiadis et al., 2009). Accordingly, it has recently been demonstrated that the gene expression of the ABA catabolic enzyme, CYP707A2, was positively regulated by both endogenous and exogenous nitrate (Matakiadis et al., 2009). Thus, the positive effect of nitrate on dormancy alleviation is presumably mediated by affecting ABA metabolism.
NITRIC OXIDE, THE KEY SIGNALING ELEMENT MEDIATING NITRATE RESPONSE IN SEEDS?
Nitric oxide is a gaseous diatomic free radical detected at low levels in the atmosphere. It is also present in the soils at a concentration depending on the micro-biotic environment (Simontacchi et al., 2007). Moreover, nitrogen fertilization was shown to increase NO release from the soils and proposed to account for the fitness of nitrogen-fertilized plants (Lamattina et al., 2003). NO was shown to efficiently break the dormancy and / or promote germination of several orthodox seeds (Beligni and Lamattina, 2000; Bethke et al., 2004b; Sarath et al., 2006; Liu et al., 2007; Gniazdowska et al., 2010a) including in Arabidopsis thaliana (Bethke et al., 2006b).
Nitric oxide: a key mediator of seed dormancy release
Recent data disclosed that the improvement of dormant-seeds germination provided by exogenous treatments with various nitrogenous molecules, including nitrate, and nitrite, most presumably occurs through NO production (Bethke et al., 2004b, 2006a). Accordingly, the NO content in homogenates from 24 h-imbibed soybean and sorghum embryonic axes, detected by electron paramagnetic resonance (EPR)-spin trapping, increased with increasing nitrate supply during seed imbibition (Caro and Puntarulo, 1999; Simontacchi et al., 2004). This result pinpoints exogenous nitrate concentration during seed imbibition as a key determinant of NO release.
Indeed, NO is well known to release seed dormancy in numerous species (Bethke et al., 2007b). For instance, pharmacological approaches demonstrated that most known NO donors promoted dormancy alleviation and subsequent germination while NO scavengers favored dormancy maintenance and counteracted the positive effect of NO donors (Bethke et al., 2007a). In addition, it has been shown that NO may alleviate dormancy of apple embryos via a transient accumulation of ROS, leading to enhanced ethylene emission as required to terminate germination sensu stricto (Gniazdowska et al., 2007, 2010a,b). NO also proved efficient to reverse blue light inhibition of dormant wheat seed germination, presumably acting interdependently with methyl-jasmonates in controlling reduction of ABA (Jacobsen et al., 2013).
In tomato seeds, the NO scavenger, carboxy-2-phenyl-4,4,5-tetramethylimidazole-1-oxyl 3-oxide (cPTIO), was shown to prevent germination stimulation by fluridone, an ABA synthesis inhibitor (Piterkova et al., 2012). On the contrary, exogenous sodium nitroprusside (SNP), commonly used as NO donor, enhanced the positive effect of norfluorazon, another ABA synthesis inhibitor, on dormancy release of Arabidopsis C24 seeds (Bethke et al., 2006b). Moreover, SNP was shown to reduce seed sensitivity to exogenous ABA (Bethke et al., 2006b). Taken together, these results suggest that NO can decrease ABA sensitivity. A possible effect of NO on ABA catabolism was consequently investigated. Seed treatment with NO donor enhanced CYP707A2 transcript and protein accumulation while the NO scavenger c-PTIO reduced CYP707A2 expression and reversed the NO donor effect (Liu et al., 2009). Thus, as for nitrate, NO was found to enhanced CYP707A2 gene expression (Liu et al., 2009). These results consequently reinforce the assumption that nitrate does not affect seed dormancy on its own but rather act through NO biosynthesis.
A rapid accumulation of NO, possibly in the endosperm layer, during the first stage of Arabidopsis seed imbibition is required for rapid ABA catabolism and breaking of dormancy (Liu et al., 2009). A similar NO accumulation during imbibition was also observed in germinating seeds from other species (Simontacchi et al., 2007). Recently, NO was suggested to act upstream of GAs in a signaling pathway leading to vacuolation of protein storage vacuoles in aleurone cells, a process inhibited by ABA (Bethke et al., 2007a). However, the growth of isolated embryos was unaffected by NO donor or scavengers. Thus, the endosperm layer, proposed as the primary determinant of seed dormancy in Arabidopsis, was proved to perceive and respond to NO, and suggested as its main site of synthesis and action in seeds (Bethke et al., 2007a). Apart from its effect on the hormonal balance, it has been speculated that NO might accelerate the flux towards the pentose phosphate pathway (PPP) by indirectly increasing the oxidation of nicotinamide adenine dinucleotide phosphate (NADPH; Hendricks and Taylorson, 1974; Bethke et al., 2007b). Interestingly, the oxidation of NADPH by S-nitrosoglutathione (GSNO) in the presence of thioredoxin reductase and thioredoxin was demonstrated, releasing glutathione (GSH) and NO (Nikitovic and Holmgren, 1996). In addition, the involvement of the hemoglobin/NO in the oxidation of NADPH has been proposed (Igamberdiev and Hill, 2004). An increase in glucose catabolism via PPP could in turn promote dormancy release (Roberts and Smith, 1977).
As a conclusion, NO is a likely player of a signaling pathway that promotes loss of dormancy and has been suggested to behave as an endogenous regulator of this process. However, the direct targets of NO in seeds remain unclear. Nonetheless, some consequences of NO accumulation on seed metabolism have been highlighted and pinpoint an implication in the regulation of ABA metabolism.
Reactive oxygen species and no crosstalk in the control of seed dormancy and germination
In parallel to NO, ROS have emerged as key players in the control of seed dormancy and germination (Bailly, 2004; Bailly et al., 2008). In cells, ROS can be generated by specific enzymatic reactions or as by-products of the metabolism. Depending on their concentration, ROS may have positive signaling effects including the promotion of dormancy release and germination or detrimental consequences (Liu et al., 2010; Leymarie et al., 2012). Accordingly, it has been proposed that the amount of ROS generated upon seed imbibition should fall within a defined “oxidative window” for germination to occur (Bailly et al., 2008). Below this window, ROS levels would be too low to promote dormancy alleviation while above, oxidative damages would be predominant.
Recently, it has been proposed that ROS might coordinate the reduction of ABA-imposed dormancy with the onset of GA-stimulated germination (Liu et al., 2010). More precisely, exogenous hydrogen peroxide (H2O2) was shown to enhance ABA catabolism and GA biosynthesis during seed imbibition. As NO scavenger efficiently reversed H2O2-mediated induction of CYP707A genes but had no effect on the stimulation of GA biosynthesis, NO was proposed to act downstream of H2O2 in enhancing ABA catabolism. In vivo, both H2O2 and NO appeared to accumulate rapidly and concomitantly upon imbibition and to precede the induction of ABA catabolism/GA biosynthesis (Liu et al., 2010).
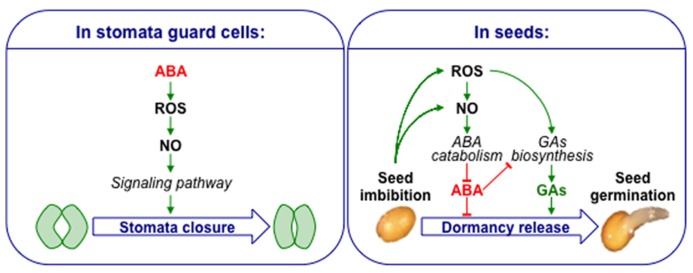
In stomatal guard cells, one of the well-established signaling pathway for ABA-induced stomatal closure involve the successive accumulation of ROS and NO, acting as secondary messengers of ABA signal (Neill et al., 2008; Simontacchi et al., 2013). Even though similar actors are present in seeds, the picture is obviously quite different as both ROS and NO counteract ABA-inhibition of seed germination. This clear distinction highlights the specificity of seed physiology (Figure 1). The exact interplay between reactive nitrogen and oxygen species is always difficult to interpret due to the non-enzymatic reactions susceptible to occur and the molecular consequences they might have.
FIGURE 1.
Contrasting models showing ROS, NO, and ABA crosstalk in stomata guard cells and seeds. ABA increases ROS and NO level in guard cells leading to ABA-dependent stomatal closure. Seed imbibition leads to ROS and NO accumulation. ROS up-regulate ABA catabolism through NO, and also GA biosynthesis. A high concentration of ABA also inhibits GA biosynthesis, but a balance of these two hormones jointly controls seed dormancy and germination.
Nitric oxide and germination vigor
Nitric oxide is well known to play a dual role in stress responses in plants (Corpas et al., 2011). In particular, NO can directly scavenge certain ROS such as superoxide anions and lipid-derived radicals and was shown to stimulate antioxidant enzymes thereby limiting oxidative damages. However, uncontrolled NO accumulation referred to as nitrosative stress can have detrimental consequences.
