Abstract
The prevalence of antibiotic resistance has resulted in the need for new approaches to be developed to combat previously easily treatable infections. The main aim of this work was to establish the potential of the synthetic α-diimine chromium(III) and ruthenium(II) complexes (where the α-diimine ligands are bpy = 2,2-bipyridine, phen = 1,10-phenanthroline, and dppz = dipyrido[3,2-a:2′,3′-c]-phenazine) like [Cr(phen)3]3+, [Cr(phen)2(dppz)]3+, [Ru(phen)3]2+, and [Ru(bpy)3]2+ as antibacterial agents by generating oxidative stress. The [Cr(phen)3]3+ and [Cr(phen)2(dppz)]3+ complexes showed activity against Gram positive and Gram negative bacteria with minimum inhibitory concentrations (MICs) ranging from 0.125 μg/mL to 1 μg/mL, while [Ru(phen)3]2+ and [Ru(bpy)3]2+ do not exhibit antimicrobial activity against the two bacterial genera studied at the concentration range used. When ciprofloxacin was combined with [Cr(phen)3]3+ for the inhibition of Staphylococcus aureus and Escherichia coli, an important synergistic effect was observed, FIC 0.066 for S. aureus and FIC 0.064 for E. coli. The work described here shows that chromium(III) complexes are bactericidal for S. aureus and E. coli. Our results indicate that α-diimine chromium(III) complexes may be interesting to open new paths for metallodrug chemotherapy against different bacterial genera since some of these complexes have been found to exhibit remarkable antibacterial activities.
1. Introduction
Since their discovery, antibiotics have played an important role in health care. However, the increasing emergence of antibiotic resistance among a variety of microbial pathogens stimulates intensive research efforts with the aim of identifying alternative therapeutic approaches [1].
The chemistry of metallic complexes with heterocyclic ligands has attracted the interest of both inorganic and bioinorganic chemists in recent years, and the field of coordination chemistry of metallic complexes has undergone a remarkable growth during the past few decades and became a growing class of research [2]. This enormous growth is attributed to the synthesis of a large number and variety of synthetic ligands (chelates) which behave as coordinating agents for metal ions [3]. It is known that 2,2′-bipyridine and 1,10-phenanthroline chelators act as potential antitumor agents [4, 5] and several metal chelates possess antibacterial, antifungal, antiviral and anticancer activities. However, in several cases, it has been found that metal chelates have more antimicrobial activity than the chelating agents themselves [6].
The study of the chromium complexes in coordination chemistry is of great significance due to their various applications in biological processes. In vitro studies have demonstrated that chromium in the proper ligand environment can lead to DNA damage, plasmid cleavage, and protein cleavage [7–9]. Some reports showed chromium complexes as the cause of oxidation of DNA by binding with guanine [10, 11]. Additionally, octahedral transition metal complexes containing three bipyridine or phenanthroline ligands have been shown to be groove binders or possible partial DNA intercalators [12, 13], whereas some chromium(III) Schiff base complexes prefer nonintercalative mode of binding to DNA [7]. On the other hand, it has been reported that the bovine serum albumin (BSA) protein damage brought about by chromium(III) complexes depends on the nature of the coordinated ligand and the nature of the metal complex [9].
As part of an ongoing program to investigate the interaction of transition metal with double-helical DNA, we found that the α-diimine chromium(III) complexes are associated with DNA by noncovalent interactions. The chromium(III) complexes are capable of producing plasmidic DNA photodamage with high efficiency of photocleavage [14–16]. This leads to consider these complexes as attractive photosensitizing agents to be used in photodynamic therapy (PDT).
On the other hand, coordination complexes of transition metals have been widely studied due to their antimicrobial properties [17–21]. The importance of chromium as an environmental pollutant and its involvement in the toxicity in animals and plants have made the study of the interactions of derivatives of this metal with microbial cells in a fertile field for both basic research and applied science [22]. In this sense, it was reported that chromium(III) complexes containing oxalohydrazide and quinoxaline as ligands would have microbiological activity [23, 24], reducing virulence and antibiotic resistance, causing loss of the plasmid of Shigella dysenteriae that contains genes encoding virulence factors responsible for this organism's ability to penetrate and multiply into the cells [25]. Furthermore, it has been reported that chromium(III) complexes containing amino acids as ligands have antibacterial and antifungal activities [26], and the authors suggest that the mechanism of antibacterial activity of this complexes would be due to oxidative DNA damage. Emerging reports on the biotoxicity of chromium(III) complexes have emphasized the effects of the coordinated ligand on metal activity [27].
Diverse substances affect the reactive oxygen species (ROS) generation in bacterial cells, and they have the capacity to undergo redox cycling, resulting in the generation of superoxide anion (O2 −) and hydrogen peroxide (H2O2). We have described that several antibiotics induce oxidative stress in bacteria, among them ciprofloxacin [28–31].
The aim of this investigation was to study the potential antimicrobial activity of certain chromium(III) and ruthenium(II) complexes and their combination with ciprofloxacin, and this will provide an understanding of the potential enhancing of the activity of this antibiotic by generating oxidative stress in two different bacterial genera.
