Abstract
A fast and simple method to control variations in carbohydrate composition of Saccharomyces cerevisiae, baker's yeast, during fermentation was developed using mid-infrared (mid-IR) spectroscopy. The method allows for precise and accurate determinations with minimal or no sample preparation and reagent consumption based on mid-IR spectra and partial least squares (PLS) regression. The PLS models were developed employing the results from reference analysis of the yeast cells. The reference analyses quantify the amount of trehalose, glucose, glycogen, and mannan in S. cerevisiae. The selection and optimization of pretreatment steps of samples such as the disruption of the yeast cells and the hydrolysis of mannan and glycogen to obtain monosaccharides were carried out. Trehalose, glucose, and mannose were determined using high-performance liquid chromatography coupled with a refractive index detector and total carbohydrates were measured using the phenol–sulfuric method. Linear concentration range, accuracy, precision, LOD and LOQ were examined to check the reliability of the chromatographic method for each analyte.
Figure.

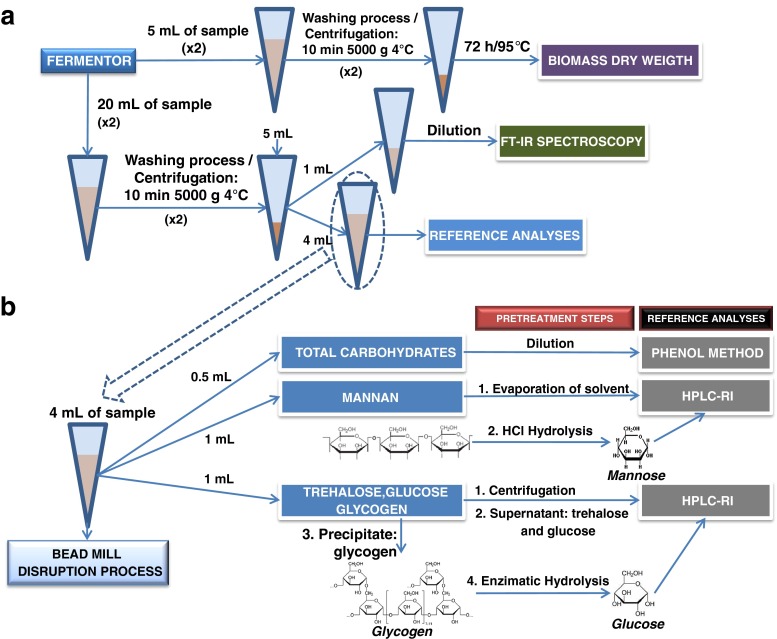
Comparison of workflows for carbohydrate determination in S.cerevisiae by FT-IR spectroscopy and HPLC-RI
Electronic supplementary material
The online version of this article (doi:10.1007/s00216-013-7239-9) contains supplementary material, which is available to authorized users.
Keywords: FT-IR spectroscopy, Partial least squares regression, Saccharomyces cerevisiae, Carbohydrates, HPLC-RI, Reference analysis
Introduction
The commercial exploitation of different microorganisms to convert chemical substrates to products of higher values is very important for several industries such as agricultural, pharmaceutical, and food industry. Yeast is the most widely used microorganism in industry and is especially important to brewing, wine production, distilling, and baking [1, 2]. The analysis of cells is of great scientific interest in the field of quality control of industrial processes. The variations in carbohydrate content of yeast suggest that these compounds play important roles during the yeast cycle [3, 4] and the control and quantification of these compounds is important because their concentrations are related to the metabolism of the yeast cells. The most common methods for studying the carbohydrate composition (mannan, trehalose, glucose, glycogen, etc.) are based on enzymatic and chromatography techniques (e.g., gas chromatography or high-performance liquid chromatography (HPLC)) [5–8]. In these techniques, tedious pretreatment steps of the cells are necessary to carry out carbohydrate determinations.
Fourier transform infrared (FT-IR) spectroscopy in the mid-IR range (400–4,000 cm−1) can be applied as a useful tool for the analysis of entire yeast cells providing a fast, effective, reagent-free, and simple method [9, 10]. Since Beer's law is obeyed, the observed absorptions are proportional to analyte concentrations. In complex samples, linear regression between absorption band areas and analyte concentrations determined by a reference method are not enough to provide robust calibration; here, multivariate methods, like partial least squares (PLS) regression, have to be employed. Wold et al. [11] give a comprehensive introduction to PLS regression models, also discussing the validity and the predictive power of the PLS models. In the case of biological cells, a multitude of components contribute to the recorded mid-IR spectra, which leads to strong overlapping of the absorption bands. PLS regression, however, can extract information on contributing components from the recorded spectra. In the last decades, several applications using FT-IR spectroscopy and multivariate chemometric tools have been published to monitor yeast metabolism taking advantage of the high sensitivity to small changes in the composition of cells [12–15]. FT-IR spectroscopy is a powerful tool for the time resolved and noninvasive measurement of multi-substrate/product interactions in complex metabolic networks such as the oscillating glycolysis in yeast extract identifying glycolytic intermediates [12]; the autolysis of Saccharomyces cerevisiae cells in a base wine studying partial hydrolysis of proteins; increase of peptides, free nucleotides, lipids, mannans, and beta-1,3 glucans [13, 14]; and the metabolic alterations of yeast cells induced by four chemical compounds: ethanol, sodium hypochlorite, sodium chloride, and sulfur dioxide taking into account five different spectral regions (fatty acids from 3,000 to 2,800 cm−1, amides from 1,800 to 1,500 cm−1, mixed region from 1,500 to 1,200 cm−1, carbohydrates from 1,200 to 900 cm−1, and typing region from 900 to 700 cm−1) [15]. Kuligowski et al. [10] developed a rapid method for the discrimination of entire Pichia pastoris cells grown under different conditions based on the direct acquisition of mid-IR spectra in the fingerprint region (900–1,800 cm−1) and partial least squares discriminate analysis.