In seeds, pharmacological experiments highlighted that NO did not significantly influence the germination of non-dormant (fully after-ripened) Arabidopsis seeds under optimal conditions (Bethke et al., 2006a). However, in rice seed, NO was proved to enhance germination by stimulating the transcription of the plasma membrane intrinsic protein (PIP) genes encoding water channels (Liu et al., 2007). In addition, several studies suggested that NO could participate in the tolerance to abiotic stresses during seed germination (Sirova et al., 2011). In particular, NO was demonstrated to delay programmed cell death of barley aleurone cells by promoting the activity of antioxidant enzymes (Beligni et al., 2002). In addition, SNP, commonly used as NO-donor, was shown to alleviate heavy metal stress during seed germination of wheat (Hu et al., 2007), lupin (Kopyra and Gwóźdź, 2003) and rice (Péres da Rocha Oliveiros Marciano et al., 2010). Seed pre-incubation (seed priming) with SNP was also proved to increase salt stress tolerance in wheat (Duan et al., 2007; Zheng et al., 2009). Finally, two recent papers on Arabidopsis reported an enhanced sensitivity of mutants with reduced NO accumulation (atnoa and nia1nia2) to salt and osmotic stress (Zhao et al., 2007; Lozano-Juste and Leon, 2010). In the few cases where it was examined, stress tolerance was associated to increased antioxidant activity. NO could therefore play a key role in germination vigor that could result from its crosstalk with ROS. NO and superoxide rapidly combine to form peroxynitrite (ONOO-), a selective oxidant able to reacts with most biological molecules. Peroxynitrite modifies protein tyrosine to create nitrotyrosines, leaving a footprint detectable in vivo (Vandelle and Delledonne, 2011). However, up to now, only indirect evidences support this assumption in Arabidopsis seeds and none investigated the underlying mechanisms associated to the increased tolerance observed. Overall, NO could play a pivotal role in the sensing of environmental conditions appropriate for seed germination.
CONSEQUENCES OF NITRIC OXIDE CHEMISTRY ON THE INVESTIGATION ON ITS SIGNALING IN SEEDS
SPECIFICITIES OF NITRIC OXIDE CHEMISTRY AND SIGNALING
Nitric oxide (NO•) is an uncharged, gaseous and lipophilic free radical that can readily diffuses across biological membranes. Thus, NO can interact with numerous distinct molecules in plant cells and therefore acts as a signaling element. Free NO• is a transient compound displaying a high reactivity toward other free radicals (e.g., superoxide anion) and transition metal ions (e.g., iron; Wink and Mitchell, 1998). Thus, upon production, released NO can adjust to the cellular redox environment leading to the formation of diverse biologically active compounds collectively referred to as reactive nitrogen species (RNS). RNS include nitrosonium (NO+) and the nitroxyl anion (NO-), respectively resulting from a gain or loss of one electron by NO and peroxynitrite (ONOO-) product of the reaction of NO with superoxide anion radical ( ; Stamler et al., 1992b). Oxidation reactions in the presence of molecular oxygen (O2) can also lead to nitrogen dioxide (NO2), nitrous anhydride (N2O3), and generation. All these molecules differ in reactivity toward the range of NO biological targets. Their differential production can thus orient and/or alter the message mediated by NO. Under physiologic conditions, a strict control of NO content is required to maintain proper cellular functions. High accuracy in signaling events can only be achieved through a tight spatio-temporal control of the intracellular levels of the messengers. Therefore, the balance between NO production and elimination (conversion or storage) is of major importance in determining the biological effects of this radical (Besson-Bard et al., 2008; Moreau et al., 2010; Baudouin, 2011). As for ROS, the chemical reactivity of NO (and associated RNS), make it a particular signal element which can readily interact with a wide range of targets (e.g., proteins, lipids) rather than interact with “dedicated” receptors (Kalyanaraman, 2004; Besson-Bard et al., 2008). The signal mediated by NO can belongs to transduction pathways or be associated with nitrosative stress depending on the biological environment.
NITRIC OXIDE SYNTHESIS AND HOMEOSTASIS IN PLANT SEEDS
Distinct pathways have been proposed to account for NO generation in plant cells (Reviewed in Gupta et al., 2011a). However, the reactions and enzymes involved are still a matter of debate and the relative contribution of these NO biosynthesis pathways remains unclear in seeds (Reviewed in Arc et al., 2013). For instance, NR-catalyzed reduction of nitrite into NO in the cytosol is presumably the most documented reaction but its relevance in seeds is controversial. Instead, nitrite reduction was suggested to occur either via non-enzymatic reactions especially within the apoplasm possibly next to the endosperm layer (Bethke et al., 2004a) or in hypoxic mitochondria (Igamberdiev et al., 2010; Gupta and Igamberdiev, 2011). Alternatively, NO synthesis could result from oxidative reactions from hydroxylamine, polyamines or L-arginine (L-Arg) pathways. NO can also be “stored” through its interaction with diverse molecules. Indeed, NO can react with reduced GSH or thiol groups leading to the reversible formation of S-nitrosothiols (e.g., GSNO, S-nitrosylated proteins). GSNO was suggested to constitute a storage and transport form for NO, even in seeds (Sakamoto et al., 2002; Catusse et al., 2008).
NITRIC OXIDE DETOXIFICATION BY NON-SYMBIOTIC HEMOGLOBINS
Hemoglobins are well known in the animal kingdom for their role as oxygen carrier. In plants, non-symbiotic hemoglobins (nsHb) are divided into two main classes with distinct properties. Class 2 nsHb are the only proteins with an affinity for oxygen fitting with a direct role in oxygen storage and supply (Spyrakis et al., 2011; Vigeolas et al., 2011). Contrarily, the very high affinity for oxygen (in the order of 1–2 nM) displayed by class 1 nsHb is not compatible with such function (Dordas, 2009; Gupta et al., 2011b; Hill, 2012). The plant nsHb1 can act as NADPH-dependent dioxygenase metabolizing NO into nitrate (Igamberdiev and Hill, 2004; Perazzolli et al., 2004). Under hypoxia, NO can be generated from nitrite by deoxyhemeproteins within the mitochondria. Then, nsHb1 and NR can allow the NADPH-dependant re-oxidation of NO into nitrite in the cytosol. As NO can reversibly inhibits cytochrome c oxidase, the reaction between NO and nsHb1 is part of a dynamic equilibrium allowing a tight adjustment of the cellular energy and redox state to oxygen availability (Hebelstrup et al., 2007). These reactions constitute the so-called hemoglobin-NO cycle (Igamberdiev et al., 2010). Furthermore, nsHb1 protein also participates in NO scavenging and therefore NO homeostasis. Accordingly, modulation of nsHb1 expression in plants was shown to directly impact NO levels at distinct developmental stages including seeds (Hebelstrup and Jensen, 2008; Thiel et al., 2011) and in diverse environmental conditions (Dordas, 2009; Cantrel et al., 2011). Thus, despite putative other functions, like CO binding (Hill, 2012), the use of transgenic lines with altered AHb1 expression proved to be a valuable tool to highlight NO implication in physiological processes and stress tolerance.
The over-expression of Arabidopsis nsHb1, AHb1 (also named GLB1 or AtHb1 in other studies; At2g16060) in seeds resulted in a pre-adaptation to stress with the repression of energy consuming pathways, modulation of hormone metabolisms (ABA, SA, auxin, ethylene⋯) and reduced NO emission under transient hypoxia (Thiel et al., 2011). Overall, this leaded to a more efficient allocation of energy resources in seeds resulting in higher weight of mature transgenic seeds (Thiel et al., 2011). Thus, this study highlighted an impact of AHb1 over-expression on the nitrosative stress induced by hypoxia and possibly on NO mediated signaling during seed maturation. However, in dry mature wild-type (WT) seeds, neither nsHb1 protein nor the corresponding mRNA has been detected so far, instead both accumulated during seed imbibition suggesting a crosstalk between nsHbs1 and NO in the germination process (Duff et al., 1998; Ross et al., 2001; Hebelstrup et al., 2007; Matilla and Rodriguez-Gacio Mdel, 2013). Indeed, the NO dioxygenase activity of nsHb1 may also have a significant impact on seed physiology. Importantly, NO accumulation upon water uptake seems to precede nsHb1 induction (Hebelstrup et al., 2007; Liu et al., 2009; Arabidopsis EFP-browser dataset, Winter et al., 2007). The shift between the induction of NO release and nsHb1 accumulation could delimit a short time window during which NO-mediates its effect on ABA catabolism thereby allowing dormancy release before the re-establishment of NO homeostasis by nsHb1 as required to avoid nitrosative stress.
