Abstract
Recent developments in pulmonary cell biology have shown that the maintenance of protein concentrations, proteostasis, is an integral process of all biologic systems. The balance of available protein is the sum total of three key elements of cell metabolism: production by transcription and translation, compartmentalization through processing and sorting, and proteolytic degradation of proteins at any stage of their life-span. Considerable advances are constantly made in each of these three essential fields, and our appreciation for the diversity of mechanisms of protein degradation has expanded greatly in the last decade. The ubiquitin proteasome system (UPS) has emerged as the predominant protein degradation pathway in eukaryotes, with the large cullin-RING family of E3 ligases responsible for ubiquitination of a broad array of proteins to be degraded. The Skip-Cullin-F-box (SCF) ubiquitin E3 ligase superfamily is the largest family of cullin-RING ligases, with interchangeable F-box proteins orchestrating the trafficking proteins for ubiquitination and degradation. We will discuss the best characterized and most recent developments in the role of this intriguing family of proteins in normal physiology and disorders of the lungs.
Keywords: Protein degradation, Proteolysis, Lung, Inflammation
1. Introduction
For vertebrates to survive, they must have oxygen. In land dwelling vertebrates, oxygen is taken from ambient air that is exchanged through an elegant pulmonary system, where it serves in a critical role for the generation of chemical energy in various tissues. The lungs are an interface between the bloodstream and the outside world, and their ability to function dependably and efficiently over many decades in most individuals without catastrophic failure is one of nature's wonders. Like the skin and the gut, the lungs are constantly exposed to environmental particulate matter, pathogens, and other noxious stimuli; the maintenance of normal respiration without succumbing to infection or activation of robust innate or adaptive immune responses with consequent inflammatory lung damage is typical and essential to maintain homeostasis. In many human lung disorders, this delicate balance is tipped and the consequences can be dire, with respiratory failure and death among the common outcomes in disorders such as chronic obstructive pulmonary disease (COPD), cystic fibrosis (CF), the acute respiratory distress syndrome (ARDS) or acute lung injury (ALI), and pneumonia. Given the severity of these diseases, many insights into the molecular pathophysiology of lung disease have emerged over the past several years in areas including biochemistry, genetics, and cell biology to name a few. In this review, we will highlight one such area of new discovery, namely the selective regulation of protein degradation in the lung by the ubiquitin proteasome system, specifically the SCF (Skp1-Cullin1-F-box) ubiquitin E3 ligase complex, and how this apparatus at the protein level affects critical functions of lung cells with profound ramifications sufficient to affect the vitality of the organism. We will explore new insights and recent findings in this field, and discuss the burgeoning area of drug development as it relates to the ubiquitin proteasome system (UPS).
1.1. Selective protein degradation and cellular function
Maintenance of any healthy tissue requires stringent quality control at protein and cellular levels. All cellular proteins undergo normal turnover in the cell. This quality control mechanism prevents misfolded or dysfunctional proteins from being improperly active or accumulating unnecessarily after a discreet event or signal that may have led to their upregulation. It also functions as a means to change critical protein concentrations in the cell in response to chemical signals or for important cellular events, such as cell division. The major cellular systems involved in this regulation are the lysosome and the ubiquitin (Ub) proteasome [1], with the latter being the more prevalent modulator of proteins, in which target proteins that have been ubiquitinated are recognized and degraded by the large 26S proteasome protein complex, which is composed of 20S and two 19S proteolytic subunits. Ubiquitination of targets occurs in an ATP-dependent fashion, through an elaborate enzymatic cascade that adds the ubiquitin protein to first E1, then E2, and last to the specific target proteins by action of a ubiquitin E3 ligase. Usually a chain of four or more Ub monomers is added to the target, determining its recognition by the proteasome and resulting in target degradation. Overall, this process consumes large amounts of cellular energy, is represented by the largest family of enzymes present in eukaryotes, and accounts for ∼5% of the genome. The targeting, ubiquitination, and degradation of proteins occur in a highly regulated and specific manner, with the E3 ligase orchestrating the ubiquitination of target proteins and shuttling them for degradation usually after the target protein has undergone some post-translational modification that generates a specific structural motif, termed a ‘degron’ [2]. The two major E3 ligase types are the HECT (homologous to the E6-AP carboxy terminus) domain proteins and the RING (really interesting new gene) families. These proteins are represented by hundreds of genes in humans, and the E3 ligases are highly represented in all eukaryotes. The RING E3 ligases far outnumber the HECT family members, and RING E3 ligases can be single enzymes, or part of a multi-subunit E3 ligase complex with adapter proteins mediating interaction between the substrate and the E2 ligase, as characterized by the anaphase-promoting complex (APC) or the Skp1-cullin-F box (SCF) subfamilies.