2. Materials and Methods
2.1. Reagents
1,10-Phenanthroline (phen), tris(1,10-phenanthroline)ruthenium(II) chloride ([Ru(phen)3]Cl2), and tris(2,2′-bipyridine)ruthenium(II) chloride ([Ru(bpy)3]Cl2) were purchased from Sigma and used as received. Tris-phenanthroline chromium (III) perchlorate ([Cr(phen)3](ClO4)3) was obtained from previous studies, synthesized according to the literature, and recrystallized at least three times. The ligand dppz was synthesized and purified according to the literature [32]. Bis(1,10-phenanthroline)dipyrido[3,2-a:2′,3′-c]-phenazine chromium(III) triflate ([Cr(phen)2(dppz)](CF3SO3)3) was synthesized with slight modifications according to a previously reported procedure [33]. The stock solutions [Cr(phen)3]3+ and [Cr(phen)2(dppz)]3+ were prepared in phosphate buffer saline (PBS, pH 7.4), and the concentration of chromium complexes was calculated using molar extinction coefficients of ε 354 = 4200 M-1 cm−1 and ε 360 = 13900 M-1 cm−1, respectively [33]. CrCl3 · 6H2O (Fluka, purity 98%) was used without further purification. Millipore Milli Q water was used for preparing buffer solutions. All other chemical reagents were of analytical grade.
2.2. Minimal Inhibitory Concentration (MIC) Determination
The antimicrobial activity of the compounds in Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 was evaluated by using the standard tube dilution method on Mueller Hinton broth (MH, Britania). Strains coming from cultures of 24 h in Mueller Hinton medium were diluted to 106 colony-forming units (CFU) per mL and incubated for 10 min at 37°C; the compounds were then added at different concentrations (from 0.004 to 2 μg/mL). Bacterial growth was observed at 18 h of incubation, following the indications of the Clinical and Laboratory Standards Institute [34]. For comparison, chromium(III) and ruthenium(II) complexes were tested. In addition, the MICs of the ligands (bpy, phen and dppz) and CrCl3 · 6H2O were calculated. Serial dilutions in phosphate buffer solution (PBS), pH 7.4 of the complexes (0.5 mL), were made. The concentrations of the compounds were ranged from 0.004 to 2 μg/mL. Dilutions of a standard antibiotic (from 0.008 to 4 μg/mL of ciprofloxacin) were also prepared in the same manner. An overnight culture of each microorganism was diluted to achieve a cell density in the range from 105 to 107 CFU/mL. The cell suspension (0.5 mL) was inoculated into each tube to give a total volume of 1 mL. The lowest concentration of each complex that prevented bacterial growth was considered to be the MIC. Inoculated complexes-free broths were used as negative controls. Viable bacterial counts were obtained for control samples by plating serial dilutions on MH agar plates, followed by aerobic incubation at 37°C for 18 h for E. coli and S. aureus.
2.3. Time-Kill Study
The bactericidal activity was explored by the time-kill method according to CLSI [34]. Kill curves were constructed by plotting the CFU/mL surviving at each time point in the presence and absence of [Cr(phen)3]3+ in E. coli ATCC 25922 and S. aureus ATCC 29213.
2.4. Checkerboard Assay
The interactions between the chromium(III) complexes and ciprofloxacin were evaluated by the checkerboard method [35]. Bacterial growth inhibition resulting from the interactions was determined by the microdilution test. The concentrations of each tested agent used in the combinations corresponded to serial 2-fold dilutions from their MIC values. The fractional inhibitory concentration (FIC) was calculated using the MIC from the checkerboard assay and the MIC of each compound alone, obtained in parallel in the same assay, according to the following formula: FIC = MIC of antimicrobial agent in combination/MIC of antimicrobial agent alone. Then, the synergistic effect was evaluated by calculating the FICindex (FICI) for each combination, by adding the individual FIC values. An FICI of 0.5 indicated synergy for the combination. When it fell between 0.5 and 1, it was defined as an additive effect, and between 1.0 and 4.0 it was classified as “no interaction.” Finally, an FICI > 4.0 indicated antagonism between the components in the combination.
2.5. Determination of ROS by Chemiluminescence (CL) Assay
Oxidative stress of S. aureus ATCC 29213 and E. coli ATCC 25922 was investigated in a BioOrbit luminometer. Bacteria were cultured in triptein soy broth (TSB) at 37°C for 24 h. 0.1 mL of bacterial suspension was incubated with 0.1 mL of 75 μg/mL lucigenin (bis-N-methylacridinium dinitrate), 0.1 mL of PBS, pH 7.2 and 0.1 mL of ciprofloxacin (from 0.033 to 4 μg/mL) and 0.1 mL of [Cr(phen)3]3+ (from 0.033 to 4 μg/mL). Finally, the reaction was triggered with 0.1 mL of dimethylsulfoxide (DMSO) in the moment of determination. Controls of basal production of ROS were performed with bacteria in absence of antibiotic. The light emitted by ROS was expressed as relative light unities (RLU) at different times in seconds.
3. Results
3.1. Minimal Inhibitory Concentration (MIC) Determination
Four chemically macrocyclic complexes and their ligands (Figure 1) were screened for their in vitro antibacterial activity against two bacteria, the Gram positive bacterium S. aureus and the Gram negative bacterium E. coli. Despite the apparent similarities of the compounds, the preliminary experiments showed that they present different antimicrobial activities.
Figure 1.