Previous studies relying on modulation of nsHb1 expression in seeds mainly focused on seed maturation (Thiel et al., 2011; Vigeolas et al., 2011). Yet, to date, the link between the NO-related AHb1 function and physiology of seed germination (dormancy, germination vigor, longevity) has never been addressed.
DETECTION AND STUDY OF NITRIC OXIDE IN SEEDS
The investigations on the mode of action of NO in plant cells still suffer from several technical limitations. Indeed, the improvement of NO detection and quantification, pharmacological approaches and biochemical assay for the analysis of NO-induced PTMs are still required.
Pharmacological experiments
Most of the known implications of NO in plant physiology were first highlighted through pharmacological experiments employing NO donors and/or NO scavengers (Bethke et al., 2011). Indeed, due to the toxicity, reactivity, and gaseous state of NO, direct application is not easy to carry out in the laboratory. Thus, a plethora of compounds known to generate NO are preferentially used instead. All these molecules differ by their characteristics of NO release (kinetic, amount, light-dependency) and can thus lead to contrasted results (Planchet and Kaiser, 2006b). Used in aqueous solutions, NO donors can lead to nitrogen oxides production. In addition, certain of these chemicals are complex molecules with potential side products. For instance, the photolysis of SNP was proved to release more cyanide than NO. Indeed, cyanide may actually be the active compound when applying SNP to seeds (Bethke et al., 2006a). Conversely, the widely used derivatives of PTIO such as c-PTIO are thought to be relatively specific NO scavengers (Akaike et al., 1993): PTIO + NO → PTI + NO2. However, the reaction products including PTI may have undesirable side effects in cells (Planchet and Kaiser, 2006a). In a general way, when using NO donors or scavengers, the potential effect of all generated compounds should always be taken into account. The demonstration of opposite effects of NO donors and NO scavengers in a given physiological process is usually considered as a reliable evidence of NO implication.
Methods available for the detection and quantification of nitric oxide in seeds
In animal cells, the absence of nitrate reduction pathways allows the use of assay based on nitrogen oxides, especially nitrite, quantification to evaluate NO production (nitrate and nitrite being considered as by-products of NO production and subsequent oxidation). In plant, such methodology is excluded due to the existence of an active nitrate assimilation pathway responsible for most of nitrite production. Consequently, distinct other methodologies have been applied including fluorescent probes based detection, EPR spectroscopy, electrochemistry, ozone based chemiluminescence, laser photoacoustic, mass spectrometry and the oxyhemoglobin assay. A short discussion on some of these techniques is provided below, for a complete review refer to (Vandelle and Delledonne, 2008; Bethke et al., 2011; Mur et al., 2011).
Several distinct fluorescent probes can be used to investigate NO biosynthesis or release by a given tissue. The diaminofluoresceins (DAF; DAF-FM, 4-amino-5-methylamino-2′,7′-difluorofluorescein) or the diaminorhodamine 4M (DAR-4M) and their cell permeable forms DAF diacetate (DAF-2DA, DAF-FM DA) and DAR-4M acetoxymethyl ester (DAR-4M AM) are the most commonly used (Kojima et al., 1998, 2001). These probes are sensitive (up to the nM range) but suffer from a serious lack of specificity. Indeed, they do not directly react with NO but with its main oxidation product N2O3. Thus, the fluorescence intensity could also depend on the rate of NO oxidation. As the non-enzymatic oxidation of NO requires oxygen, these fluorescent probes cannot be used under anoxia. Finally, numerous distinct compounds were reported to affect DAF-T fluorescence in vivo including ascorbate and dehydroascorbate (Vandelle and Delledonne, 2008). Nonetheless, N2O3 detection with DAF-FM was successfully applied on Arabidopsis seeds but required to remove the seed testa (Liu et al., 2009).
Electron paramagnetic resonance spectroscopy is a more specific method that can be applied to the direct detection of radical species including NO both in vitro and in vivo. However, in order to increase its sensitivity, EPR spectroscopy is often associated with the use of spin-traps, molecules that can react with NO and enhance its EPR signal. This technique has been successfully applied to the detection and quantification of NO in embryonic axes homogenates from soybean and sorghum (Caro and Puntarulo, 1999; Simontacchi et al., 2004). However, NO detection from intact seed tissues, eventually supplemented with a spin trap remains a technical challenge as it would require a sufficiently high production to reach the sensitivity threshold.
Another widely used approach is based on the chemiluminescent reaction between gaseous NO and ozone. This technique can allow the direct quantification of NO release from a tissue placed in a sealed compartment under a gaseous flux driving the gas released in the environment to an analyser. NO-specific electrodes are also available but are also difficult to apply to the study of the tiny Arabidopsis seeds. They could only be useful to assess the amount of NO released by the seeds in their environment.
Overall, despites all the existing techniques, an accurate detection and quantification of NO generation in plant tissue remain difficult. In addition, most techniques require preparation steps or experimental conditions that can lead to undesirable signal. Thus, as for the pharmacological experiments, a cross validation with at least two distinct quantification methods is highly recommended (Gupta and Igamberdiev, 2013). In case of Arabidopsis seeds, the size and characteristics of the mature seeds represents significant technical constraints to an accurate and specific detection/quantification of NO levels by the methodologies currently available.
Genetic resources for the study of nitric oxide production and signaling
The genetic resources available to investigate NO signaling remain restricted due to our limited actual knowledge of NO biosynthesis pathways in plants. Thus, most of the studies rely either on a pharmacological approach (as discussed previously) or on mutants affected in NO availability although their NO levels are not always explained. Some mutants somehow related to NO homeostasis in plants (e.g., nia1nia2, gsnor, atnoa1) have been associated to seed phenotypes. However, the interpretation of these phenotypes is often difficult and requires a lot of caution.
Nitrate reductase, being the only identified enzyme proven to be directly involved in NO biosynthesis, NR-deficient mutants has been extensively used, especially the G′4–3 mutant in Arabidopsis (Wilkinson and Crawford, 1993). However, NR-deficiency causes important perturbation of nitrogen metabolism and a significant nitrate accumulation resulting in a pleiotropic phenotype (Alboresi et al., 2005). Consequently, it is difficult to establish a direct link between nitrate-related phenotypes and reduced NO production by NR-NiR activity. Moreover, the high nitrate levels could lead to an enhanced NO-independent nitrate-mediated signaling (Alboresi et al., 2005). Contradictory results have been published regarding G′4–3 seeds physiology (Alboresi et al., 2005; Lozano-Juste and Leon, 2010).
Several other mutants known as affected in NO levels have also been used to investigate NO signaling in Arabidopsis. Mutants associated to reduced NO levels include NO-Associated 1 (atnoa1, At3g47450; Guo et al., 2003) and prohibitin 3 (phb3; At5g40770; Wang et al., 2010) while one mutant with enhanced endogenous NO levels was identified as the phosphoenolpyruvate/phosphate translocator chlorophyll a/b binding protein underexpressed 1/NO overproducer 1 (cue1/nox1; At5g33320; He et al., 2004). The exact relation between the function of the corresponding proteins and the NO levels in these mutants has not been clearly elucidated yet. Most of these mutants have strong phenotypes but, phb3 and cue1/nox1 have not been investigated for seed phenotypes. However, the atnoa1 mutant has been more studied as it was first proposed as encoding a NO synthase (NOS)-like protein based on sequence similarity with an hypothetical snail NOS and subsequent characterization of a corresponding mutant displaying reduced NOS activity in leaves and lower NO levels in roots (Guo et al., 2003). However, further experiments excluded a direct role for this protein in NO synthesis. Instead, it was later identified as a GTPase. The atnoa1 mutant seeds were associated with a slightly increased dormancy and a hypersensitivity to salt and osmotic stresses (Zhao et al., 2007; Lozano-Juste and Leon, 2010).
Alternative strategies have been developed to get around the known limitations and pursue the investigations on NO signaling in plants. Promising examples include the use of transgenic lines with altered hemoglobin expression (Perazzolli et al., 2004) and the over-expression of rat neuronal NOS in Arabidopsis (Shi et al., 2012). Both strategies already led to significant results even thought all putative side consequences, apart from NO levels alteration, must be considered with extreme caution.