One example of E3 ligase molecular regulation related to respiration is the oxygen-sensing role of the hypoxia inducible factor (HIF) 1α, which is a potent transcriptional activator of many stress response proteins including chemokines, growth factors, and proteases. In normoxic conditions HIF-1α is hydroxylated forming its degron, and then recognized by the von Hippel–Lindau protein (vHL), an E3 ligase that mediates HIF-1α polyubiquitination and disposal by the proteasome [3]. During hypoxia, however, HIF-1α protein hydroxylation is reduced leading to its accumulation in oxygen starved cells. The net effect of HIF-1α stabilization is rapid activation of stress and survival responses. This is important in normal growth, but has also been well characterized in neoplasia where cancer cells with limited local blood supplies augment their growth potential resulting in local tissue invasion [4]. In translational biomedical research, roles of the vHL protein and HIF1α signaling axis predominantly focus on its role in oncogenesis, but mutations of the vHL cause increased HIF-1α signaling, leading to polycythemia and pulmonary arterial hypertension in afflicted patients, who universally suffer from respiratory insufficiency. In addition to regulation of HIF1α protein concentrations to modulate cell growth and survival responses, vHL also controls edema formation during lung injury. In severe lung injury associated with hypoxia, vHL mediates degradation of Na-K-ATPase required for lung fluid clearance from the airways [5]. Here, it appears that reactive oxygen species generated during lung injury are important to activate vHL that targets the Na-K-ATPase needed to maintain proper lung fluid clearance [6]. As a whole, these observations suggest that components within the UPS are sensitive to low oxygen tension that in turn regulate key UPS physiologic effectors involved in maintaining lung structure and function.
1.2. The importance of the proteasome and ubiquitin E3 ligases in lung homeostasis and disease
As was recently discussed [7], the proteasome has broad activities in acute lung injury, with diverse biologic roles becoming more specialized during this acute response. For example, tumor necrosis factor (TNF) or interferon release from pro-inflammatory cells leads to the conversion of 19S elements in the proteasomal machinery to form an ‘immunoproteasome’, which produces 8–10 residue peptides that are trafficked preferentially through antigen processing machinery and ultimately to the type I major histocompatibility complex to be presented to T-lymphocytes that bolster immunity to pathogens.
In inflammation associated with tobacco exposure, some smokers have a constitutively active inflammatory phenotype and many of them develop COPD. Part of this dysfunction is now known to be secondary to ubiquitin dependent degradation of the genetic regulator histone deacetylase (HDAC) 2 after phosphorylation or adduction of peroxynitrite that is generated by reactive oxygen and nitrogen species created by smoking [8]. This loss of HDAC2 results in aberrant transcription of pro-inflammatory genes, most notably the neutrophil chemokine interleukin 8, causing feed-forward inflammation in some smokers with HDAC2 levels correlating inversely with COPD severity [9].
In a study of acute lung injury, the E3 ligase Cblb was shown to regulate signaling through the Toll-like receptor (TLR) axis [10], which senses pathogen associated molecular patterns (like bacterial flagellin, lipopolysaccharide (LPS), or peptidoglycan, as well as viral capsid proteins or double stranded RNA) and initiates an inflammatory response. Loss of Cblb potentiated the inflammatory response with increased expression of inflammatory genes, persistence of the TLR surface expression, and worsened tissue inflammation leading to decreased survival in LPS challenged animals.
Recent studies have detected proteasomes and activation of the ubiquitin E3 ligase system outside the cellular environment in many organ systems, including the blood, CSF, gut lumen, and lung alveolar fluid. Such proteasomes in lung fluid are only shown to have the 20S subunit, which nonselectively cleaves what proteins it encounters without prior processing or ubiquitination [11]. There is some controversy in the literature regarding the origin of these extracellular proteasomes, as some authors believe that it is a consequence of cell lysis and the spillage of intracellular contents into the extracellular space; often concentrations of proteasomes in lung fluid correlated with concentrations of the intracellular protein lactate dehydrogenase (LDH). Others assert that there is packaging and exocytosis (i.e. active secretion) of intact proteasomes. Regardless of the specific mechanisms of release to the extracellular space, these extracellular proteasomes are increased in the setting of acute infection and inflammation, and they may play an important role in processing extracellular pathogen proteins for antigen presentation by macrophages and dendritic cells to activate immunity against the viruses, bacteria, fungi, or parasites that the host encounters [12].