The main objective of this study was to investigate the applicability of FT-IR spectroscopy as a rapid, precise, and accurate method not requiring sample preparation, for the determination and quantification of carbohydrate composition of S. cerevisiae. As a prerequisite, robust reference analysis methods for carbohydrates are needed. Only when reliable reference methods are available, a robust calibration for the different carbohydrates with respect to the recorded FT-IR spectra of the cells can be established. S. cerevisiae was selected because it is the most studied yeast due to the easy cultivation and short generation time and serves as a model of a eukaryotic cell. The main carbohydrates of yeast such as mannan, a cell wall polysaccharide composed of mannose units, and the intracellular carbohydrates, trehalose, a disaccharide of glucose, and glycogen, a polysaccharide of glucose, were studied. For reference analysis, the optimization of the cell disruption and hydrolysis processes and the validation of quantification methods are essential to avoid alterations in the final compositional results. The phenol–sulfuric method, a well-known method to determine the total carbohydrates, was used and a separation technique, liquid chromatography with refractive index detector, was used to determine individual carbohydrates, trehalose, glucose, glycogen, and mannan. The developed method uses a single sample with low sample volume requirement, generating reliable and precise results without interferences. These robust reference data are the basis for building PLS regression models for the individual carbohydrates. Robust models, indicated by low cross-validation errors (root mean square errors of cross validation (RMSECV), to be more precise), show the suitability of FT-IR spectroscopy for simple and reagent-free determination of carbohydrates in yeast cells.
Materials and methods
Chemicals and reagents
All reagents were of analytical grade. For stock standard solutions, d-(+)-glucose monohydrate supplied by Fluka (Steinheim, Germany), trehalose and d-(+)-mannose obtained from Merck (Darmstadt, Germany), and glycogen desiccate type II from Oyster and mannan from S. cerevisiae purchased from Sigma (Saint Louis, USA) were used.
For the total carbohydrate method, phenol and sulfuric acid 95–98 % were supplied by Sigma-Aldrich (Saint Louis, USA). For glycogen hydrolysis, amyloglucosidase from Aspergillus niger were obtained from Fluka (Steinheim, Germany), and for mannan hydrolysis, hydrochloric acid 37 % and sodium hydroxide ≥98 % were purchased from Sigma-Aldrich (Saint Louis, USA).
All of the solutions were prepared in high purity deionized Milli-Q water (Millipore, Molshem, France).
Apparatus and materials
For acquisition of FT-IR spectra of dried yeast samples, a Bruker Optics Hyperion 3000 FT-IR microscope connected to a Bruker Optics Tensor 37 spectrometer (both Ettlingen, Germany) was used. It was equipped with a liquid nitrogen-cooled single point mercury–cadmium–telluride detector and a 15-fold Cassegrain objective. The dedicated software OPUS 7 (Bruker Optics) was used for recording spectra and instrument control. A Genesys 20 Visible spectrophotometer (Thermo Scientific, Waltham, USA) was used for all the photometric measurements, a heating oven (Binder, Tuttlingen, Germany) for biomass dry weight determination, and a heating plate (Ika Labortechnik, Staufen, Germany) and a Vortex (Yellow Line TTS2, Wilmington, USA) for shaking the samples. An Eppendorf Mini Spin centrifuge for microcentrifuge tubes and an Eppendorf 5810R centrifuge (Hamburg, Germany) for centrifuge tubes were used. The pH measurements were carried out using the portable WTW pH meter 3400i (Weilheim, Germany). Fermentations were carried out in a 15-L autoclavable, fully automated and controlled-stirred bioreactor (Infors, Switzerland) coupled to the integrated process control and management system Lucullus (SecureCell AG, Schlieren, Switzerland).
Microorganism and culture conditions
In order to optimize the experimental conditions for carbohydrate measurements, commercial pressed baker's yeast (S. cerevisiae, “Mautner Markhof”—Austria Hefe AG, Austria) were used. The yeast was stored at 8 °C and was taken out just before running the experiment to avoid contamination. The yeast cells were suspended in deionized water following centrifugation at 6,575×g for 10 min. The supernatant was decanted and, subsequently, the cells were resuspended in deionized water and vortexed to ensure good mixing. The washing procedure was repeated four times. After that, different amounts of cells were resuspended in 4 mL of water and used as standards.
For yeast fermentation, yeast peptone glucose (YEPG) medium was prepared as preculture medium (40 g L−1 glucose, 5 g L−1 peptone, and 5 g L−1 yeast extract). The YEPG medium was autoclaved at 121 °C for 20 min. One milliliter of frozen stock of S. cerevisiae (strain, CBS 8340) was added to 200 mL of YEPG medium under sterile conditions and subsequently incubated for 24 h at 30 °C and 220 rpm in shake flasks.
Six liters of batch medium was prepared using 20 g L−1 glucose, 5 gL−1 ammonium sulfate, 3 g L−1 potassium hydrogen phosphate, and 0.5 g L−1 magnesium sulfate heptahydrate. Additionally, 2.67 mL L−1 of sterile vitamin and trace element solutions, respectively, as well as 0.1 mL L−1 of sterile antifoam were added after autoclavation (121 °C, 20 min). For the subsequent fed-batch fermentation, 4 L of feed medium containing 100 g L−1 glucose, 3 g L−1 potassium hydrogen phosphate, 0.5 g L−1 magnesium sulfate heptahydrate, and 2.67 mL L−1 of sterile vitamin and trace element solutions, respectively, as well as 0.1 mL L−1 of sterile antifoam was made. The conditions of the batch fermentation were 6 L min−1 (1 vvm) of airflow, 800 rpm speed of agitation, temperature was set to 30 °C, and the pH automatically adjusted to 4.8 by addition of 2 M NaOH. All reagents for yeast media were supplied by Carl Roth GmbH (Karlsruhe, Germany). The batch process was started by inoculation with 600 mL of preculture (10 % of batch volume). During the fed-batch phase, conditions were the same as for the batch phase except that the airflow was adjusted to remain at 1 vvm, i.e., increased with culture volume.