MOLECULAR TARGETS OF NITRIC OXIDE IN SEEDS
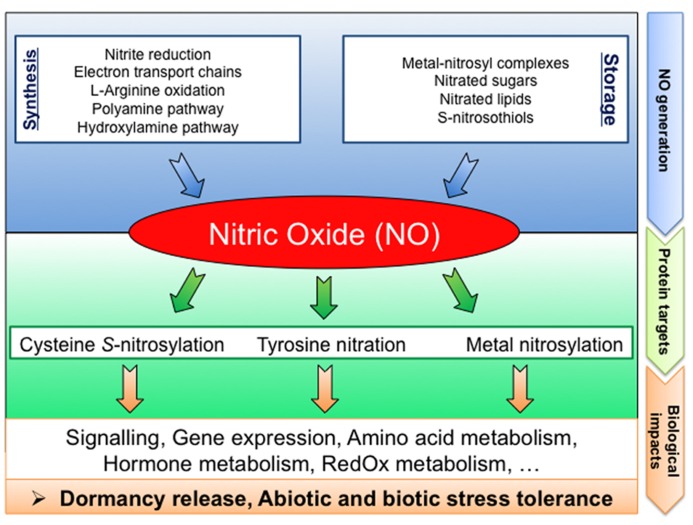
Aside from the long lasting question concerning the relevant NO sources in seeds, the re-constitution of NO signaling pathways require the identification of the NO biological targets. Yet, direct molecular targets of NO remain poorly documented in plants. NO could regulate physiological processes by affecting gene transcription. Indeed, several NO-regulated genes, involved in different functional and biological processes, have previously been described (Huang et al., 2002; Polverari et al., 2003; Parani et al., 2004; Grun et al., 2006; Palmieri et al., 2008; Besson-Bard et al., 2009). Furthermore, NO can bind to transition metals of metalloproteins (metal nitrosylation) or cause protein PTMs such as cysteine S-nitrosylation or tyrosine nitration (Figure 2; Moreau et al., 2010; Arc et al., 2011).
FIGURE 2.
Schematic diagram summarizing NO signaling in seeds: from generation to targets. NO can be produced by various biosynthesis pathways or released from NO-storage compounds. Proteins are preferentially targeted by NO resulting in PTMs (cysteine S-nitrosylation, tyrosine nitration, and metal nitrosylation). These NO mediated PTMs modulate the protein functions, leading to strong impacts on cell metabolism thereby affecting seed physiology.
PROTEIN S-NITROSYLATION IN SEEDS
Nitric oxide-mediated S-nitrosylation of cysteine thiol groups within polypeptide chains is a likely mechanism by which NO may function in signaling processes (Stamler et al., 1992a; Jaffrey et al., 2001). S-nitrosylation consists in the covalent attachment of a NO moiety to a reactive cysteine thiol resulting in the formation of a S-nitrosothiol group (S-NO). In animal systems, regulation of specific proteins by S-nitrosylation is an intensively investigated PTM. This PTM, which is thought to be particularly labile, is associated with a precise spatio-temporal regulation and can potentially result in the activation or inactivation of targeted proteins (Hess et al., 2005). It occurs mainly through non-enzymatic reactions being dependent on the physiochemical environment of the protein cysteinyl residues and the proximity of susceptible proteins to NO production sites in cells (Lindermayr and Durner, 2009). On the contrary, protein de-nitrosylation seems to be catalyzed by several enzymes, such as thioredoxins (Trxs) or Cu/Zn superoxide dismutases, as well as by reducing metals and intracellular reducing agents (Lindermayr and Durner, 2009). In fact, because of its selectivity toward protein targets, S-nitrosylation may represent a general pathway for modulating protein structure/function, analogs to protein phosphorylation (Spickett et al., 2006). Up to now, only few intracellular S-nitrosylated proteins have been identified in plants (Lindermayr et al., 2005; Tanou et al., 2009; Astier et al., 2011; Lounifi et al., 2012). A recent and promising example is the NO-mediated modulation of auxin signaling through the S-nitrosylation of the TIR1 auxin receptor. This PTM of TIR1 promotes its interaction with Aux/IAA repressors thereby facilitating their degradation (Terrile et al., 2012). Moreover, NO and ethylene act antagonistically in fruit ripening through inhibition of enzymes involved in ethylene production by S-nitrosylation (Manjunatha et al., 2012). In contrast, NO and ethylene act synergistically in seed dormancy release but the underlying molecular mechanisms are still unknown (Gniazdowska et al., 2010b; Arc et al., 2013). Due to the limited permeability of most of their outer layers, seeds can experience hypoxia (Borisjuk and Rolletschek, 2009). Consequently, a fine regulation of oxygen consumption is necessary. This seems to be achieved through NO-mediated inhibition of seed mitochondrial activity (Borisjuk et al., 2007). Consequently, NO-related protein modifications are likely to be increased in seed mitochondria and therefore to play an important role in regulating the activity of these organelles. Many S-nitrosylated proteins identified in plants are implicated in metabolic processes (Lindermayr et al., 2005; Abat et al., 2008; Romero-Puertas et al., 2008; Abat and Deswal, 2009; Tanou et al., 2009; Palmieri et al., 2010) suggesting that NO could participate in the regulation of the energy status of the seeds. In agreement, a β-subunit of the mitochondrial ATP synthase complex was found to be S-nitrosylated in dry Arabidopsis seeds (Arc et al., 2011). Since a homologous protein was shown to be inactivated by S-nitrosylation in alcoholic fatty liver of rats (Moon et al., 2006) and more recently in pea leaves mitochondria (Camejo et al., 2013), the seed mitochondrial ATP synthase activity might be inhibited by this NO-mediated PTM. Further experiments are required to assess this hypothesis.
In wheat seeds, a parallel increase in NO and protein S-nitrosylation was reported during sensu stricto germination (Sen, 2010). Noteworthy, seed treatments with NO promoted desiccation tolerance, in the recalcitrant species Antiaris toxicaria, by limiting protein carbonylation and enhancing protein S-nitrosylation (Bai et al., 2011).
PROTEIN NITRATION IN SEEDS
Tyrosine nitration consists in the addition of a nitro group (–NO2) resulting in an alteration of diverse protein functions. The very fast reaction between NO and O2- gives rise to peroxynitrite (ONOO-) which is considered as a potent oxidizing and nitrating agent (Ducrocq et al., 1999; Abello et al., 2009). Tyrosine nitration is consequently predominantly observed in states prone to the concomitant release of NO and ROS. Until recently, tyrosine nitration was considered as being irreversible suggesting that the presence of nitrotyrosine in proteins represents a footprint of nitrosative stress. However, increasing evidence suggests the existence of a de-nitration mechanism in vivo (Abello et al., 2009). Protein nitration can result in an alteration of diverse protein functions (Alvarez et al., 2011; Melo et al., 2011; Jacques et al., 2012) and could enhance protein sensibility to proteolytic degradation via the proteasome (Abello et al., 2009). Thus, protein nitration would be more than a biological marker of nitrosative stress and could participate in protein turnover or signal transduction in plants (Corpas et al., 2008, 2009; Ischiropoulos, 2009). A single study has been carried out on seeds, more precisely on sorghum embryonic axes (Jasid et al., 2008). This work revealed the appearance of several nitrated proteins upon seed imbibition. A recent study based on immunoprecipitation with an anti-3-nitrotyrosine antibody and subsequent analysis by shotgun liquid chromatography–mass spectrometry (LC-MS/MS) led to the identification of 127 proteins putatively targeted by this PTM in protein extracts from Arabidopsis seedlings (Lozano-Juste et al., 2011). Among this important list, a few candidates were further confirmed by additional experiments. Among these numerous putative targets of tyrosine nitration were a few proteins with known implications in seed physiology. For instance, the molybdenum cofactor (MoCo) sulfurase ABA3 (At1g16540) was among these candidates. ABA3 is involved in the last step of ABA synthesis (Mendel, 2007). Thus, the inactivation of ABA synthesis by this PTM might contribute to the control of dormancy release and germination vigor. Overall, nitration may be more than a biological marker of nitrosative stress and could participate in protein turnover or signal transduction in plants (Corpas et al., 2009; Ischiropoulos, 2009). In seeds, the concomitant generation of NO and ROS upon imbibition could lead to enhanced peroxynitrite formation thereby improving tyrosine nitration. Therefore, protein tyrosine nitrations appear likely to occur in this context and in lights of the discussed examples could be of paramount importance.
CONCLUSIONS AND PROSPECTS
Most of the analysis published up to date pinpoint ABA content as a major determinant of dormancy release or maintenance. It appears that the decision to pursue the transition toward germination or maintain a dormant state can be taken during seed imbibition depending on environmental parameters. Thus, the control of ABA levels and sensitivity during early imbibition appears of paramount importance. During this phase, both NO and ROS accumulation has been reported. The intensity of the generation of these radicals could depend on both endogenous and environment cues. In turn, the interplay between ROS and RNS would determine both the extent of ABA catabolism (via the regulation of CYP707A2 expression for instance) and the sensitivity to this hormone. As a result, theses reactive species could determine the kinetics of ABA degradation and the threshold below which ABA content should fall for germination to occur. As the de novo protein synthesis is low during the first hours upon imbibition these effects could be mainly modulated via non-enzymatic protein PTMs such as carbonylation, nitration and/or S-nitrosylation. Still, both ROS and RNS accumulation can also lead to detrimental damages. Thus, we believe that the concept of “oxidative window” for seed germination should be extended to include NO and associated RNS.