1.3. The SCF ligase and the role of F-box proteins
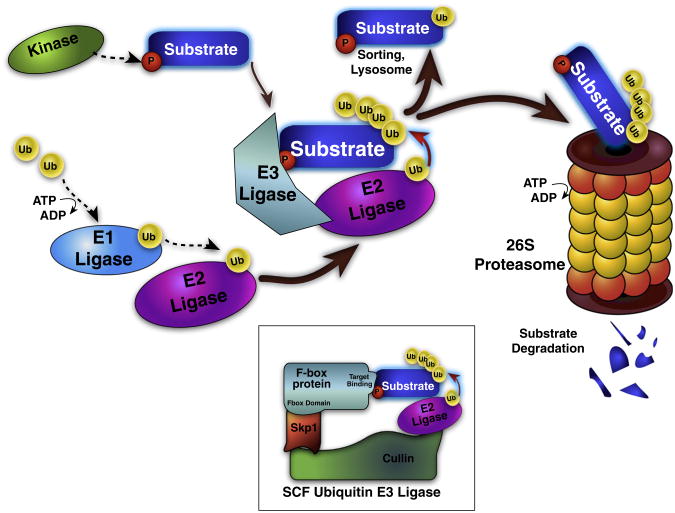
The largest family of cullin-RING E3 ligases is the Skp1-Cullin-F-box protein (SCF) family, a multi-module complex that mediates ubiquitination of post-translationally modified target proteins or substrates often for proteasomal degradation. In this family, the F-box proteins (FBPs) are responsible for substrate specificity, recognizing the degron motif of substrates (usually phosphorylated or otherwise modified) with a target binding domain, and tethering the substrate protein to the rest of the complex via specific interaction between the F-box domain and the Cullin protein [13]. The loaded E3 ligase then ubiquitinates the substrate through activity of a ubiquitin conjugating (E2) enzyme. If the ubiquitin monomer is attached to the substrate by the Ub Lys 48 residue, and four or more Ub residues are added, degradation by the proteasome proceeds (Fig. 1).
Fig. 1.
Schematic of the ubiquitin proteasome system and SCF E3 Ligase. Protein ubiquitination is a regulated, multi-step process. E1 ligases are loaded with ubiquitin (Ub) in an ATP-dependent fashion, and then transferred to E2 ligases. The same E2 ligase can bind many E3 ligases, which in turn can bind multiple target substrate proteins and coordinate Ub transfer from the E2 ligase to the substrate. E3 ligases bind specific substrate proteins based on substrate degron motifs usually consisting of a post-translational modification, such as phosphorylation. Ub addition tags the substrate for sorting, lysosomal destruction, or proteolytic cleavage and degradation by the 26S ubiquitin proteasome. Inset: the Skp1-cullin-F box (SCF) E3 Ub ligase represents the largest family of E3 ligase enzymes, with 68 identified human F-box proteins that interchangeably bind Skp1 and cullin; each F box selectively binds degron-containing substrate proteins, and mediates ubiquitination.
The F-box proteins are thus vital for affecting changes in cellular states including responses to exogenous stimuli or cell cycle progression. Each of these FBPs brings about degradation of a specific set of target or substrate proteins based on the degron structure that it binds. Some 68 FBPs have now been identified in humans, and are designated based on the primary structure of their presumed target binding domains as FBXL (containing a leucine-rich domain), FBXW (containing a WD-40 domain), or FBXO (containing neither leucine rich nor WD-40 domains) [14]. The precise action of each of these molecules is only now beginning to be characterized, with multiple laboratories actively investigating the specific details of the diverse and critical role that these proteins play in substrate binding, regulation, and cell physiology.
2. F-box proteins in inflammatory lung disease
A few FBPs are well described for their essential roles in cell cycle progression, but the physiologic consequences of these proteins in the lung are only being recognized, and our laboratory among several others have recently uncovered potentially important roles of FBPs' by their ability to target key substrates linked to critical pathways of pulmonary homeostasis and disease. Perhaps the most prominently implicated signaling axis in pulmonary inflammation is through the activity of the nuclear factor of kappa light polypeptide gene enhancer in B-cells, NF-κB [15]. This master regulator of inflammation serves as a transcription factor for all classes of inflammatory mediators, and its activity leads to expression of cytokines, chemokines, adhesion molecules, matrix metalloproteases, leukocyte growth factors, and generators of reactive oxygen species. The negative regulator of NFκB is IκB, which binds and sequesters NFκB in the cytosol under normal, noninflammatory conditions [16]. IκB is degraded by the ubiquitin proteasome via one of the first FBPs identified, β-transducin repeat containing protein (β-Trcp, now designated FBXW1). When IκB is phosphorylated by the IκB kinase, it is recognized by SCFFBXW1 to mediate its degradation, leaving NFκB unrestricted to initiate the inflammatory cascade. IκB phosphorylation is in turn regulated by the activity of multiple kinases, which are each activated in response to receptor-associated intermediate protein second messengers, such as the TNF-receptor associated factor proteins (TRAFs; see Section 2.3).