Sampling and pretreatment
During the fermentations, several samples from the bioreactor were taken. Each sample was split up for duplicate biomass dry weight determination (5 mL in pre-weighted test tubes) and a pellet sample for carbohydrate determinations by FT-IR spectroscopy of whole cells and by HPLC after a disruption process (20 mL). A schematic of sampling and pretreatment steps can be seen in Fig. 1a. All the samples were centrifuged for 10 min at 5,000×g and 4 °C and, afterwards, the medium (supernatant) was discarded. The yeast cells were resuspended in distilled water and centrifuged again for 10 min to rinse the cells. The washing procedure was repeated two times in order to eliminate potential interferences of the medium. The supernatants were removed carefully. During the whole process, the samples were stored on ice to avoid and stop further reactions. The pellet samples for carbohydrate determination were stored in the freezer (−20 °C).
Fig. 1.
Diagram of the sampling and pretreatment steps of real samples (a); experimental process to carry out the reference analyses (b)
For determination of biomass dry weight, the yeast cells in the pre-weighted test tubes were dried in a heating oven for 72 h at 105 °C. The cell dry weight was determined gravimetrically.
For carbohydrate measurements, each sample (pellet) was resuspended in 5 mL Milli-Q water and split off to carry out FT-IR measurements (1 mL) and reference analyses (4 mL).
FT-IR spectroscopy of dried yeast samples
One milliliter of the washed and resuspended fermentation sample (pellet) was used for FT-IR spectroscopy. The measurement procedure was guided by Kuligowski et al. [10]. Absorbance spectra were recorded in the spectral range between 4,000 and 600 cm−1 with a spectral resolution of 4 cm-1 as the co-addition of 32 scans in transmission mode. Dilution rates were adjusted between 1:10 and 1:20 depending on the initial biomass concentration of the respective sample. Three spots of 3 μL each were pipetted onto a zinc selenide (ZnSe) slide and the H2O was allowed to evaporate in the dry air-purged sample compartment of the microscope. Built-in shutters were used to limit the area of acquisition to 100 × 100 μm. A background spectrum was acquired on a clean part of the ZnSe slide and 40 to 50 spectra of each yeast sample spot were recorded at different positions. Measurement time for each sample spot was around 3 min. A mean spectrum of each sample spot was calculated for further analysis, thus three spectra per sample were available.
Reference analyses
To obtain reference results for FT-IR spectroscopy, the carbohydrates of yeast cells were measured using several pretreatment steps and different methods as can be seen in Fig. 1b. For reference analyses, it was necessary to rupture the cells using 6 g acid-washed glass beads (diameter: 0.45-0.55 mm) from Sigma (St. Louis, USA) as abrasive and 4 mL cell suspension (dry weight from 20 mL fermentation sample) in a glass tube. The cell disruptions were carried out using a bench-top vortex homogenizer at 2,500 rpm 12 times for 1 min each, with 1 min of pauses on ice bath between vortexing.
After the disruption, the sample was split to measure total carbohydrates (0.5 mL), mannan (1 mL), glucose, trehalose, and glycogen (1 mL). The aliquots were kept on ice at a temperature below 4 °C to avoid the hydrolysis of trehalose to glucose by enzymes released by cell disruption.
For total carbohydrate measurements, the phenol–sulfuric acid method [16] using glucose as standard was used. One milliliter of suspension containing between 20 and 200 μg of carbohydrates was mixed with 1 mL of 5 % (w/v) phenol solution. Subsequently, 5 mL of sulfuric acid was added and carefully mixed using a vortex. The absorbance at 488 nm after incubation for 10 min at room temperature and 20 min at 30 °C is proportional to the initial carbohydrate concentration of the sample. A calibration curve using a series of standards of glucose between 0 and 200 mg L−1 was constructed. Several dilutions were carried out in function of the concentration of yeast to obtain suitable absorbance values. For yeast cell concentrations from 0 to 5 g L−1, dilution factor (DF) was 10, from 5 to 25 g L−1 DF was 50, for 25–50 g L−1 DF was 125, for 50–100 g L−1 DF was 250, and for 100–150 g L-1 DF was 375.
The measurement of the individual carbohydrates was achieved using an ion exchange chromatographic method. An Ultimate 3000 Dionex HPLC equipment (Thermo Scientific, USA) coupled with a refractive index detector (Shodex® RI-101, Shoko Scientific, Japan) was used. The Chromeleon Chromatography Management Software from Dionex was used for chromatogram acquisition and data analysis. The column used to achieve the complete separation of carbohydrates was the Recex RCM-Monosaccharide Ca2+ column with a size of 300 × 7.8 mm (Phenomenex, Germany). In order to protect the analytical column, a Security Guard Cartridge (Carbo Ca2+, 4 × 3 mm, Phenomenex) was used. The stationary phase of both, the guard and the analytical column, was an anion exchange resin with Ca2+ counter ions, with a particle size of 8 μm and 8 % cross-linkage. The experimental conditions summarized in Table 1 have been published by Mazaverica et al. [17] and Kuligowski et al. [18]. This separation method is, however, only applicable to disaccharides and monosaccharides. Therefore, it was necessary to carry out pretreatments of the sample to obtain mannose and glucose from the mannan and glycogen, respectively. In the case of mannan, a hydrolysis process was utilized. One milliliter of suspension sample after the disruption process was dried at 95 °C during 24 h and then resuspended in 3 mL of 2 M HCl and heated in sealed glass tubes at 100 °C for 7 h [6]. The tubes were cooled on ice and the hydrosylate was neutralized with 2 M NaOH. Several standards using mannan from S. cerevisiae (333–2,570 mg L−1) were prepared to relate the mannan concentration with the real samples. No interferences were found because the only polysaccharide containing mannose was mannan. Finally, the last aliquot, 1 mL of disrupted cell suspension, was used to determine trehalose, glucose, and glycogen. Glycogen is insoluble in water; therefore, the suspension was centrifuged at 6,575×g for 10 min at 4 °C to separate the supernatant, containing trehalose and glucose, and precipitate, containing glycogen (Fig. 1b). The supernatant was directly determined using the chromatographic separation. The precipitate, however, needed a pretreatment step to hydrolyze the glycogen to glucose for chromatographic measurements. Since trehalose and glucose were removed first, the enzyme could only digest glycogen, therefore avoiding interferences and the overestimation of the glycogen content. The glycogen was hydrolyzed using the amyloglucosidase enzyme. One milliliter of solution containing 1 mg mL−1 of amyloglucosidase was added to the disrupted cells and incubated for 2 h at 60 °C. Several standards using glycogen from Oyster (240-3,000 mg L−1) were prepared to relate the glycogen concentration with real samples.