However, despite a general consensus regarding NO importance in seed physiology, the pathways involved in its biosynthesis remain uncertain. This observation presumably reflects the complexity of the regulation of NO biosynthesis in plants. Indeed, multiple different endogenous sources all potentially depending on environmental and/or molecular parameters may contribute to NO accumulation in seeds. Moreover, the relevant reactions in seeds may be significantly different from those described at other physiological stages including the non-enzymatic reactions that may occur in the apoplast next to the aleurone layer (Bethke et al., 2004a). To discriminate between the relative contribution of the distinct known NO sources, accurate determination of NO content in seeds and especially during imbibition appears absolutely required. However, the relatively low amount of NO released under physiological conditions and the drawbacks of the techniques currently available makes NO measurement a very challenging issue. In any case, an unambiguous confirmation of NO accumulation in the seed endosperm and/or embryo appears as a priority to consolidate the available evidences and determine the seed NO content.
In a similar way, we are firmly convinced that NO-related PTMs, namely tyrosine nitration and cysteine S-nitrosylation, can explain the effect of NO in seeds though this assumption is not totally confirmed yet. Indeed, the detection and identification methods for both cysteine S-nitrosylation and tyrosine nitration proved difficult to apply on seeds most presumably due to the low abundance of modified proteins and/or the limited stability of the modifications. Nonetheless, these two PTMs represent very seducing models to explain the roles ascribed to NO in seeds. The characterization of NO-targeted proteins in various seed physiology context will undoubtedly reveal new area of research to explore for understanding the control of germination.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a doctoral and post-doctoral fellowship to Erwann Arc from the French Ministry of Higher Education and Research and from European Commission funding (EcoSeed project, KBBE), respectively. The post-doctoral fellowship to Marc Galland was funded by the French Ministry of Industry (FUI NUTRICE).
REFERENCES
- Abat J. K., Deswal R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9 4368–4380 10.1002/pmic.200800985 [DOI] [PubMed] [Google Scholar]
- Abat J. K., Mattoo A. K., Deswal R. (2008). S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata-ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275 2862–2872 10.1111/j.1742-4658.2008.06425.x [DOI] [PubMed] [Google Scholar]
- Abello N., Kerstjens H. A., Postma D. S., Bischoff R. (2009). Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8 3222–3238 10.1021/pr900039c [DOI] [PubMed] [Google Scholar]
- Akaike T., Yoshida M., Miyamoto Y., Sato K., Kohno M., Sasamoto K., et al. (1993). Antagonistic action of imidazolineoxyl N-oxides against endothelium-derived relaxing factor/.NO through a radical reaction. Biochemistry 32 827–832 10.1021/bi00054a013 [DOI] [PubMed] [Google Scholar]
- Alboresi A., Gestin C., Leydecker M. T., Bedu M., Meyer C., Truong H. N. (2005). Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 28 500–512 10.1111/j.1365-3040.2005.01292.x [DOI] [PubMed] [Google Scholar]
- Ali-Rachedi S., Bouinot D., Wagner M. H., Bonnet M., Sotta B., Grappin P., et al. (2004). Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219 479–488 10.1007/s00425-004-1251-4 [DOI] [PubMed] [Google Scholar]
- Alvarez C., Lozano-Juste J., Romero L. C., Garcia I., Gotor C., Leon J. (2011). Inhibition of Arabidopsis O-acetylserine(thiol)lyase A1 by tyrosine nitration. J. Biol. Chem. 286 578–586 10.1074/jbc.M110.147678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc E., Chibani K., Grappin P., Jullien M., Godin B., Cueff G., et al. (2012). Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. J. Proteome Res. 11 5418–5432 10.1021/pr3006815 [DOI] [PubMed] [Google Scholar]
- Arc E., Galland M., Cueff G., Godin B., Lounifi I., Job D., et al. (2011). Reboot the system thanks to protein post-translational modifications and proteome diversity: how quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11 1606–1618 10.1002/pmic.201000641 [DOI] [PubMed] [Google Scholar]
- Arc E., Sechet J., Corbineau F., Rajjou L., Marion-Poll A. (2013). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63 10.3389/fpls.2013.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier J., Rasul S., Koen E., Manzoor H., Besson-Bard A., Lamotte O., et al. (2011). S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci. 181 527–533 10.1016/j.plantsci.2011.02.011 [DOI] [PubMed] [Google Scholar]
- Bai X., Yang L., Tian M., Chen J., Shi J., Yang Y., et al. (2011). Nitric oxide enhances desiccation tolerance of recalcitrant Antiaris toxicaria seeds via protein S-nitrosylation and carbonylation. PLoS ONE 6:e20714 10.1371/journal.pone.0020714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C. (2004). Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 14 93–107 10.1079/SSR2004159 [DOI] [Google Scholar]
- Bailly C., El-Maarouf-Bouteau H., Corbineau F. (2008). From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C. R. Biol. 331 806–814 10.1016/j.crvi.2008.07.022 [DOI] [PubMed] [Google Scholar]
- Batak I., Dević M., Gibal Z., Grubišić D., Poff K. L., Konjević R. (2002). The effects of potassium nitrate and NO-donors on phytochrome A- and phytochrome B-specific induced germination of Arabidopsis thaliana seeds. Seed Sci. Res. 12 253–259 10.1079/SSR2002118 [DOI] [Google Scholar]
- Baudouin E. (2011). The language of nitric oxide signalling. Plant Biol. 13 233–242 10.1111/j.1438-8677.2010.00403.x [DOI] [PubMed] [Google Scholar]
- Beligni M. V., Fath A., Bethke P. C., Lamattina L., Jones R. L. (2002). Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 129 1642–1650 10.1104/pp.002337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni M. V., Lamattina L. (2000). Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210 215–221 10.1007/PL00008128 [DOI] [PubMed] [Google Scholar]
- Bentsink L., Koornneef M. (2008). Seed dormancy and germination. Arabidopsis Book 6:e0119 10.1199/tab.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson-Bard A., Astier J., Rasul S., Wawer I., Dubreuil-Maurizi C., Jeandroz S., et al. (2009). Current view of nitric oxide-responsive genes in plants. Plant Sci. 177 302–309 10.1016/j.plantsci.2009.06.006 [DOI] [Google Scholar]
- Besson-Bard A., Pugin A., Wendehenne D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59 21–39 10.1146/annurev.arplant.59.032607.092830 [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Badger M. R., Jones R. L. (2004a). Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16 332–341 10.1105/tpc.017822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P. C., Gubler F., Jacobsen J. V., Jones R. L. (2004b). Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219 847–855 10.1007/s00425-004-1282-x [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Libourel I. G., Aoyama N., Chung Y. Y., Still D. W., Jones R. L. (2007a). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143 1173–1188 10.1104/pp.106.093435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P. C., Libourel I. G. L., Jones R. L. (2007b). “Nitric oxide in seed dormancy and germination,” in Annual Plant Reviews, Vol. 27 Seed Development, Dormancy and Germination eds Bradford K. J., Nonogaki H. (New Jersey: Blackwell Publishing Ltd; ) 153–175 [Google Scholar]
- Bethke P. C., Libourel I. G., Reinohl V., Jones R. L. (2006a). Sodium nitroprusside, cyanide, nitrite, and nitrate break Arabidopsis seed dormancy in a nitric oxide-dependent manner. Planta 223 805–812 10.1007/s00425-005-0116-9 [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Libourel I. G., Jones R. L. (2006b). Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 57 517–526 10.1093/jxb/erj060 [DOI] [PubMed] [Google Scholar]
- Bethke P. C., Libourel I. G., Vitecek J., Jones R. L. (2011). Nitric oxide methods in seed biology. Methods Mol. Biol. 773 385–400 10.1007/978-1-61779-231-1_22 [DOI] [PubMed] [Google Scholar]
- Bewley J. D. (1997). Seed germination and dormancy. Plant Cell 9 1055–1066 10.1105/tpc.9.7.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J. D., Black M. (1994). Seeds: Physiology of Development and Germination. New York: Plenum Press [Google Scholar]