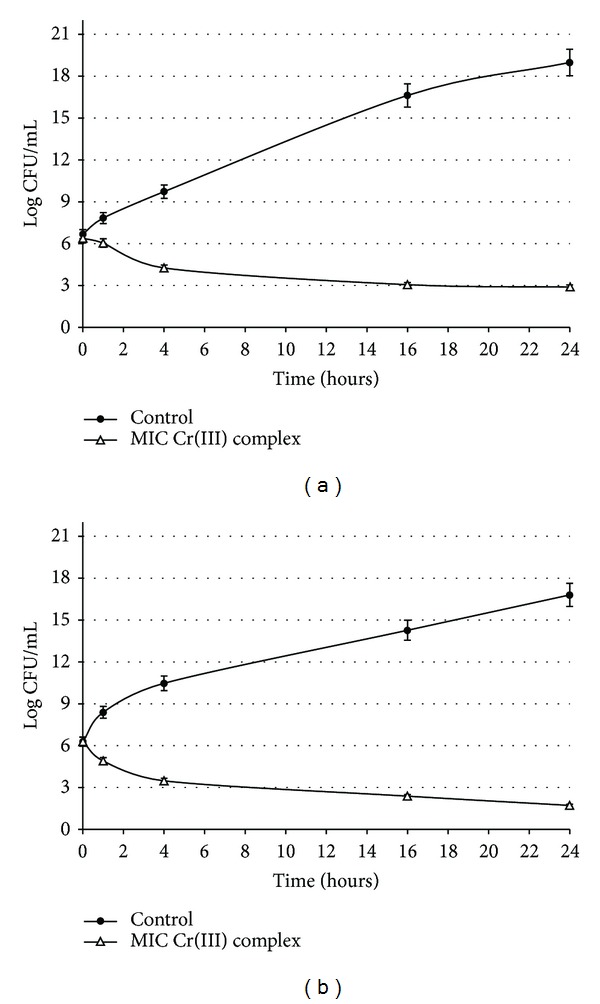
Time-kill kinetics of [Cr(phen)3]3+ complex at minimum inhibitory concentration (MIC) (Δ) and control (•). (a) Staphylococcus aureus ATCC 29213. (b) Escherichia coli ATCC 25922.
Chromium(III) complexes demonstrated antimicrobial activity against Gram positive and Gram negative bacteria. The Ru(II) complexes showed less activity than the Cr(III) complexes (Table 1). The bpy, phen, dppz, and CrCl3 · 6H2O (Cr3+ as free ion) show poor activity in comparison with that of the metal complexes against the two bacterial strains. Among all complexes of the series under test for determination of MIC (Figure 1), [Cr(phen)2(dppz)]3+ was found to be the most potent. It registered MIC as shown by ciprofloxacin against two bacterial strains.
Table 1.
Minimum inhibitory concentrations (MICs) shown by complexes and ligands against test bacteria.
| MIC in µg/mL (mmol/L) | ||
|---|---|---|
| Staphylococcus aureus | Escherichia coli | |
| [Cr(phen)3]3+ | 1 (0.0011) | 0.25 (0.0003) |
| [Cr(phen)2(dppz)]3+ | 0.5 (0.0004) | 0.125 (0.0001) |
| CrCl3·6 H2O | No activity | No activity |
| [Ru(phen)3]2+ | >1024 (>1.437) | >1024 (>1.437) |
| [Ru(bpy)3]2+ | >1024 (>1.599) | >1024 (>1.599) |
| Dipyrido[3,2-a:2′,3′-c]-phenazine | No activity | No activity |
| 1,10-Phenanthroline | No activity | No activity |
| 2,2′-Bipyridine | No activity | No activity |
| Ciprofloxacin | 0.5 (0.0013) | 0.125 (0.0003) |
E. coli was found to be more susceptible to [Cr(phen)2(dppz)]3+ than S. aureus, with an MIC for E. coli of 0.125 μg/mL and an MIC for S. aureus of 0.5 μg/mL while the MIC for E. coli was 0.25 μg/mL and for S. aureus 1 μg/mL for [Cr(phen)3]3+. Unlike those found for the ligands or Cr3+ alone, synthesized metal complexes of chromium(III) showed high antimicrobial activities against E. coli and S. aureus.
3.2. Checkerboard Assay
When ciprofloxacin was combined with [Cr(phen)3]3+ for the inhibition of S. aureus and E. coli, an important synergistic effect was observed (Tables 2 and 3). A notable synergism was observed in E. coli (FIC 0.032) when the concentration of the ciprofloxacin was decreased to 0.016 μg/mL, equivalent to 15 times less than its MIC (Table 3). When the [Cr(phen)3]3+ was combined with ciprofloxacin, a surprising synergistic effect (FICIC+A = 0.096) was observed, even when the concentration of the former was decreased 4 times below its MIC and of the antibiotic 31 times below its individual MIC (Figures 2 and 3).
Table 2.
Minimum inhibitory concentrations (MICs) in µg/mL and fractional inhibitory concentration indices (FICIs) of [Cr(phen)3]3+ in combination with ciprofloxacin against Staphylococcus aureus ATCC 29213*.
| Ciprofloxacin (MIC = 0.5) | |||
|---|---|---|---|
| [Cr(phen)3]3+ (MIC = 1) | MICA−C | FIC | |
| MICC−A | FIC | 0.033 | 0.066 |
| 0.5 | 0.5 | FICIC+A = 0.566 | |
| 0.25 | 0.25 | FICIC+A = 0.316 | |
| 0.125 | 0.125 | FICIC+A = 0.191 | |
*MICC−A: MIC of [Cr(phen)3]3+ in combination with ciprofloxacin. MICA−C: MIC of ciprofloxacin in combination with [Cr(phen)3]3+. FIC: fractional inhibitory concentration. FICIC+A: fractional inhibitory concentration index (FIC of [Cr(phen)3]3+ plus FIC of ciprofloxacin).