Table 1.
HPLC conditions to separate carbohydrates with HPLC and RI detection
| Experimental conditions | |
|---|---|
| Mobile phase | Milli-Q Water |
| Chromatographic mode | Isocratic |
| Flow rate | 0.6 mL |
| Temperature | 80 °C |
| Injection | 20 μL |
| Detector temperature | 35 °C |
Partial least squares regression
The PLS toolbox 6.2.1 (Eigenvector Research, Wenatchee, WA, USA) and Matlab 7.10.0 (Mathworks, Natick, MA, USA) were used for data treatment and building PLS regression models, correlating the FT-IR spectra with mannan, trehalose, and glycogen concentrations found with the reference methods described above. To compensate for differences in sample thickness, spectra were normalized (min–max) to the amide I band (1,740–1,575 cm−1) since the protein content of the cells remained nearly constant over the course of the fermentation (determined using the bicinchoninic acid assay, data not shown). Appropriate spectral regions for calibration were chosen with consideration of pure analyte spectra. For all three analytes, the wavenumbers chosen were in the carbohydrate region of C–O and C–O–C stretching (900–1,200 cm−1) [19]. To test the PLS regression models for their ability to predict new, unseen data, i.e., their ability to predict analyte concentrations from spectra not included in the calibration set, cross-validation was performed using random subsets. For this purpose, the calibration set was divided into a set number of subsets and a PLS regression model excluding one of the subsets was built. The excluded subset was used to validate the PLS model, i.e., the concentrations predicted by the PLS model (y i) for these data were compared to the values determined by reference analysis . One iteration of the process corresponded to repeating the process until all subsets had been used for validation of models built excluding these. The RMSECV, i.e., the normalized sum square of the residuals, which is calculated as
was calculated using these data; it is a measure of the error one can expect when predicting analyte concentrations from unknown spectra using the PLS model, a low RMSECV is therefore desirable. Here, eight data splits and three iterations were used for the calculation of the RMSECV. Different data preprocessing methods were tested and the methods leading to the lowest RMSECV were chosen. For all three analytes, calculation of second derivative (Savitzky–Golay, polynomial order 2, window size 15) and mean centering of data led to the best results.
Results and discussion
Reference analyses
Several processes of reference analyses such as the disruption method, mannan hydrolysis, and glycogen hydrolysis were selected and optimized in order to get precise and reliable results. Moreover, the validity of the reference analyses was tested by carrying out repeatability, reproducibility, and linearity studies; results are shown in the section below.
Selection of disruption method
For the purpose of determining the intracellular carbohydrate content of yeast, the cells must be disintegrated by mechanical means or non-mechanical methods (physical, chemical, or enzymatic) [20, 21]. The method of choice needs to be compatible with the measurements of the chemical content of the yeast without interferences. Furthermore, a high percentage of cell disruption, adequate for small-scale applications with a high throughput and of course short times is desirable. The mechanical methods are most efficient for cell disruption at small and large scale [22]. Several methods/protocols based on mechanical methods were tested and optimized: ultrasound, homogenization, boiling, freeze–thawing, and bead milling disruption (details see Electronic Supplementary Material). The best results were achieved when the bead milling method was applied. This procedure is particularly efficient in the disruption of yeasts which have rigid cell walls constituted by polysaccharides (80–90 %) [21]. Factors governing the performance of the bead mill process including impeller speed, cell density, amount, and size of the glass beads were optimized [21]. Percentages of cell breakage higher than 80 % were achieved using this modified Ricci-Silva et al. method [21]. Disruptions were carried out in glass tubes with a ratio of 1 mL of suspension/1.5 g of glass beads (diameter, 0.45–0.5 mm) for 12 cycles, each using a bench-top vortex at 2,500 rpm with 1 min intervals on ice (see “Materials and methods”).
Selection and optimization of mannan hydrolysis
Mannan, composed mainly of d-mannose, is a major constituent of the yeast cell wall, accounting for approximately 13 % (w/w) [23]. This polysaccharide possesses a backbone of α-(1–6)-linked d-mannose units which are substituted by side chains containing (1–2) and (1–3)-linked d-mannose units [24]. Two methods to hydrolyze mannan to mannose were tested. First of all, α-mannosidase from Canavalia ensiformis (Jack bean) (Sigma-Aldrich) was used; 0.5 mL of 0.45 mg L−1 α-mannosidase solution was added to 1.5 mg of mannan in presence of 1.5 mL of 20 mM sodium acetate (pH = 8). The final concentration of mannan was 750 mg L−1. The solution was kept overnight at room temperature. No peak was obtained when the sample was analyzed by HPLC-refractive index (RI) method. Therefore, HCl hydrolysis was selected to hydrolyze mannan samples based on the method published by Dallies et al. [6]. The dried mannan samples were mixed with 3 mL of 2 M HCl and heated in sealed tubes at 100 °C for 1–10 h. Then, the tubes were cooled on ice and the hydrolysate was neutralized with 2 M NaOH. The effect of the heating time (1 to 10 h) of the glass tubes was studied and 7 h was selected for achieving the best yield of disruption of mannan.