In addition to FBXW1, recent studies from our laboratory have identified and characterized mechanistic roles of other F-box proteins (FBXL19, FBXL2, and FBXO3) relevant to pulmonary inflammation at the level of cytokine receptor stability, surfactant homeostasis, and inflammatory signaling that will be discussed below.
2.1. SCFFBXL19 regulation of the IL-33 receptor
Interleukin 33 (IL-33) is a central mediator of the inflammatory response during asthma and acute lung injury. Zhao and colleagues have shown that the IL-33 receptor ST2 is phosphorylated by the kinase glycogen synthase kinase β (GSK3β), resulting in formation of a phosphodegron motif within the receptor [17]. After phosphorylation, the IL-33 receptor, ST2L, is avidly bound by a previously uncharacterized SCF-based subunit, F box protein FBXL19, that within the SCFFBXL19 E3 ligase complex was sufficient and required to mediate STL2 site-specific polyubiquitination leading to its degradation. Upregulation of GSK3β or overexpression of FBXL19 decreased ST2 availability and downstream mitogen activated protein (MAP) kinase signaling through the IL-33 axis. Importantly, the authors demonstrated that ectopically expressed FBXL19, by mediating ST2 disposal, effectively blunted IL-33 bioactivity in mice evidenced by reduced pro-inflammatory responses, improved lung pathology, and improved survival in animal models of Pseudomonas aeruginosa pneumonia and LPS-mediated ALI [17].
2.2. SCFFBXW1 regulation of Lpcat
Proper respiration requires functional alveolar surface characteristics, with surfactant being a critical component in the tissue's structure and function; lysophosphatidylcholine acyltransferase (Lpcat) is a necessary enzyme for surfactant biosynthesis that is phosphorylated by GSK-3β and then subsequently bound by FBXW1 for ubiquitination [18,19]. In lung injury, loss of Lpcat function by exogenous FBXW1 causes decreased surfactant, worsened lung pathology, and shorter survival.
2.3. SCFFBXL2 regulation of CCTα
FBXL2 was originally described as a geranylgeranylated host F-box protein that was required for hepatitis C virus RNA replication [20]. However, its authenticity as an SCF component and identity of its substrate(s) until recently were unknown. The first description of an FBXL2 substrate was CTP: phosphocholine cytidylyltransferase (CCTα), a rate-regulatory and rate-limiting lipogenic enzyme needed for surfactant phospholipid synthesis in the lung, and thus essential for maintenance of the airway surface dynamics. Chen and colleagues demonstrated that calcium influx after P. aeruginosa infection led to rapid lysosomal degradation of CCTα after its monoubiquitination catalyzed by the SCFFBXL2 E3 ligase complex [21]. Of note, unlike phosphodegron recognition signatures needed for recruitment of other F box proteins to their substrates, the FBXL2 binding site on CCTα was identified within a canonical IQ domain of CCTα; IQ domains are expressed in a great number of diverse proteins, and are typical of calmodulin binding sites. Not surprisingly, calmodulin also bound CCTα within this IQ motif to stabilize the enzyme and displayed intermolecular competition with FBXL2 for occupancy within CCTα. Cellular depletion of FBXL2 also stabilized CCTα protein levels and stimulated surfactant biosynthesis whereas depletion of intracellular cal-modulin destabilized CCTα, underscoring a unique mode of regulation of this surfactant enzyme by calcium and the SCFFBXL2 complex.
Moreover, adenoviral gene transfer of calmodulin lessened the severity of lung injury in a Pseudomonas pneumonia mouse model. Hence, FBXL2 may play an important role in lung homeostasis by regulating concentrations of a key enzyme involved in surfactant metabolism. The findings do not exclude other critical molecular targets for SCFFBAL2.