Table 3.
Minimum inhibitory concentrations (MICs) in µg/mL and fractional inhibitory concentration indices (FICIs) of [Cr(phen)3]3+ in combination with ciprofloxacin against Escherichia coli ATCC 25922*.
| Ciprofloxacin (MIC = 0.25) | |||
|---|---|---|---|
| [Cr(phen)3]3+ (MIC = 0.125) | MICA−C | FIC | |
| MICC−A | FIC | 0.016 | 0.064 |
| 0.064 | 0.512 | FICIC+A = 0.576 | |
| 0.032 | 0.256 | FICIC+A = 0.320 | |
| 0.016 | 0.128 | FICIC+A = 0.192 | |
| 0.008 | 0.064 | FICIC+A = 0.128 | |
| 0.004 | 0.032 | FICIC+A = 0.096 | |
*MICC−A: MIC of [Cr(phen)3]3+ in combination with ciprofloxacin. MICA−C: MIC of ciprofloxacin in combination with [Cr(phen)3]3+. FIC: fractional inhibitory concentration. FICIC+A: fractional inhibitory concentration index (FIC of [Cr(phen)3]3+ plus FIC of ciprofloxacin).
Figure 2.
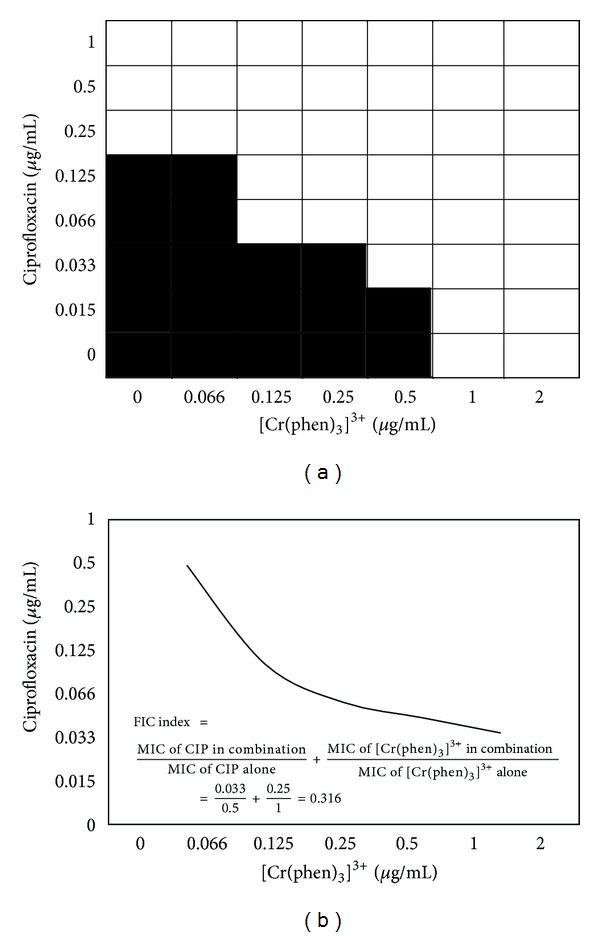
The fractional inhibitory concentration (FIC) index of [Cr(phen)3]3+ in Staphylococcus aureus ATCC 29213 using the checkerboard technique.
Figure 3.
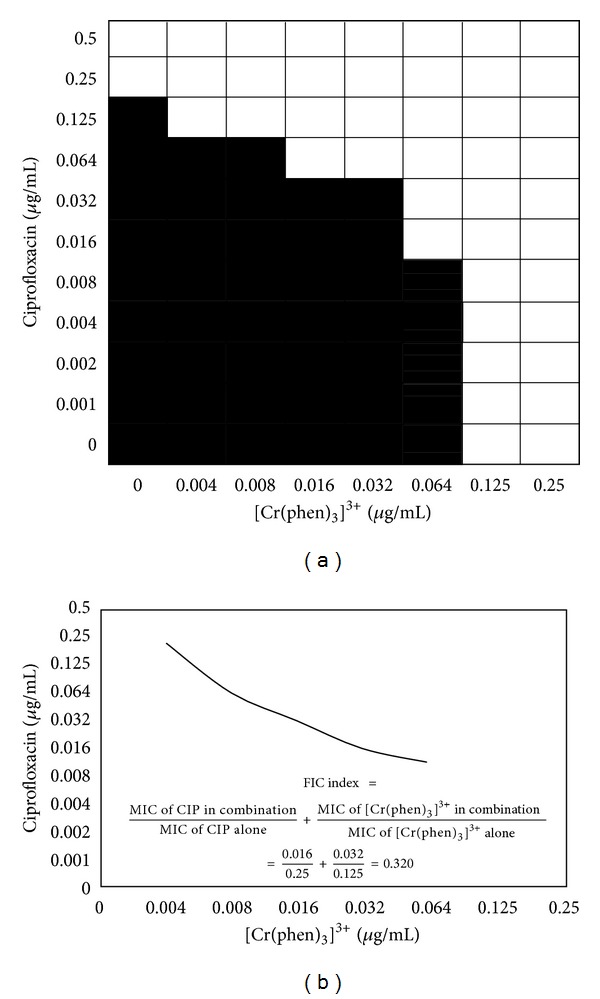
The fractional inhibitory concentration (FIC) index of [Cr(phen)3]3+ in Escherichia coli ATCC 25922 using the checkerboard technique.