Selection and optimization of glycogen hydrolysis
Glycogen is a branched polymer of α-(1–4) and α-(1–6)-glycoside bonds. The hydrolysis of both types of bonds is achieved using the enzyme amyloglucosidase from A. niger which is thus well suited for complete degradation of glycogen [7]. Several publications found that the enzyme degraded 97 % of glycogen [7, 25]. The optimized hydrolysis is based on the Bernfeld method [26]. Parameters such as the concentration of the enzyme, temperature, and exposure time of the hydrolysis process were optimized to obtain the maximum percentage of glucose for the same amount of glycogen (see Electronic Supplementary Material Figure S1). The diluted glycogen standard samples (DF 50) were injected into the chromatographic system after the hydrolysis process. The area of the glucose peak was calculated and used to monitor the hydrolysis reaction and hence the yield of this reaction could easily be evaluated. The optimal conditions are described in section “Materials and methods.”
Analytical features of HPLC-RI
The characteristic parameters of the analytical method obtained under the optimum conditions were evaluated. Mannan and glycogen samples after the hydrolysis process and standard of trehalose and glucose were used. The respective LODs and LOQs were determined and the precision of the method was checked in terms of repeatability and intermediate precision (reproducibility) (details see Electronic Supplementary Material and Table S1). Table 2 shows the LOD and LOQ values for each analyte. The calibration curve was checked by analyzing standard solutions of each analyte at eight different levels. A linear regression equation with a regression coefficient near to the unit was obtained (Table 2). To check the accuracy of the proposed method, several synthetic mixtures containing the analytes in variable amounts were prepared and analyzed in triplicate using the HPLC-RI method. Quantification was achieved in terms of peak areas and the results were expressed as mean value and recoveries were calculated. In all cases, recoveries were very close to 100 % and relative deviations were between 0.1 and 6.3 %.
Table 2.
Figures of merit for trehalose and glucose standards and for glucose and mannose after hydrolysis process using a HPLC with RI detection
| Analyte | Linear regression equationa | Linear range (mg L−1) | R 2 | LOD (mg L−1) | LOQ (mg L−1) |
|---|---|---|---|---|---|
| Mannoseb | y = (0.0016 ± 0.0000)x + (0.5204 ± 0.0409) | 333–2,570 | 0.991 | 26 | 87 |
| Trehalose | y = (0.0036 ± 0.0000)x − (0.0013 ± 0.0040) | 50–5,000 | 0.999 | 12 | 39 |
| Glucose | y = (0.0036 ± 0.0000)x − (0.0337 ± 0.0689) | 50–5,000 | 0.999 | 12 | 39 |
| Glycogenb | y = (0.0017 ± 0.0000)x + (0.2278 ± 0.0447) | 240–3,000 | 0.992 | 25 | 82 |
aCalibration curve obtained with standards. Concentration versus peak area
bAfter hydrolysis process
Analysis of baker's yeast standards
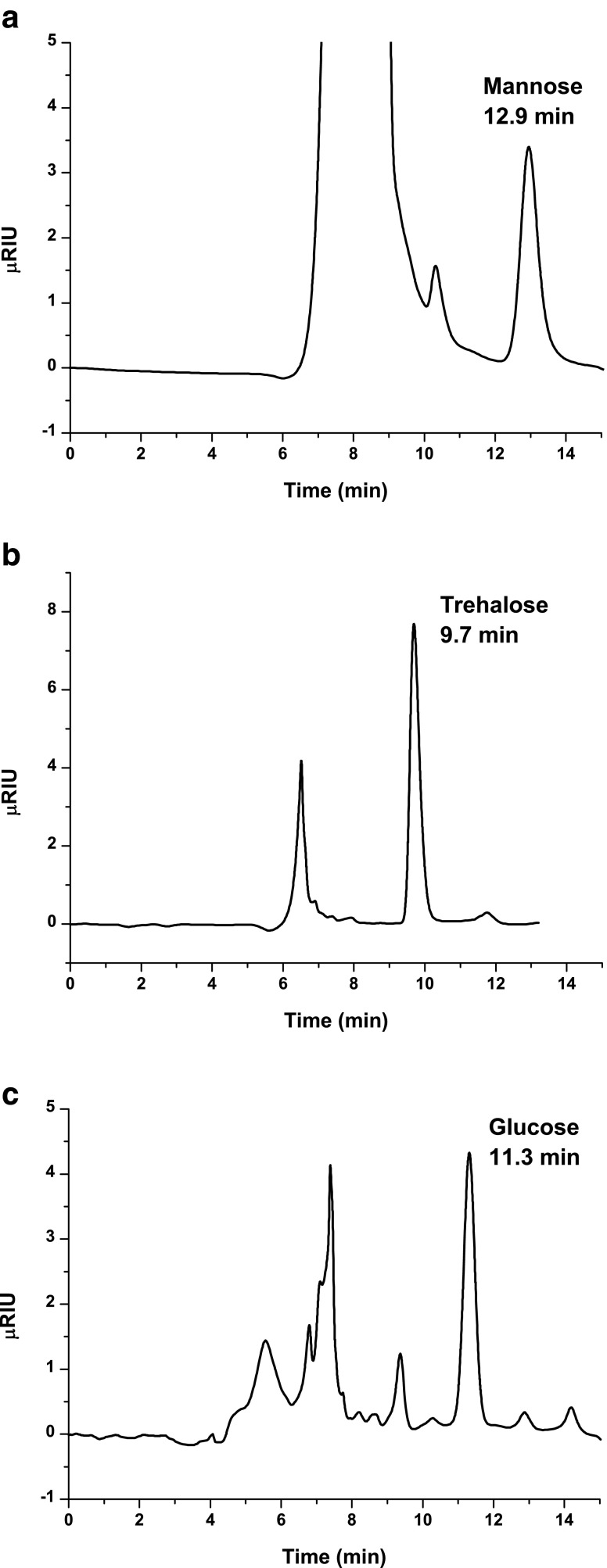
In order to demonstrate the applicability of the proposed method, six samples with different concentrations of baker's yeast were prepared as standard samples. First of all, the dry weight was determined to calculate the initial concentration of yeast, and then the concentration of each carbohydrate was determined using the respective methods. A chromatogram of 850 mg−1 mannan standard after the hydrolysis process is shown in Fig. 2a. The experimental signal for 0.6 g L−1 standard of yeast after applying the trehalose and glucose method is shown in Fig. 2b. A peak of trehalose can be observed but the area of the glucose peak is not enough to be quantified. Figure 2c corresponds to a 34.5 g L−1 standard of yeast after applying the glycogen method. The area of the glucose peak can be related to the glycogen concentration of yeast. Finally, the percentages of each carbohydrate were calculated by dividing the concentration of carbohydrate by the concentration of yeast and multiplying by 100. The cell concentration in the samples ranged from 10 to 35 g L−1. Table 3 lists the results for carbohydrate content relative to biomass dry weight (%DW) of the respective samples. They are in nice agreement with each other, indicating that dilution factors were correct and the method is valid for the range of cell concentrations. The %RSD values for all methods were between 2.7 and 7.1 %.