2.4. Emerging roles of FBXL2: a pivotal regulator of inflammation
Other activities of FBXL2 are emerging and are particularly relevant to cell proliferation and inflammation. The most recently discovered target proteins of FBXL2 are the TNF-receptor associated factors (TRAFs1-6), which also possess a calmodulin binding motif. These proteins are critical adaptor elements that link cell surface receptors to transduction of intracellular signals that may ultimately trigger NF-κB dependent pro-inflammatory gene transcription. Hence, some, but not all receptors that activate TRAF proteins can lead to the synthesis of new inflammatory proteins depending on the cell type and circumstances of receptor ligation and TRAF expression pattern. Using this paradigm, Chen et al. showed that FBXL2 acts as a sentinel feedback inhibitor of inflammation by targeting TRAF1-6 proteins for their polyubiquitination and degradation [22]. Interestingly, similar to the IQ motif identified within CCTα, the authors identified a calmodulin binding site with a critical tryptophan residue conserved among all TRAF family members required for FBXL2 binding. FBXL2 ectopic plasmid expression in a U937 monocyte cell line was sufficient to suppress LPS-induced secretion of inflammatory cytokines, while shRNA knockdown of FBXL2 increased inflammatory cytokine levels in the cells. These new findings have potentially significant implications, suggesting that pharmacologic potentiation or enhancement of the SCFFBXL2 activity might modulate TRAF function and exert a global blunting of inflammatory activity (see Section 4.2.1), in addition to the suppression of cell proliferation (see Section 3.1).
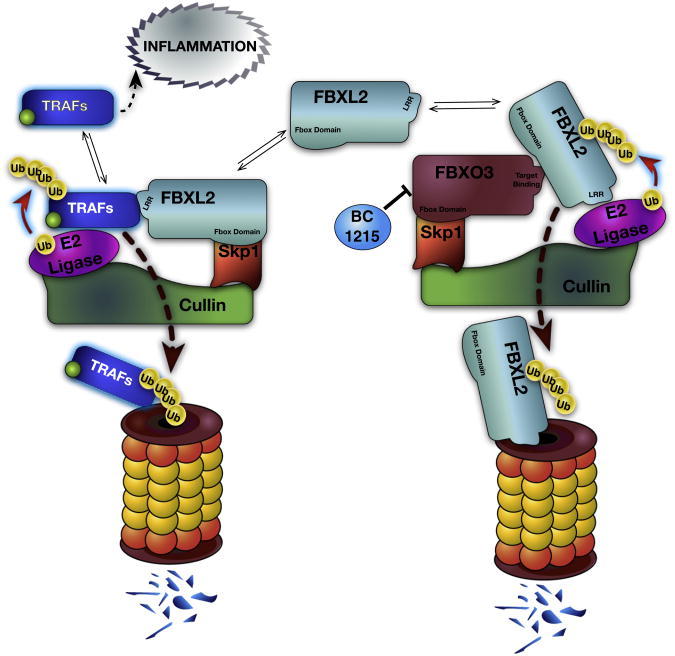
2.4.1. SCFFBXO3 regulation of FBXL2
Generally, components within the SCF machine are targeted by the ubiquitin ligase anaphase-promoting complex or by auto-ubiquitination within their own SCF complex [23,24]. In the process of identifying a mechanism for regulation of the FBXL2 protein, Chen et al., by using a F-box protein expression library for screening of FBXL2 degradation, discovered that another SCF E3 ligase complex, SCFFBXO3, was sufficient to mediate FBXL2 ubiquitination (Fig. 2) [22]. There is limited data regarding the expression and molecular behavior of FBXO3. The transcripts for FBXO3 are increased during malignancy and are elevated in the synovium of rheumatoid arthritis subjects compared to healthy subjects [25]. In leukemia FBXO3 was shown to mediate degradation of HIPK2 and p300 components within the promyelocytic leukemia (PML) protein complex that promotes p53 tumor suppressor gene transcriptional activity [26].
Fig. 2.
Dynamic regulation of inflammatory TRAF proteins by FBXL2 and FBXO3. TNF receptor associated factor (TRAF) proteins have variable signaling effects and act to mediate inflammatory cell activation when active. SCFFBXL2 binds TRAFs, facilitating their ubiquitination under normal circumstances. In inflammatory disease, FBXO3 is upregulated, and leads to ubiquitination of FBXL2 by SCFFBXO3, with elimination of FBXL2 by the UPS. TRAF accumulation directly correlates with FBXO3 levels, with activation of many inflammatory pathways. The FBXO3 inhibitor BC-1215 prevents FBXL2 degradation, restoring TRAF disposal, and blunting inflammation.
SiRNA knockdown of FBXO3 caused accumulation of FBXL2 and predictably decreased TRAF1-6 protein levels in lung epithelia with blunting of LPS-induced inflammatory cytokine release in vitro. Conversely, ectopic expression of FBXO3 plasmid in cells stimulated TRAF-mediated cytokine release in response to endotoxin [22]. These observations provide a unique model of innate immunity in which these two F-box proteins are counter-regulatory for inflammation by acting via TRAF protein abundance.