- Borisjuk L., Macherel D., Benamar A., Wobus U., Rolletschek H. (2007). Low oxygen sensing and balancing in plant seeds: a role for nitric oxide. New Phytol. 176 813–823 10.1111/j.1469-8137.2007.02226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L., Rolletschek H. (2009). The oxygen status of the developing seed. New Phytol. 182 17–30 10.1111/j.1469-8137.2008.02752.x [DOI] [PubMed] [Google Scholar]
- Botto J. F., Sanchez R. A., Whitelam G. C., Casal J. J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykova N. V., Hoehn B., Rampitsch C., Hu J., Stebbing J. A., Knox R. (2011a). Thiol redox-sensitive seed proteome in dormant and non-dormant hybrid genotypes of wheat. Phytochemistry 72 1162–1172 10.1016/j.phytochem.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Bykova N. V., Hoehn B., Rampitsch C., Banks T., Stebbing J. A., Fan T., et al. (2011b). Redox-sensitive proteome and antioxidant strategies in wheat seed dormancy control. Proteomics 11 865–882 10.1002/pmic.200900810 [DOI] [PubMed] [Google Scholar]
- Camejo D., Romero-Puertas Mdel C., Rodriguez-Serrano M., Sandalio L. M., Lazaro J. J., Jimenez A., et al. (2013). Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteomics 79 87–99 10.1016/j.jprot.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Cantrel C., Vazquez T., Puyaubert J., Reze N., Lesch M., Kaiser W. M., et al. (2011). Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 189 415–427 10.1111/j.1469-8137.2010.03500.x [DOI] [PubMed] [Google Scholar]
- Caro A., Puntarulo S. (1999). Nitric oxide generation by soybean embryonic axes. Possible effect on mitochondrial function. Free Radic. Res. 31(Suppl) S205–S212 10.1080/10715769900301521 [DOI] [PubMed] [Google Scholar]
- Catusse J., Strub J. M., Job C., Van Dorsselaer A., Job D. (2008). Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U.S.A. 105 10262–10267 10.1073/pnas.0800585105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F., Orsel M., Dorbe M. F., Chardon F., Truong H. N., Miller A. J., et al. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19 1590–1602 10.1105/tpc.107.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Chaki M., Leterrier M., Barroso J. B. (2009). Protein tyrosine nitration: a new challenge in plants. Plant Signal. Behav. 4 920–923 10.4161/psb.4.10.9466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Del Rio L. A., Barroso J. B. (2008). Post-translational modifications mediated by reactive nitrogen species: nitrosative stress responses or components of signal transduction pathways? Plant Signal. Behav. 3 301–303 10.4161/psb.3.5.5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F. J., Leterrier M., Valderrama R., Airaki M., Chaki M., Palma J. M., et al. (2011). Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 181 604–611 10.1016/j.plantsci.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Dordas C. (2009). Nonsymbiotic hemoglobins and stress tolerance in plants. Plant Sci. 176 433–440 10.1016/j.plantsci.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Duan P., Ding F., Wang F., Wang B. S. (2007). Priming of seeds with nitric oxide donor sodium nitroprusside (SNP) alleviates the inhibition on wheat seed germination by salt stress. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33 244–250 [PubMed] [Google Scholar]
- Ducrocq C., Blanchard B., Pignatelli B., Ohshima H. (1999). Peroxynitrite: an endogenous oxidizing and nitrating agent. Cell. Mol. Life Sci. 55 1068–1077 10.1007/s000180050357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff S. M. G., Guy P. A., Nie X., Durnin D. C., Hill R. D. (1998). Haemoglobin expression in germinating barley. Seed Sci. Res. 8 431–436 10.1017/S0960258500004396 [DOI] [Google Scholar]
- Finch-Savage W. E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171 501–523 10.1111/j.1469-8137.2006.01787.x [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W. E. (2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 108 20236–20241 10.1073/pnas.1116325108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gniazdowska A., Dobrzynska U., Babanczyk T., Bogatek R. (2007). Breaking the apple embryo dormancy by nitric oxide involves the stimulation of ethylene production. Planta 225 1051–1057 10.1007/s00425-006-0384-z [DOI] [PubMed] [Google Scholar]
- Gniazdowska A., Krasuska U., Debska K., Andryka P., Bogatek R. (2010a). The beneficial effect of small toxic molecules on dormancy alleviation and germination of apple embryos is due to NO formation. Planta 232 999–1005 10.1007/s00425-010-1214-x [DOI] [PubMed] [Google Scholar]
- Gniazdowska A., Krasuska U., Bogatek R. (2010b). Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 232 1397–1407 10.1007/s00425-010-1262-2 [DOI] [PubMed] [Google Scholar]
- Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35 1769–1786 10.1111/j.1365-3040.2012.02542.x [DOI] [PubMed] [Google Scholar]
- Grun S., Lindermayr C., Sell S., Durner J. (2006). Nitric oxide and gene regulation in plants. J. Exp. Bot. 57 507–516 10.1093/jxb/erj053 [DOI] [PubMed] [Google Scholar]
- Guo F. Q., Okamoto M., Crawford N. M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302 100–103 10.1126/science.1086770 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Fernie A. R., Kaiser W. M, Van Dongen J. T. (2011a). On the origins of nitric oxide. Trends Plant Sci. 16 160–168 10.1016/j.tplants.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Hebelstrup K. H., Mur L. A., Igamberdiev A. U. (2011b). Plant hemoglobins: important players at the crossroads between oxygen and nitric oxide. FEBS Lett. 585 3843–3849 10.1016/j.febslet.2011.10.036 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Igamberdiev A. U. (2011). The anoxic plant mitochondrion as a nitrite: NO reductase. Mitochondrion 11 537–543 10.1016/j.mito.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Igamberdiev A. U. (2013). Recommendations of using at least two different methods for measuring NO. Front. Plant Sci. 4:58 10.3389/fpls.2013.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Tang R.-H., Hao Y., Stevens R. D., Cook C. W., Ahn S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305 1968–1971 10.1126/science.1098837 [DOI] [PubMed] [Google Scholar]
- Hebelstrup K. H., Igamberdiev A. U., Hill R. D. (2007). Metabolic effects of hemoglobin gene expression in plants. Gene 398 86–93 10.1016/j.gene.2007.01.039 [DOI] [PubMed] [Google Scholar]
- Hebelstrup K. H., Jensen E. O. (2008). Expression of NO scavenging hemoglobin is involved in the timing of bolting in Arabidopsis thaliana. Planta 227 917–927 10.1007/s00425-007-0667-z [DOI] [PubMed] [Google Scholar]
- Hendricks S. B., Taylorson R. B. (1974). Promotion of seed germination by nitrate, nitrite, hydroxylamine, and ammonium salts. Plant Physiol. 54 304–309 10.1104/pp.54.3.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005). Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell. Biol. 6 150–166 10.1038/nrm1569 [DOI] [PubMed] [Google Scholar]
- Hilhorst H. W., Karssen C. M. (1988). Dual effect of light on the gibberellin- and nitrate-stimulated seed germination of Sisymbrium officinale and Arabidopsis thaliana. Plant Physiol. 86 591–597 10.1104/pp.86.2.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R. D. (2012). Non-symbiotic haemoglobins – what’s happening beyond nitric oxide scavenging? AoB Plants 2012 pls004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.-D., Hu L.-Y., Li Y.-H., Zhang F.-Q., Zhang H. (2007). Protective roles of nitric oxide on germination and antioxidant metabolism in wheat seeds under copper stress. Plant Growth Regul. 53 173–183 10.1007/s10725-007-9216-9 [DOI] [Google Scholar]
- Huang X., Von Rad U., Durner J. (2002). Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215 914–923 10.1007/s00425-002-0828-z [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U., Bykova N. V., Shah J. K., Hill R. D. (2010). Anoxic nitric oxide cycling in plants: participating reactions and possible mechanisms. Physiol. Plant 138 393–404 10.1111/j.1399-3054.2009.01314.x [DOI] [PubMed] [Google Scholar]
- Igamberdiev A. U., Hill R. D. (2004). Nitrate, NO and haemoglobin in plant adaptation to hypoxia: an alternative to classic fermentation pathways. J. Exp. Bot. 55 2473–2482 10.1093/jxb/erh272 [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H. (2009). Protein tyrosine nitration–an update. Arch. Biochem. Biophys. 484 117–121 10.1016/j.abb.2008.10.034 [DOI] [PubMed] [Google Scholar]
- Jacobsen J. V., Barrero J. M., Hughes T., Julkowska M., Taylor J. M., Xu Q., et al. (2013). Roles for blue light, jasmonate and nitric oxide in the regulation of dormancy and germination in wheat grain (Triticum aestivum L.). Planta 238 121–138 10.1007/s00425-013-1878-0 [DOI] [PubMed] [Google Scholar]