Fig. 2.
Chromatograms of mannose using a mannan standard solution (a); trehalose and glucose (b) and glucose from glycogen (c) using a yeast standard sample
Table 3.
Percentages of carbohydrates from baker's yeast samples after disruption process by the developed methods, phenol method for total carbohydrates, and trehalose, glucose, glycogen, and mannose using the HPLC-RI (n = 3)
| Analyte | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Average ± SD |
|---|---|---|---|---|---|---|---|
| Total carbohydrates | 48.1 ± 0.2 | 45.6 ± 0.7 | 43.2 ± 0.3 | 42.3 ± 0.5 | 42.6 ± 0.1 | 39.9 ± 0.1 | 43.6 ± 2.9 |
| Mannosea | 8.9 ± 0.1 | 9.4 ± 0.1 | 8.4 ± 0.1 | 8.6 ± 0.1 | 9.5 ± 0.3 | 9.8 ± 0.0 | 9.1 ± 0.6 |
| Trehalose | 8.7 ± 0.1 | 9.3 ± 0.2 | 9.4 ± 0.1 | 9.2 ± 0.1 | 9.3 ± 0.1 | 9.1 ± 0.0 | 9.2 ± 0.2 |
| Glucose | 2.0 ± 0.2 | 2.4 ± 0.2 | 2.2 ± 0.1 | 2.1 ± 0.1 | 2.2 ± 0.1 | 2.2 ± 0.1 | 2.2 ± 0.1 |
| Glycogena | 1.9 ± 0.1 | 2.0 ± 0.1 | 2.1 ± 0.0 | 2.0 ± 0.0 | 2.3 ± 0.1 | 2.2 ± 0.0 | 2.1 ± 0.1 |
| Sum | 21.5 ± 0.5 | 23.1 ± 0.6 | 22.1 ± 0.3 | 21.9 ± 0.3 | 23.3 ± 0.5 | 23.3 ± 0.1 | 22.5 ± 0.8 |
aAfter hydrolysis process
Analysis of yeast samples from fed-batch fermentations
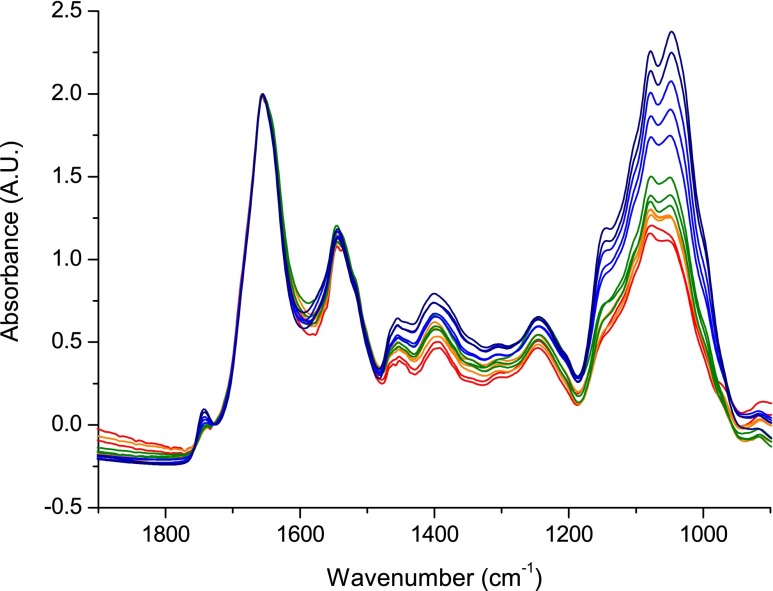
The outlined method has been used to study the carbohydrate content of S. cerevisiae in nitrogen-limited fed-batch fermentation. After 24 h of batch cultivation, the conditions changed to a nitrogen-limited fed-batch fermentation. A medium deficient in nitrogen lead to the accumulation of storage carbohydrates such as trehalose and glycogen and structural modifications. The change of the carbohydrate composition could be studied using the proposed method and reference analyses using several samples taken from the fermentor during the fed-batch process. The FT-IR spectrum of each sample was measured in triplicate for both fermentations. In Fig. 3, the average mid-IR absorbance spectra acquired from samples at different time points between 1,900 and 900 cm−1 are shown for fed-batch fermentation 1. In the depicted region, the main spectral contributions are derived from proteins (amide I (1,660 cm−1) and amide II (1,540 cm−1) bands), the region from 1,500 to 1,250 cm−1 with less intense and strongly overlapping bands which correspond to proteins, lipids, and storage carbohydrates and the region between 1,250 and 900 cm−1 for carbohydrates with a minor contribution of phosphate bands from DNA, RNA, and phospholipids. The data were normalized to the amide I band and significant changes over fermentation time can be observed. To get an estimate of the total carbohydrate content of the cells, the area under the carbohydrate bands (1,186–936 cm−1) was calculated and normalized by biomass. The carbohydrate content relative to protein content increased with time and spectral features in the carbohydrate region changed, indicating a change in the carbohydrate constitution of the cells. These results were compared to data obtained by the phenol method and the sum of values obtained by HPLC-RI for each carbohydrate (mannan, trehalose, and glycogen). Figure 4 shows the normalized total carbohydrate content in function of time for each method. In accordance with the data obtained from FT-IR spectra, both the phenol method and the sum of carbohydrates determined by HPLC-RI increase over the course of the fermentation. The carbohydrate content found by calculation of the sum of HPLC-RI measurements of the respective analytes and by FT-IR spectroscopy are in good agreement with each other over the course of the fermentation. The total carbohydrate content was practically constant until 4.4 h, and a linear increase was observed for times greater than 4.8 h. The results obtained from the phenol method, however, show a different development: an almost sinusoidal development was observed. When only comparing the values found at the beginning and at the end of the fermentation, the same trend as for the other data was found: an increase of the total carbohydrate content. It should be noted that the standard deviations of experimental data for FT-IR method are lower than those of the two other methods. The low reproducibility of values obtained by the HPLC-RI and the phenol method, respectively, are most likely due to differences in cell disruption efficiency and the necessary but error-prone sample handling. Similar results were obtained for fed-batch fermentation 2 (data not shown).