2.4.2. Genetic protection by a nonfunctional human FBXO3 polymorphism
Given the association of F-box proteins with inflammation, a genetic screen of FBXO3 was performed revealing a SNP allele with a 6% frequency that is nonsynonymous, changing the protein primary structure with a valine ➔ isoleucine substitution at residue 221. In human peripheral blood mononuclear cells (PBMCs), this allele was associated with decreased SCFFBXO3 E3 ligase activity, with increased immunoreactive FBXL2 levels, decreased TRAF levels, and decreased inflammatory cytokine release in response to LPS [22]. Accordingly, FBXL2 polyubiquitination was decreased in vitro by FBXO3V221I compared to wild-type FBXO3 (FBXO3WT). In vivo, mice treated with lentiviral vectors containing FBXO3V221I had decreased airway inflammation in a murine pseudomonal pneumonia model compared to FBXO3WT. When studied in a small series of clinical samples from humans hospitalized for sepsis, those with the FBXO3V221I genotype had lower levels of inflammatory cytokines secreted in plasma compared to their wild-type counterparts [22].
3. F-box proteins in cancer biology: how much do we know about lung cancer?
There has been much research in the past decade validating the critical role of the UPS in cell proliferation and, by extension, cancer pathobiology. Many proteins in the ubiquitin pathways and the SCF family have been evaluated in the context of cancer biology, and proteasome inhibitors represent a new class of antineoplastic agents, whose clinical utility is now a cornerstone of cancer chemotherapy for some hematologic malignancies (see Section 4.1). A general discussion of these complex processes is well described elsewhere [27] and beyond the scope of this review. Among the best-characterized F-box proteins in malignancy are the cancer-promoting proteins β-Trcp a.k.a. FBXW1, and SKP2 (FBXL1) [28]. As discussed above, β-Trcp drives inflammatory NFκB activity resulting in increased expression of cell-activating cytokines, growth factors, and proteases, all of which can contribute to dysregulated physiology underlying cancer proliferation and tissue invasion. β-Trcp also targets the β-catenin protein, which is critical for cell proliferation and differentiation through the Wnt signaling pathway. Thus, loss of β-catenin could impair cell differentiation, typical of aggressive malignancies [29]. In addition, SKP2 facilitates degradation of multiple tumor suppressor proteins including p27, Fox01, p21, and p57 and is thus generally considered a proto-oncoprotein. FBXW7, however, is p53 induced for degradation of oncoprotein transcription factors (c-jun, c-Myc), and is considered a cancer-protective F-box; mutations in FBXW7 are associated with poor prognosis in cancer [30].
In lung cancer specifically, SKP2 has been implicated as a pro-neoplastic factor. In a small Canadian study from 2004 [31], nonsmall cell lung cancer cell lines and primary tumor analysis revealed significant upregulation of SKP2 in squamous cell cancers with worse outcomes seen when both oncogenic RAS mutation and SKP2 upregulation were present. These findings were corroborated by two Japanese clinicopathologic studies [32,33] showing that increased SKP2 protein levels in biopsy specimens are a poor prognostic factor associated with increased metastasis. Moreover, decreased p27 in SKP2hi specimens is robustly correlated with shorter survival.
3.1. FBXL2 as a regulator of cell proliferation and mitosis
In a new series of studies from our group, the range of FBXL2 targets has been expanded. As described above, FBXL2 binds the calmodulin-binding IQ domain of multiple proteins involved in cellular replication. For example, FBXL2 regulates proteins critical to cell cycle progression by binding cyclin D2 and cyclin D3 proteins involved in G1/S or G2/M phase transition, respectively; the F box protein also impacts mitotic division by regulating Aurora B concentrations. Overexpression of FBXL2 depletes cyclin D3 and prevents unchecked cell growth in epithelial cells [34], while it impairs cellular proliferation and induces apoptosis by mediating destabilization of cyclin D2 in a lymphoblastic leukemia cell line [35]. FBXL2 also targets Aurora B for its degradation by recognizing an IQmotif [36]. Calmodulin competes for the IQ domain present on cyclinD2, cyclin D3, and Aurora B, and stabilizes these proteins to promote cell division. When FBXL2 protein levels are increased or cal-modulin concentrations are decreased, the cyclin proteins are degraded, causing cell cycle arrest, and Aurora B levels are likewise depleted resulting in tetraploidy, mitotic failure, and apoptosis of the tumorigenic cells. These studies indicate that FBXL2 is protective by acting as a functional growth inhibitor in settings of cellular hyperproliferation as observed in cancer.