3.3. Time-Kill Study
When [Cr(phen)3]3+ was tested against S. aureus (Figure 1) at MIC, a difference of 3 log10 in the number of viable cells compared to the control was observed after 16 h of incubation. When this complex was assayed against E. coli, the treatment at MIC reached differences of 3 log10 with respect to the control after 8 h of exposure. At this time, a complete bactericidal effect was observed.
3.4. Determination of ROS by Chemiluminescence (CL) Assay
Chemiluminescence assays indicated a higher degree of oxidative stress in S. aureus ATCC 29213 when it was treated with a combination of [Cr(phen)3]3+ at 1 μg/mL with ciprofloxacin at 0.5 μg/mL, and the maximum value of RLU obtained was 1.167 ± 0.016, representing an increase of 2-fold with respect to the control without antibiotic or the complex (Figure 4), while for the combination of [Cr(phen)3]3+ at 4 μg/mL (concentration supra MIC) with ciprofloxacin at 0.5 μg/mL, the increase in RLU was 3.9-fold with respect to the control (data not shown).
Figure 4.
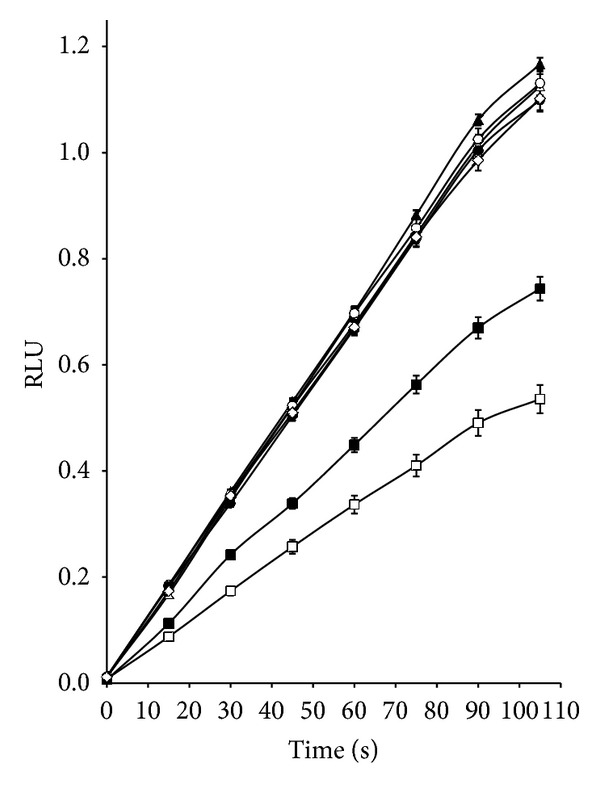
ROS generation in Staphylococcus aureus ATCC 29213 incubated with [Cr(phen)3]3+ at MIC concentration determined by chemiluminescence assay. Control (□); treated with 1 μg/mL [Cr(phen)3]3+ and 0.125 μg/mL ciprofloxacin (■); treated with 1 μg/mL [Cr(phen)3]3+ and 0.25 μg/mL ciprofloxacin (Δ); treated with 1 μg/mL [Cr(phen)3]3+ and 0.5 μg/mL ciprofloxacin (▲); treated with 1 μg/mL [Cr(phen)3]3+ and 1 μg/mL ciprofloxacin (◦); treated with 1 μg/mL [Cr(phen)3]3+ and 2 μg/mL ciprofloxacin (•); treated with 1 μg/mL [Cr(phen)3]3+ and 4 μg/mL ciprofloxacin (◊).
When E. coli was treated with a combination of [Cr(phen)3]3+ at 0.125 μg/mL with ciprofloxacin at 0.5 μg/mL, the maximum value of RLU obtained was 0.779 ± 0.040, representing an increase of 2-fold with respect to the control (Figure 5), while for the combination with lower concentrations of [Cr(phen)3]3+ and ciprofloxacin, 0.03 and 0.06 μg/mL, respectively, the increase of RLU with respect to the control was 4.8-fold.
Figure 5.
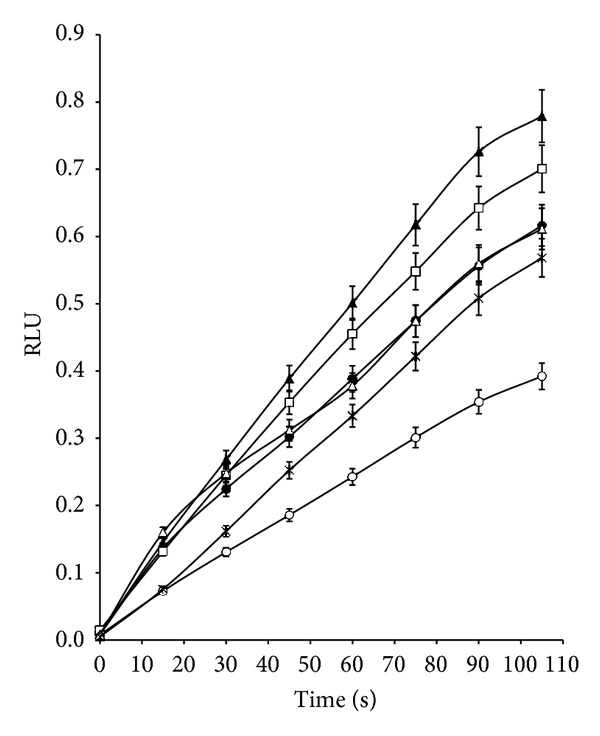
ROS generation in Escherichia coli ATCC 25922 incubated with [Cr(phen)3]3+ at MIC concentration determined by chemiluminescence assay. Control (◦); treated with 0.125 μg/mL [Cr(phen)3]3+ and 0.06 μg/mL ciprofloxacin (•); treated with 0.125 μg/mL [Cr(phen)3]3+ and 0.125 μg/mL ciprofloxacin (□); treated with 0.125 μg/mL [Cr(phen)3]3+ and 0.5 μg/mL ciprofloxacin (▲); treated with 0.125 μg/mL [Cr(phen)3]3+ and 1 μg/mL ciprofloxacin (Δ); treated with 0.125 μg/mL [Cr(phen)3]3+ and 2 μg/mL ciprofloxacin (∗).