- Jacques S., Ghesquiere B., Van Breusegem F., Gevaert K. (2012). Plant proteins under oxidative attack. Proteomics 13 932–940 10.1002/pmic.201200237 [DOI] [PubMed] [Google Scholar]
- Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001). Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell. Biol. 3 193–197 10.1038/35055104 [DOI] [PubMed] [Google Scholar]
- Jasid S., Simontacchi M., Puntarulo S. (2008). Exposure to nitric oxide protects against oxidative damage but increases the labile iron pool in sorghum embryonic axes. J. Exp. Bot. 59 3953–3962 10.1093/jxb/ern235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B. (2004). Nitrated lipids: a class of cell-signaling molecules. Proc. Natl. Acad. Sci. U.S.A. 101 11527–11528 10.1073/pnas.0404309101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall S. L., Hellwege A., Marriot P., Whalley C., Graham I. A., Penfield S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23 2568–2580 10.1105/tpc.111.087643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Hirotani M., Nakatsubo N., Kikuchi K., Urano Y., Higuchi T., et al. (2001). Bioimaging of nitric oxide with fluorescent indicators based on the rhodamine chromophore. Anal. Chem. 73 1967–1973 10.1021/ac001136i [DOI] [PubMed] [Google Scholar]
- Kojima H., Nakatsubo N., Kikuchi K., Kawahara S., Kirino Y., Nagoshi H., et al. (1998). Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal. Chem. 70 2446–2453 10.1021/ac9801723 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5 33–36 10.1016/S1369-5266(01)00219-9 [DOI] [PubMed] [Google Scholar]
- Kopyra M., Gwóźdź E. A. (2003). Nitric oxide stimulates seed germination and counteracts the inhibitory effect of heavy metals and salinity on root growth of Lupinus luteus. Plant Physiol. Biochem. 41 1011–1017 10.1016/j.plaphy.2003.09.003 [DOI] [Google Scholar]
- Krouk G., Crawford N. M., Coruzzi G. M., Tsay Y. F. (2010). Nitrate signaling: adaptation to fluctuating environments. Curr. Opin. Plant Biol. 13 266–273 10.1016/j.pbi.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Lamattina L., Garcia-Mata C., Graziano M., Pagnussat G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54 109–136 10.1146/annurev.arplant.54.031902.134752 [DOI] [PubMed] [Google Scholar]
- Leymarie J., Vitkauskaite G., Hoang H. H., Gendreau E., Chazoule V., Meimoun P., et al. (2012). Role of reactive oxygen species in the regulation of Arabidopsis seed dormancy. Plant Cell Physiol. 53 96–106 10.1093/pcp/pcr129 [DOI] [PubMed] [Google Scholar]
- Lindermayr C., Durner J. (2009). S-nitrosylation in plants: pattern and function. J. Proteomics 73 1–9 10.1016/j.jprot.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Durner J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137 921–930 10.1104/pp.104.058719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y., Yu X., Cui D. Y., Sun M. H., Sun W. N., Tang Z. C., et al. (2007). The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 17 638–649 10.1038/cr.2007.34 [DOI] [PubMed] [Google Scholar]
- Liu Y., Shi L., Ye N., Liu R., Jia W., Zhang J. (2009). Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183 1030–1042 10.1111/j.1469-8137.2009.02899.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Ye N., Liu R., Chen M., Zhang J. (2010). H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 61 2979–2990 10.1093/jxb/erq125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Mclachlin D. T., Chait B. T., Chua N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 317–328 10.1046/j.1365-313X.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Lounifi I., Arc E., Molasiotis A., Job D., Rajjou L., Tanou G. (2012). Interplay between protein carbonylation and nitrosylation in plants. Proteomics 13 568–578 10.1002/pmic.201200304 [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J., Colom-Moreno R., Leon J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62 3501–3517 10.1093/jxb/err042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J., Leon J. (2010). Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152 891–903 10.1104/pp.109.148023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludidi N., Gehring C. (2003). Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 278 6490–6494 10.1074/jbc.M210983200 [DOI] [PubMed] [Google Scholar]
- Manjunatha G., Gupta K. J., Lokesh V., Mur L. A., Neelwarne B. (2012). Nitric oxide counters ethylene effects on ripening fruits. Plant Signal. Behav. 7 476–483 10.4161/psb.19523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C., Wong J. H., Buchanan B. B. (2003). Thioredoxin and germinating barley: targets and protein redox changes. Planta 216 454–460 [DOI] [PubMed] [Google Scholar]
- Matakiadis T., Alboresi A., Jikumaru Y., Tatematsu K., Pichon O., Renou J. P., et al. (2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149 949–960 10.1104/pp.108.126938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla A. J, Rodriguez-Gacio Mdel C. (2013). Non-symbiotic hemoglobins in the life of seeds. Phytochemistry 87 7–15 10.1016/j.phytochem.2012.11.016 [DOI] [PubMed] [Google Scholar]
- Melo P. M., Silva L. S., Ribeiro I., Seabra A. R., Carvalho H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157 1505–1517 10.1104/pp.111.186056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel R. R. (2007). Biology of the molybdenum cofactor. J. Exp. Bot. 58 2289–2296 10.1093/jxb/erm024 [DOI] [PubMed] [Google Scholar]
- Moon K. H., Hood B. L., Kim B. J., Hardwick J. P., Conrads T. P., Veenstra T. D., et al. (2006). Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology 44 1218–1230 10.1002/hep.21372 [DOI] [PubMed] [Google Scholar]
- Moreau M., Lindermayr C., Durner J., Klessig D. F. (2010). NO synthesis and signaling in plants – where do we stand? Physiol. Plant. 138 372–383 10.1111/j.1399-3054.2009.01308.x [DOI] [PubMed] [Google Scholar]
- Muller K., Tintelnot S., Leubner-Metzger G. (2006). Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47 864–877 10.1093/pcp/pcj059 [DOI] [PubMed] [Google Scholar]
- Mur L. A., Mandon J., Cristescu S. M., Harren F. J., Prats E. (2011). Methods of nitric oxide detection in plants: a commentary. Plant Sci. 181 509–519 10.1016/j.plantsci.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20 55–67 10.1017/S0960258510000012 [DOI] [Google Scholar]
- Neill S., Barros R., Bright J., Desikan R., Hancock J., Harrison J., et al. (2008). Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 59 165–176 10.1093/jxb/erm293 [DOI] [PubMed] [Google Scholar]
- Nikitovic D., Holmgren A. (1996). S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271 19180–19185 10.1074/jbc.271.32.19180 [DOI] [PubMed] [Google Scholar]
- Nonogaki H. (2006). Seed germination – the biochemical and molecular mechanisms. Breed. Sci. 56 93–105 10.1270/jsbbs.56.93 [DOI] [Google Scholar]
- Nonogaki H., Chen F., Bradford K. J. (2007). “Mechanisms and genes involved in germination sensu stricto,” in Annual Plant Reviews, Vol. 27 Seed Development, Dormancy and Germination eds Bradford K. J., Nonogaki H. (New Jersey: Blackwell Publishing Ltd; ) 264–304 10.1002/9780470988848.ch11 [DOI] [Google Scholar]
- Palmieri M. C., Lindermayr C., Bauwe H., Steinhauser C., Durner J. (2010). Regulation of plant glycine decarboxylase by S-nitrosylation and glutathionylation. Plant Physiol. 152 1514–1528 10.1104/pp.109.152579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M. C., Sell S., Huang X., Scherf M., Werner T., Durner J., et al. (2008). Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J. Exp. Bot. 59 177–186 10.1093/jxb/erm345 [DOI] [PubMed] [Google Scholar]
- Parani M., Rudrabhatla S., Myers R., Weirich H., Smith B., Leaman D. W., et al. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2 359–366 10.1111/j.1467-7652.2004.00085.x [DOI] [PubMed] [Google Scholar]
- Perazzolli M., Dominici P., Romero-Puertas M. C., Zago E., Zeier J., Sonoda M., et al. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16 2785–2794 10.1105/tpc.104.025379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péres da Rocha Oliveiros Marciano D., Toledo Ramos F., Neiva Alvim M., Ronaldo Magalhaes J., Giovanni Costa França M. (2010). Nitric oxide reduces the stress effects of aluminum on the process of germination and early root growth of rice. J. Plant Nutr. Soil Sci. 173 885–891 10.1002/jpln.200900312 [DOI] [Google Scholar]