4. The ubiquitin proteasome system as a drug target
4.1. Nonselective proteasome inhibitors
Bortezomib is the first FDA approved drug directed toward the proteasome. This reversible inhibitor of the 20S proteasome, causes cell cycle derangement, and is marketed by the trade name Velcade. Bortezomib has revolutionized multiple myeloma (MM) treatment, with a survival prolongation to greater than a decade in many cases, compared to 50% mortality at 20 months after diagnosis before routine use of novel therapies like thalidomide and bortezomib [37,38]. In 2012, Carfilzomib became the second FDA-approved proteasome inhibitor, and is a second line therapy for multiple myeloma and non-Hodgkins lymphoma patients whose cancer progresses after treatment with first line therapy.
4.2. New proteasome inhibitors
Bortezomib and Carfilzomib have set the stage for proteasome inhibition as a drug target. Many second generation proteasome inhibitors including Carfilzomib were recently discussed [39], with each agent targeting similar processes (i.e. cell proliferation), and being tested preclinically and in phase I and II trials against hematologic malignancy and solid tumors.
Other drugs that target upstream events in proteasome biology are also under development. MLN4924 targets the NEDD8 activating enzyme, which is necessary for function of the cullin RING proteins [40]; ubiquitination through cullin RING ligases (including SCF E3 ligases and vWF) is therefore disabled by the drug. In vitro antitumor activity of this compound is promising, promoting apoptosis or autophagy of these cells, and MLN4924 safety testing is now being conducted in a phase I trial.
CC0651 is the first small molecule inhibitor of an E2 ligase, with high potency and specificity for the E2 ligase Cdc 34 [41]; this drug suppresses ubiquitination through all E3 ligases that depend on Cdc34, which includes all of the cullin RING ligases, and was identified by assaying FBXL1 activity. Based on this high degree of overlap, CC0651 and MLN4924 would theoretically result in similar biologic changes via different targets, and may therefore be synergistic. Tosyl-L-Arginine Methyl Ester (TAME) was described as a small molecule inhibitor of the anaphase promoting complex/cyclosome, a RING containing E3 ligase (that is not among the cullin RING family) whose activity is required for dismantling the spindle assembly check point and completion of mitotic division [42]. TAME was identified from a chemical library using high throughput screening, and prevents depletion of cyclin B1 thereby leading to mitotic arrest in metaphase.
The next putative selective target of the ubiquitination-proteasome pathways is to target individual subunits of the E3 ligases. Although no drugs with this activity have entered clinical trials, there have been some recent advances in this area, which we will discuss next.
4.2.1. F-box protein specific inhibitors
The first report of an E3 ligase-targeting drug was published in 2010, where the authors discovered SCF-I2 by small molecule interrogation of the SCFCdc4 molecular complex for drugs that would displace the SCF from its phosphodegron in a budding yeast system. This molecule seemed to work by allosteric inhibition of the WD40 β-propeller domain, making it unable to bind the substrate protein. This molecule antagonized SCFCdc4, but not SCF complexes with related FBPs nor the human ortholog, FBXW7 [43].
Another important development in this arena is the development of a small molecule inhibitor of the FBXO3 molecule that targets its c-terminal ApaG domain. This compound, BC1215, blocks FBXO3 activity to decrease TRAFs and IL-1β production in vitro in PBMCs treated with LPS [22]. In vivo, BC-1215 was administered intraperitoneally in animal models of pseudomonal pneumonia and systemic sepsis with lung injury [22]. BC-1215 treatment reduced the inflammatory phenotype and prolonged survival in mice. This represents the first small molecule E3 ligase antagonist to display efficacy in pulmonary and systemic inflammation. Moreover, it is the first targeted, rationally designed, and specific inhibitor to a single F-box protein; efficacy of the drug requires validation, but may set the stage for a new genus of anti-inflammatory drugs.
5. Conclusions
In summary, the UPS and SCF biology has proven to be a fundamental area of discovery in recent years with many new insights into specific lung physiology, and the above discussion leaves behind many valuable contributions. Most F-box proteins' activities are completely unknown, so many discoveries await us in the years to come, however, it is becoming clear that protein processing via the UPS plays a central role in most of the principle disease types of the lungs. Along with these newly described mechanisms of pathology come significant advances in our strategies for intervention and expansion of our arsenal of potential therapies (Section 4). While treatment with UPS-targeting medications has thus far been limited to hematologic malignancy, the use of E3 ligase or F-box specific drugs as anti-inflammatory agents may become a commonplace in the next decade, especially as more selective compounds with fewer side effects are identified. It is difficult to know at this early stage what drugs may become important for which diseases, but as we expand our knowledge about this captivating biology, we hope that our capacity for devising therapies for an array of diseases will become a focused and effective enterprise.