These results were in agreement with the level of susceptibility of each strain to the combination of the complex with the antibiotic, since S. aureus was more resistant than E. coli, and this could have been related to less generation of ROS occurring in the Gram positive bacteria.
4. Discussion
Despite the range of antibacterial agents currently available, the development of new therapeutic alternatives to deal with pathogens is of great interest for researchers and pharmaceutical industries. In our biological experiments, by using chromium(III) complexes we have observed high biological activity against Gram negative and Gram positive bacteria. It may be concluded that the chromium(III) complexes used in the present work inhibit the growth of bacteria to a greater extent compared to ciprofloxacin, the reference drug. This fact contrasts with what was found for others chromium(III) complexes containing amino acids as a ligands against Gram negative and Gram positive bacteria, where the MIC values of the complexes are higher than the values for the standard antibiotic [26].
The efficiencies of the ligands and the chromium(III) complexes have been tested against Gram positive, S. aureus, and Gram negative, E. coli, microorganisms. It is not clear why the other compounds tested, [Ru(phen)3]2+ and [Ru(bpy)3]2+, did not show antimicrobial activity at the concentration range used. The results are presented in Table 1. The antimicrobial activity of CrCl3 · 6H2O has also been investigated. It has been found that CrCl3 · 6H2O does not exhibit antimicrobial activity at the concentration range used to assay the activity of the complexes in this work.
In general, for metal complexes showing antimicrobial activity, the following five principal factors should be considered: (i) the chelate effect, that is, ligands that are bound to metal ions in a bidentate fashion, such as the nitrogen-donor ligands phen and bpy, show higher antimicrobial efficiency towards complexes with unidentate nitrogen-donor ligands, for example, pyridine; (ii) the nature of the ligands; (iii) the total charge of the complex, generally the antimicrobial efficiency decreases in the order cationic > neutral > anionic complex, (iv) the nature of the ion neutralizing the ionic complex; (v) the nuclearity of the metal center in the complex; dinuclear centers are usually more active than mononuclear ones [36]. The first two of the five above-mentioned factors are present in our compounds, that is, the chelate effect provided by nitrogen-donor ligands (phen, bpy, and dppz) and the nature of the ligands.
It is evident (Table 1) that the antimicrobial activity of the chromium(III) complexes against S. aureus and E. coli is better than that of the ligands. A similar behavior was observed by Efthimiadou et al. for copper(II) complexes with nitrogen-donor heterocyclic ligands against P. aeruginosa, S. aureus, and E. coli [37]. Considering the nature of the nitrogen-donor heterocyclic ligand in the chromium(III) complexes, our results suggest that the inhibition of the growth of microorganisms increases in the order dppz > phen, where the [Cr(phen)2(dppz)]3+ complex was found to be the most potent.
Finally, when ciprofloxacin was combined with [Cr(phen)3]3+ for the inhibition of S. aureus and E. coli, an important synergistic effect was observed (Tables 2 and 3). We found that S. aureus was more resistant than E. coli to generation of ROS by both antimicrobial agents combined, ciprofloxacin and [Cr(phen)3]3+, this would be related to less increase of ROS in Gram positive bacteria (Figures 4 and 5).
The present study provides the first evidence of antimicrobial activity of the synthetic metallomolecules [Cr(phen)3]3+ and [Cr(phen)2(dppz)]3+. We have synthesized and characterized various α-diimine chromium(III) complexes with different ligands, and similar compounds of copper(II) have been recently synthesized, some of which may show a higher degree of antimicrobial activity [38]. Therefore, future studies will be directed to the antimicrobial activity of the newly synthesized complexes, as well as the activity of metal complexes on drug-resistant pathogenic bacteria.
Acknowledgments
The authors thank Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET), Secretaría de Ciencia y Técnica de la Universidad Nacional de Córdoba (SECyT), and Agencia Nacional de Promoción de la Ciencia y Técnica (ANPCYT) for financial support. María C. Becerra, Paulina L. Páez, and Gerardo A. Argüello are researchers from CONICET.