- Piterkova J., Luhova L., Hofman J., Tureckova V., Novak O., Petrivalsky M., et al. (2012). Nitric oxide is involved in light-specific responses of tomato during germination under normal and osmotic stress conditions. Ann. Bot. 110 767–776 10.1093/aob/mcs141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchet E., Kaiser W. M. (2006a). Nitric oxide (NO) detection by DAF fluorescence and chemiluminescence: a comparison using abiotic and biotic NO sources. J. Exp. Bot. 57 3043–3055 10.1093/jxb/erl070 [DOI] [PubMed] [Google Scholar]
- Planchet E., Kaiser W. M. (2006b). Nitric oxide production in plants: facts and fictions. Plant Signal. Behav. 1 46–51 10.4161/psb.1.2.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverari A., Molesini B., Pezzotti M., Buonaurio R., Marte M., Delledonne M. (2003). Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant Microbe Interact. 16 1094–1105 10.1094/MPMI.2003.16.12.1094 [DOI] [PubMed] [Google Scholar]
- Rajjou L., Belghazi M., Huguet R., Robin C., Moreau A., Job C., et al. (2006). Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 141 910–923 10.1104/pp.106.082057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L., Duval M., Gallardo K., Catusse J., Bally J., Job C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63 507–533 10.1146/annurev-arplant-042811-105550 [DOI] [PubMed] [Google Scholar]
- Rajjou L., Gallardo K., Debeaujon I., Vandekerckhove J., Job C., Job D. (2004). The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134 1598–1613 10.1104/pp.103.036293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L., Lovigny Y., Groot S. P., Belghazi M., Job C., Job D. (2008). Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol. 148 620–641 10.1104/pp.108.123141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E. H., Smith R. D. (1977). “Dormancy and the pentose phosphate pathway,” in The Physiology and Biochemistry of Seed Dormancy and Germination ed. Khan A. A. (Amsterdam: Elsevier; ) 385–411 [Google Scholar]
- Romero-Puertas M. C., Campostrini N., Matte A., Righetti P. G., Perazzolli M., Zolla L., et al. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8 1459–1469 10.1002/pmic.200700536 [DOI] [PubMed] [Google Scholar]
- Ross E. J., Shearman L., Mathiesen M., Zhou Y. J., Arredondo-Peter R., Sarath G., et al. (2001). Nonsymbiotic hemoglobins in rice are synthesized during germination and in differentiating cell types. Protoplasma 218 125–133 10.1007/BF01306602 [DOI] [PubMed] [Google Scholar]
- Saini H. S., Bassi P. K., Spencer M. S. (1985). Seed germination in Chenopodium album L.: relationships between nitrate and the effects of plant hormones. Plant Physiol. 77 940–943 10.1104/pp.77.4.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A., Ueda M., Morikawa H. (2002). Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 515 20–24 10.1016/S0014-5793(02)02414-6 [DOI] [PubMed] [Google Scholar]
- Sano N., Permana H., Kumada R., Shinozaki Y., Tanabata T., Yamada T., et al. (2012). Proteomic analysis of embryonic proteins synthesized from long-lived mRNAs during germination of rice seeds. Plant Cell Physiol. 53 687–698 10.1093/pcp/pcs024 [DOI] [PubMed] [Google Scholar]
- Sarath G., Bethke P. C., Jones R., Baird L. M., Hou G., Mitchell R. B. (2006). Nitric oxide accelerates seed germination in warm-season grasses. Planta 223 1154–1164 10.1007/s00425-005-0162-3 [DOI] [PubMed] [Google Scholar]
- Sen S. (2010). S-nitrosylation process acts as a regulatory switch for seed germination in wheat. Am. J. Plant Physiol. 5 122–132 10.3923/ajpp.2010.122.132 [DOI] [Google Scholar]
- Shi H. T., Li R. J., Cai W., Liu W., Wang C. L., Lu Y. T. (2012). Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant Cell Physiol. 53 344–357 10.1093/pcp/pcr181 [DOI] [PubMed] [Google Scholar]
- Simontacchi M., Garcia-Mata C., Bartoli C. G., Santa-Maria G. E., Lamattina L. (2013). Nitric oxide as a key component in hormone-regulated processes. Plant Cell Rep. 32 853–866 10.1007/s00299-013-1434-1 [DOI] [PubMed] [Google Scholar]
- Simontacchi M., Jasid S., Puntarulo S. (2004). Nitric oxide generation during early germination of sorghum seeds. Plant Sci. 167 839–847 10.1016/j.plantsci.2004.05.028 [DOI] [Google Scholar]
- Simontacchi M., Jasid S., Puntarulo S. (2007). “Enzymatic sources of nitric oxide during seed germination,” in Nitric Oxide in Plant Growth, Development and Stress Physiology eds Lamattina L., Polacco J. (Berlin, Heidelberg: Springer; ) 73–90 [Google Scholar]
- Sirova J., Sedlarova M., Piterkova J., Luhova L., Petrivalsky M. (2011). The role of nitric oxide in the germination of plant seeds and pollen. Plant Sci. 181 560–572 10.1016/j.plantsci.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Sliwinska E., Bassel G. W., Bewley J. D. (2009). Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 60 3587–3594 10.1093/jxb/erp203 [DOI] [PubMed] [Google Scholar]
- Spickett C. M., Pitt A. R., Morrice N., Kolch W. (2006). Proteomic analysis of phosphorylation, oxidation and nitrosylation in signal transduction. Biochim. Biophys. Acta 1764 1823–1841 10.1016/j.bbapap.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Spyrakis F., Bruno S., Bidon-Chanal A., Luque F. J., Abbruzzetti S., Viappiani C., et al. (2011). Oxygen binding to Arabidopsis thaliana AHb2 nonsymbiotic hemoglobin: evidence for a role in oxygen transport. IUBMB Life 63 355–362 10.1002/iub.470 [DOI] [PubMed] [Google Scholar]
- Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., et al. (1992a). S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U.S.A. 89 444–448 10.1073/pnas.89.1.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J. S., Singel D. J., Loscalzo J. (1992b). Biochemistry of nitric oxide and its redox-activated forms. Science 258 1898–1902 10.1126/science.1281928 [DOI] [PubMed] [Google Scholar]
- Tanou G., Job C., Rajjou L., Arc E., Belghazi M., Diamantidis G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60 795–804 10.1111/j.1365-313X.2009.04000.x [DOI] [PubMed] [Google Scholar]
- Terrile M. C., Paris R., Calderon-Villalobos L. I., Iglesias M. J., Lamattina L., Estelle M., et al. (2012). Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 70 492–500 10.1111/j.1365-313X.2011.04885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J., Rolletschek H., Friedel S., Lunn J. E., Nguyen T. H., Feil R., et al. (2011). Seed-specific elevation of non-symbiotic hemoglobin AtHb1: beneficial effects and underlying molecular networks in Arabidopsis thaliana. BMC Plant Biol. 11:48 10.1186/1471-2229-11-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelle E., Delledonne M. (2008). Methods for nitric oxide detection during plant–pathogen interactions. Methods Enzymol. 437 575–594 10.1016/S0076-6879(07)37029-8 [DOI] [PubMed] [Google Scholar]
- Vandelle E., Delledonne M. (2011). Peroxynitrite formation and function in plants. Plant Sci. 181 534–539 10.1016/j.plantsci.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Vigeolas H., Huhn D., Geigenberger P. (2011). Nonsymbiotic hemoglobin-2 leads to an elevated energy state and to a combined increase in polyunsaturated fatty acids and total oil content when overexpressed in developing seeds of transgenic Arabidopsis plants. Plant Physiol. 155 1435–1444 10.1104/pp.110.166462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ries A., Wu K., Yang A., Crawford N. M. (2010). The Arabidopsis prohibitin gene PHB3 functions in nitric oxide-mediated responses and in hydrogen peroxide-induced nitric oxide accumulation. Plant Cell 22 249–259 10.1105/tpc.109.072066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K., Muller K., Leubner-Metzger G. (2011). First off the mark: early seed germination. J. Exp. Bot. 62 3289–3309 10.1093/jxb/err030 [DOI] [PubMed] [Google Scholar]
- Wilkinson J. Q., Crawford N. M. (1993). Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. 239 289–297 [DOI] [PubMed] [Google Scholar]
- Wilson I. D., Neill S. J., Hancock J. T. (2008). Nitric oxide synthesis and signalling in plants. Plant Cell Environ. 31 622–631 10.1111/j.1365-3040.2007.01761.x [DOI] [PubMed] [Google Scholar]
- Wink D. A., Mitchell J. B. (1998). Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. Med. 25 434–456 10.1016/S0891-5849(98)00092-6 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. G., Tian Q. Y., Zhang W. H. (2007). Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 144 206–217 10.1104/pp.107.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Jiang D., Liu F., Dai T., Liu W., Jing Q., et al. (2009). Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environ. Exp. Bot. 67 222–227 10.1016/j.envexpbot.2009.05.002 [DOI] [Google Scholar]