Fig. 3.
FT-IR spectra of whole S. cerevisiae. Samples were drawn from nitrogen limit fed-batch fermentation 1 at different times over the course of the fermentation
Fig. 4.
Variations of normalized total carbohydrate content for real samples from nitrogen limited fed-batch fermentation 1 of S. cerevisiae applying reference and FT-IR methods
Prediction of concentration values from FT-IR spectra by PLS regression
Parameters and results of the PLS regression models for trehalose, glycogen, and mannan are summarized in Table 4. The number of latent variables used to build the PLS regression models was seven for trehalose and mannan and five for glycogen, and the RMSECV were found to be 0.33 % DW for trehalose, 0.55 % DW for glycogen, and 1.17 % DW for mannan. Considering the concentration ranges used for calibration, these values show that FT-IR spectroscopy is a promising technique for fast, reagent-free measurement of intracellular carbohydrates. The analyte concentrations determined by cross-validated PLS regression and the reference concentrations found for the different samples drawn from the two fermentations are shown in Fig. 5. The black squares indicate the values found by reference analysis and the red circles correspond to the values predicted from cross-validated PLS regression, i.e., the average concentration predicted from the spectra of each of the three sample spots. The values are in good agreement with each other and the expected changes in carbohydrate content of the yeast cells. Generally, the data found by FT-IR spectroscopy show a smoother trend/development over time than the respective reference data, where in some cases steep slopes, i.e., big differences between consecutive samples, were found. These large increases and decreases in reference values are also the points where FT-IR data and reference data deviate strongly. Each single value obtained by reference analysis is prone to errors due to the necessary sample pretreatments and handling. When calculating a PLS regression model from these data, the erroneous values (which are both positive and negative deviations from the true value) are averaged over the whole calibration set and resulting PLS predictions show a smoother time course.
Table 4.
Parameters and root mean square error of cross-validation of PLS models for trehalose, glycogen, and mannan
| Analyte | Calibration range (% dry weight) | Spectral range (cm−1) | Latent variables | RMSECV (% dry weight) |
|---|---|---|---|---|
| Mannan | 14.40–22.90 | 1,184–895 | 7 | 1.17 |
| Trehalose | 0.55–3.31 | 1,184–956 | 5 | 0.33 |
| Glycogen | 0.91–6.10 | 1,188–925 | 4 | 0.56 |
Fig. 5.
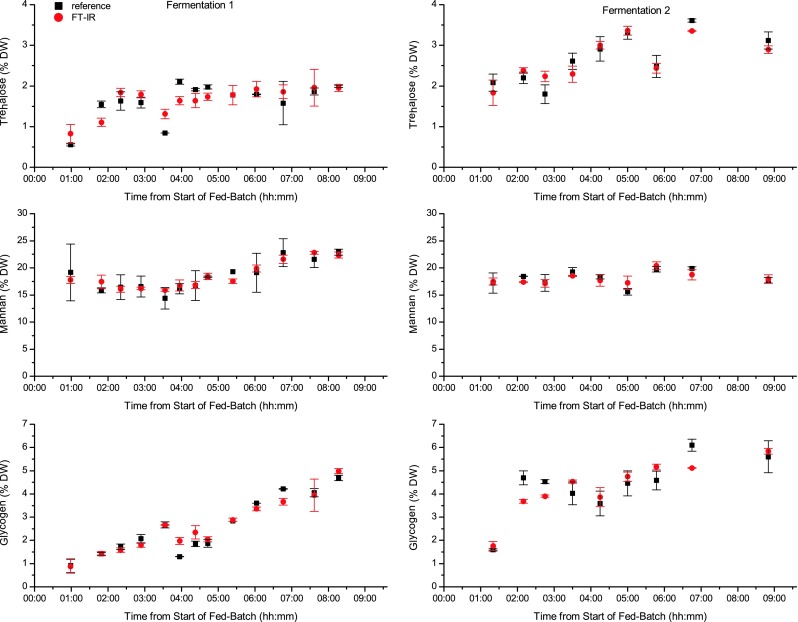
Relative trehalose, mannan, and glycogen content of the samples drawn from fermentations 1 and 2 at different times determined by reference analysis (black squares) and from FT-IR spectra by cross-validated PLS regression (red circles)
Conclusions
Most of the published articles to determine the carbohydrate composition of yeast cells describe time-consuming wet chemical assays requiring handling-intensive pretreatment steps. The proposed FT-IR spectroscopy method of whole S. cerevisiae cells presents clear advantages with respect to the other methods such as operational simplicity, lower sample consumption, and no reagent consumption, avoiding the risk of errors as a result of the labor-intense cell disruption and sample handling. The method determines the content of several polysaccharides such as mannan and glycogen and the disaccharide trehalose by cross-validated PLS regression. Results can be obtained within 30 min from sampling, including the most time-consuming step, i.e., drying of the sample. Using the instrumentation and settings in this study, acquisition of spectra took approximately 3 min per sample spot, showing that significant improvement of measurement time is possible by using other drying methods, e.g., a dessicator. The obtained results show that concentration values are in good agreement with reference values.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 409 kb).