References
- 1.Taylor PW, Stapleton PD, Paul Luzio J. New ways to treat bacterial infections. Drug Discovery Today. 2002;7(21):1086–1091. doi: 10.1016/s1359-6446(02)02498-4. [DOI] [PubMed] [Google Scholar]
- 2.Del Carmen Fernández-Fernández M, Bastida R, MacÍas A, Valencia L, Pérez-Lourido P. Different nuclearities of M(II) nitrate complexes (M = Co, Ni, Cu and Cd) with a tetrapyridyl pendant-armed hexaazamacrocyclic ligand. Polyhedron. 2006;25(3):783–792. [Google Scholar]
- 3.Chaudhary A, Bansal N, Fahmi N, Singh RV. Tetraazamacrocycles of manganese and iron metals: synthesis, spectroscopic characterization and biological evaluation. Indian Journal of Chemistry A. 2004;43(2):320–322. [Google Scholar]
- 4.Krishnamurti C, Saryan LA, Petering DH. Effects of ethylenediaminetetraacetic acid and 1,10-phenanthroline on cell proliferation and DNA synthesis of Ehrlich ascites cells. Cancer Research. 1980;40(11):4092–4099. [PubMed] [Google Scholar]
- 5.Patel RN, Singh N, Shukla KK, et al. Characterization and biological activities of two copper(II) complexes with diethylenetriamine and 2,2′-bipyridine or 1,10-phenanthroline as ligands. Spectrochimica Acta A. 2005;62(1–3):261–268. doi: 10.1016/j.saa.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava RS. Synthesis, characterization and fungitoxicity of bidentate high-spin six coordinate 3rd metal complexes with N-(5-phenyl-1,3,4-thiadiazol-2-yl) aceta/benzamidines. Inorganica Chimica Acta. 1981;55:L71–L74. [Google Scholar]
- 7.Vijayalakshmi R, Kanthimathi M, Subramanian V, Nair BU. Interaction of DNA with [Cr(Schiff base)(H2O)2]ClO4 . Biochimica et Biophysica Acta. 2000;1475(2):157–162. doi: 10.1016/s0304-4165(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 8.Shrivastava HY, Nair BU. Chromium(III)-mediated structural modification of glycoprotein: impact of the ligand and the oxidants. Biochemical and Biophysical Research Communications. 2001;285(4):915–920. doi: 10.1006/bbrc.2001.5258. [DOI] [PubMed] [Google Scholar]
- 9.Shrivastava HY, Nair BU. Protein degradation by peroxide catalyzed by chromium (III): role of coordinated ligand. Biochemical and Biophysical Research Communications. 2000;270(3):749–754. doi: 10.1006/bbrc.2000.2492. [DOI] [PubMed] [Google Scholar]
- 10.Vijayalakshmi R, Subramanian V, Nair BU. A study of the interaction of Cr(III) complexes and their selective binding with B-DNA: a molecular modeling approach. Journal of Biomolecular Structure and Dynamics. 2002;19(6):1063–1071. doi: 10.1080/07391102.2002.10506809. [DOI] [PubMed] [Google Scholar]
- 11.Joudah L, Moghaddas S, Bose RN. DNA oxidation by peroxo-chromium(v) species: oxidation of guanosine to guanidinohydantoin. Chemical Communications. 2002;(16):1742–1743. doi: 10.1039/b205407h. [DOI] [PubMed] [Google Scholar]
- 12.Zeglis BM, Pierre VC, Barton JK. Metallo-intercalators and metallo-insertors. Chemical Communications. 2007;(44):4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X-W, Li J, Li H, Zheng K-C, Chao H, Ji L-N. Synthesis, characterization, DNA-binding and photocleavage of complexes [Ru(phen)2(6-OH-dppz)]2+ and [Ru(phen)2(6- NO2-dppz)]2+ . Journal of Inorganic Biochemistry. 2005;99(12):2372–2380. doi: 10.1016/j.jinorgbio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Toneatto J, Boero RA, Lorenzatti G, Argüello GA, Cabanillas AM. Binding and photocleavage properties of [Cr(phen)2(dppz)]3+ to DNA. Implications in Phototherapy. Biocell. 2009;33:p. 64. [Google Scholar]
- 15.Toneatto J, Lorenzatti G, Cabanillas AM, Argüello GA. Novel DNA photocleavage properties of [Cr(NN)3]3+ complexes. Inorganic Chemistry Communications. 2012;15:43–46. [Google Scholar]
- 16.Toneatto J, Boero RA, Lorenzatti G, Cabanillas AM, Argüello GA. New insights in the DNA-[Cr(phen)2(dppz)]3+ binding and photocleavage properties by the complex with an intercalating ligand. Journal of Inorganic Biochemistry. 2010;104(7):697–703. doi: 10.1016/j.jinorgbio.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Kamalakannan P, Venkappayya D. Synthesis and characterization of cobalt and nickel chelates of 5-dimethylaminomethyl-2-thiouracil and their evaluation as antimicrobial and anticancer agents. Journal of Inorganic Biochemistry. 2002;90(1-2):22–37. doi: 10.1016/s0162-0134(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 18.Islam MS, Farooque MA, Bodruddoza MAK, Mosaddik MA, Alam MS. Antimicrobial and toxicological studies of mixed ligand transition metal complexes of Schiff bases. International Journal of Biological Sciences. 2002;2:797–799. [Google Scholar]
- 19.Sultana C, Rahman MAA, Al-Bari MAA, et al. In vitro Antimicrobial screening of three cadmium complexes and two addition compounds of antimony and arsenic. Pakistan Journal of Biological Sciences. 2003;6:525–527. [Google Scholar]