References
- 1.Ciechanover A. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Ravid T, Hochstrasser M. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 5.Zhou G, Dada LA, Chandel NS, Iwai K, Lecuona E, Ciechanover A, Sznajder JI. FASEB J. 2008;22:1335–1342. doi: 10.1096/fj.07-8369com. [DOI] [PubMed] [Google Scholar]
- 6.Vadasz I, Raviv S, Sznajder JI. Intensive Care Med. 2007;33:1243–1251. doi: 10.1007/s00134-007-0661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vadasz I, Weiss CH, Sznajder JI. Chest. 2012;141:763–771. doi: 10.1378/chest.11-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Am J Respir Cell Mol Biol. 2009;40:464. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, Hogg JC. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 10.Bachmaier K, Toya S, Gao X, Triantafillou T, Garrean S, Park GY, Frey RS, Vogel S, Minshall R, Christman JW, Tiruppathi C, Malik AB. Nat Med. 2007;13:920–926. doi: 10.1038/nm1607. [DOI] [PubMed] [Google Scholar]
- 11.Majetschak M, Sorell LT, Patricelli T, Seitz DH, Knoferl MW. Physiol Res. 2009;58:363–372. doi: 10.33549/physiolres.931526. [DOI] [PubMed] [Google Scholar]
- 12.Sixt SU, Peters J. Proc Am Thorac Soc. 2010;7:91–96. doi: 10.1513/pats.200906-035JS. [DOI] [PubMed] [Google Scholar]
- 13.Cardozo T, Pagano M. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 14.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Verma IM. Nat Rev Immunol. 2002;2:725. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 16.Mercurio F, Manning AM. Curr Opin Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, Zou C, Mallampalli RK, Zhao Y. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou C, Ellis BM, Smith RM, Chen BB, Zhao Y, Mallampalli RK. J Biol Chem. 2011;286:28019–28025. doi: 10.1074/jbc.M111.253385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou C, Butler PL, Coon TA, Smith RM, Hammen G, Zhao Y, Chen BB, Mallampalli RK. J Biol Chem. 2011;286:2719–2727. doi: 10.1074/jbc.M110.192377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Gale M, Jr, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Mol Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Chen BB, Coon TA, Glasser JR, Mallampalli RK. Mol Cell Biol. 2011;31:1905–1920. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen BB, Coon T, McVerry BJ, Zhao J, Zou C, Ellis BM, Sciurba FC, Zhang Y, Zhao Y, Glasser JR, Mallampalli RK. Nat Immunol. 2013;14:470–479. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galan JM, Peter M. Proc Natl Acad Sci U S A. 1999;96:9124–9129. doi: 10.1073/pnas.96.16.9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 25.Masuda K, Masuda R, Neidhart M, Simmen BR, Michel BA, Muller-Ladner U, Gay RE, Gay S. Arthritis Res. 2002;4:R8. doi: 10.1186/ar427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I. Mol Cell Biol. 2008;28:7126–7138. doi: 10.1128/MCB.00897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani A, Gelmann EP. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 28.Frescas D, Pagano M. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs SY, Spiegelman VS, Kumar KS. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 30.Tan HL, Regamey N, Brown S, Bush A, Lloyd CM, Davies JC. Am J Respir Crit Care Med. 2011;184:252–258. doi: 10.1164/rccm.201102-0236OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu CQ, Blackhall FH, Pintilie M, Iyengar P, Liu N, Ho J, Chomiak T, Lau D, Winton T, Shepherd FA. Clin Cancer Res. 2004;10:1984–1991. doi: 10.1158/1078-0432.ccr-03-0470. [DOI] [PubMed] [Google Scholar]
- 32.Osoegawa A, Yoshino I, Tanaka S, Sugio K, Kameyama T, Yamaguchi M, Maehara Y. J Clin Oncol. 2004;22:4165–4173. doi: 10.1200/JCO.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 33.Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Am J Pathol. 2004;165:175–180. doi: 10.1016/S0002-9440(10)63286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen BB, Glasser JR, Coon TA, Mallampalli RK. Oncogene. 2012;31:2566–2579. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, McDyer JF, Boyiadzis M, Mallampalli RK. Blood. 2012;119:3132–3141. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BB, Glasser JR, Coon TA, Mallampalli RK. Cell Cycle. 2011;10:3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust JA. Blood. 2008;111:2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonneveld P, Schmidt-Wolf IG, van der Holt B, el Jarari L, Bertsch U, Salwender H, Zweegman S, Vellenga E, Broyl A, Blau IW. J Clin Oncol. 2012;30:2946–2955. doi: 10.1200/JCO.2011.39.6820. [DOI] [PubMed] [Google Scholar]
- 39.Dick LR, Fleming PE. Drug Discov Today. 2010;15:243–249. doi: 10.1016/j.drudis.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 41.Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 42.Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]