Acknowledgments
European Social Fund from European Union and Junta de Comunidades de Castilla-La Mancha are gratefully acknowledged for supporting given through a Postdoctoral research contract for Maria R. Plata. Partial financial support of the Austrian Science Fund (FWF) through the stand-alone project scheme (project number, P24154), and the Austrian research funding association (FFG) under the scope of the COMET program within the research network “Process Analytical Chemistry (PAC)” (contract # 825340) is gratefully acknowledged. The authors want to express their gratitude to Bettina Lorantfy for guidance in media design and Daniela Roxana Ghiţã for her support with the experimental work.
References
- 1.Milic TV, Rakin M, Siler-Marinkovic S. Utilization of baker's yeast (Saccharomyces cerevisiae) for the production of yeast extract: effects of different enzymatic treatments on solid, protein and carbohydrate recovery. J Serb Chem Soc. 2007 [Google Scholar]
- 2.Sceni P, Palazolo GG, Vasallo MDC, et al. Thermal and surface behavior of yeast protein fractions from Saccharomyces cerevisiae. Food Sci Technol-Leb. 2009 [Google Scholar]
- 3.Kath F, Kulicke W-M. Mild enzymatic isolation of mannan and glucan from yeast Saccharomyces cerevisiae. Angew Makromol Chem. 1999 [Google Scholar]
- 4.Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prieto MA, Vázquez JA, Murado MA. Hydrolysis optimization of mannan, curdlan and cell walls from Endomyces fibuliger grown in mussel processing wastewaters. Process Biochem. 2011 [Google Scholar]
- 6.Dallies N, François J, Paquet V. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast. 1998 doi: 10.1002/(SICI)1097-0061(1998100)14:14<1297::AID-YEA310>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Schulze U, Larsen ME, Villadsen J. Determination of intracellular trehalose and glycogen in Saccharomyces cerevisiae. Anal Biochem. 1995;228:143–149. doi: 10.1006/abio.1995.1325. [DOI] [PubMed] [Google Scholar]
- 8.Parrou JL, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997 doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 9.Naumann D, Keller S, Helm D, et al. FT-IR spectroscopy and FT-Raman spectroscopy are powerful analytical tools for non-invasive characterization of intact microbial cells. J Mol Struct. 1995;347:399–405. doi: 10.1016/0022-2860(95)08562-A. [DOI] [Google Scholar]
- 10.Kuligowski J, Quintás G, Herwig C, Lendl B. A rapid method for the differentiation of yeast cells grown under carbon and nitrogen-limited conditions by means of partial least squares discriminant analysis employing infrared micro-spectroscopic data of entire yeast cells. Talanta. 2012 doi: 10.1016/j.talanta.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometr Intell Lab. 2001;58:109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
- 12.Mair T, Zimányi L, Khoroshyy P, et al. Analysis of the oscillatory kinetics of glycolytic intermediates in a yeast extract by FT-IR spectroscopy. Biosystems. 2006;83:188–194. doi: 10.1016/j.biosystems.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Burattini E, Cavagna M, Dell'Anna R, et al. A FTIR microspectroscopy study of autolysis in cells of the wine yeast Saccharomyces cerevisiae. Vib Spectrosc. 2008 [Google Scholar]
- 14.Cavagna M, Dell'Anna R, Monti F, et al. Use of ATR-FTIR microspectroscopy to monitor autolysis of Saccharomyces cerevisiae cells in a base wine. J Agr Food Chem. 2010 doi: 10.1021/jf902369s. [DOI] [PubMed] [Google Scholar]
- 15.Corte L, Rellini P, Roscini L, et al. Development of a novel, FTIR (Fourier transform infrared spectroscopy) based, yeast bioassay for toxicity testing and stress response study. Anal Chim Acta. 2010 doi: 10.1016/j.aca.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 16.DuBois M, Gilles KA, Hamilton JK, et al. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956 [Google Scholar]
- 17.Mazarevica G, Diewok J, Baena JR, et al. On-line fermentation monitoring by mid-infrared spectroscopy. Appl Spectrosc. 2004 doi: 10.1366/0003702041389229. [DOI] [PubMed] [Google Scholar]
- 18.Kuligowski J, Quintás G, Lendl B. High performance liquid chromatography with on-line dual quantum cascade laser detection for the determination of carbohydrates, alcohols and organic acids in wine and grape juice. Appl Phys BlLasers O. 2010 [Google Scholar]
- 19.Naumann D. In: Encyclopedia of analytical chemistry. Meyers RA, editor. New York: Wiley; 2000. [Google Scholar]
- 20.Geciova J, Bury D, Jelen P. Methods for disruption of microbial cells for potential use in the dairy industry—a review. Int Dairy J. 2002;12:541–553. doi: 10.1016/S0958-6946(02)00038-9. [DOI] [Google Scholar]
- 21.Ricci-Silva M, Vitolo M, Abraha J. Protein and glucose 6-phosphate dehydrogenase releasing from baker' s yeast cells disrupted by a vertical bead mill. Process Biochem. 2000;35:831–835. doi: 10.1016/S0032-9592(99)00151-X. [DOI] [Google Scholar]
- 22.Engler CR (1985) In: Moo-Young M, Cooney CL (eds.) Comprehensive biotechnology. Pergamon, Oxford
- 23.Förster J, Famili I, Fu P, et al. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 2003 doi: 10.1101/gr.234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart TS, Ballou CE. A comparison of yeast mannans and phosphomannans by acetolysis. Biochemistry. 1968;7:1855–1863. doi: 10.1021/bi00845a033. [DOI] [PubMed] [Google Scholar]
- 25.Becker JU. A method for glycogen determination in whole yeast cells. AnalBiochem. 1978;86:56–64. doi: 10.1016/0003-2697(78)90318-4. [DOI] [PubMed] [Google Scholar]
- 26.Bernfeld P. Amylases, alpha and beta. Methods Enzymol. 1955;1:149–158. doi: 10.1016/0076-6879(55)01021-5. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 409 kb).