- 20.Shakir M, Chingsubam P, Chishti H-T, Azim Y, Begum N. Synthesis and physico-chemical studies on transition metal complexes of macrocyclic ligand derived from 2,6-diacetylpyridine dihydrazone. Indian Journal of Chemistry A. 2004;43(3):556–561. [Google Scholar]
- 21.Singh DP, Kumar R, Malik V, Tyagi P. Template synthesis, spectroscopic studies and biological activities of macrocyclic complexes derived from thiocarbohydrazide and glyoxal. Journal of Enzyme Inhibition and Medicinal Chemistry. 2007;22(2):177–182. doi: 10.1080/14756360601051399. [DOI] [PubMed] [Google Scholar]
- 22.Cervantes C, Campos-García J, Devars S, et al. Interactions of chromium with microorganisms and plants. FEMS Microbiology Reviews. 2001;25:335–347. doi: 10.1111/j.1574-6976.2001.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 23.El-Asmy AA, El-Gammal OA, Radwan HA. Synthesis, characterization and biological study on Cr3+, ZrO2+, HfO2+ and UO2 complexes of oxalohydrazide and bis(3-hydroxyimino)butan-2-ylidene)-oxalohydrazide. Spectrochimica Acta A. 2010;76(5):496–501. doi: 10.1016/j.saa.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Anantha Lakshmi PV, Reddy PS, Raju VJ. Synthesis, characterization and antimicrobial activity of 3d transition metal complexes of a biambidentate ligand containing quinoxaline moiety. Spectrochimica Acta A. 2009;74(1):52–57. doi: 10.1016/j.saa.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Shrivastava HY, Devaraj SN, Nair BU. A Schiff base complex of chromium(III): an efficient inhibitor for the pathogenic and invasive potential of Shigella dysenteriae. Journal of Inorganic Biochemistry. 2004;98(2):387–392. doi: 10.1016/j.jinorgbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Sheikh C, Houssain MS, Easmin MS, Islam MS, Houssain MA, Rashid M. New coordination complexes of chromium as cytotoxic and antimicrobial agents. Pakistan Journal of Biological Sciences. 2004;7:335–339. [Google Scholar]
- 27.Shrivastava HY, Ravikumar T, Shanmugasundaram N, Babu M, Unni Nair B. Cytotoxicity studies of chromium(III) complexes on human dermal fibroblasts. Free Radical Biology and Medicine. 2005;38(1):58–69. doi: 10.1016/j.freeradbiomed.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Becerra MC, Sarmiento M, Páez PL, Argüello G, Albesa I. Light effect and reactive oxygen species in the action of ciprofloxacin on Staphylococcus aureus . Journal of Photochemistry and Photobiology B. 2004;76:13–18. doi: 10.1016/j.jphotobiol.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Becerra MC, Páez PL, Laróvere LE, Albesa I. Lipids and DNA oxidation in Staphylococcus aureus as a consequence of oxidative stress generated by ciprofloxacin. Molecular and Cellular Biochemistry. 2006;285(1-2):29–34. doi: 10.1007/s11010-005-9051-0. [DOI] [PubMed] [Google Scholar]
- 30.Páez PL, Becerra MC, Albesa I. Antioxidative mechanisms protect resistant strains of Staphylococcus aureus against ciprofloxacin oxidative damage. Fundamental and Clinical Pharmacology. 2010;24(6):771–776. doi: 10.1111/j.1472-8206.2009.00806.x. [DOI] [PubMed] [Google Scholar]
- 31.Páez PL, Becerra MC, Albesa I. Comparison of macromolecular oxidation by reactive oxygen species in three bacterial genera exposed to different antibiotics. Cell Biochemistry and Biophysics. 2011;61(3):467–472. doi: 10.1007/s12013-011-9227-z. [DOI] [PubMed] [Google Scholar]
- 32.Dickeson JE, Summers LA. Derivatives of 1,10-phenanthroline-5,6-quinone. Australian Journal of Chemistry. 1970;23:1023–1027. [Google Scholar]
- 33.Barker KD, Barnett KA, Connell SM, et al. Synthesis and characterization of heteroleptic Cr(diimine)3 complexes. Inorganica Chimica Acta. 2001;316(1-2):41–49. [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement; M100-S20 and M100 S20 Supplement. Wayne, Pa, USA, EE.UU, 2010.
- 35.Eliopoulos GM, Moellering RC., Jr. Antimicrobial combinations. In: Lorian V, editor. Antibiotics in Laboratory Medicine. 4th edition. Baltimore, Md, USA: Williams & Wilkins; 1996. pp. 432–492. [Google Scholar]
- 36.Psomas G, Dendrinou-Samara C, Philippakopoulos P, et al. CuII-herbicide complexes: structure and bioactivity. Inorganica Chimica Acta. 1998;272(1-2):24–32. [Google Scholar]
- 37.Efthimiadou EK, Katsarou ME, Karaliota A, Psomas G. Copper(II) complexes with sparfloxacin and nitrogen-donor heterocyclic ligands: structure-activity relationship. Journal of Inorganic Biochemistry. 2008;102(4):910–920. doi: 10.1016/j.jinorgbio.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Kumar RS, Arunachalam S. DNA binding and antimicrobial studies of polymer-copper(II) complexes containing 1,10-phenanthroline and l-phenylalanine ligands. European Journal of Medicinal Chemistry. 2009;44(5):1878–1883. doi: 10.1016/j.ejmech.2008.11.001. [DOI] [PubMed] [Google Scholar]


