Abstract
The genome of influenza A virus encodes a newly discovered protein that diminishes its pathogenicity.
Influenza A virus (IAV) remains a major cause of human mortality and morbidity due to its remarkable genetic variability, which limits vaccine effectiveness. IAV’s high intrinsic mutation rate is augmented by its segmented genome, which greatly facilitates beneficial interactions among its genes (1) and is likely a key to IAV’s capacity to infect multiple mammalian and avian species. IAV must accomplish much with minimal genetic information—a mere 13.5 kilo-bases of RNA. To maximize its genome, IAV uses many tricks, including gene splicing, downstream translation initiation in normal or alternative overlapping frames, and, as Jagger et al. report on page 199 of this issue (2), ribosomal frameshifting, which generates a gene product that diminishes viral pathogenicity.
Because of the degeneracy of the genetic code, each amino acid can be encoded by as many as six different nucleotide triplets (codons). High codon conservation in a given gene region can reflect multiple factors, including RNA folding, RNA interaction with viral or cell macromolecules, transfer RNA (tRNA) preference (because synonymous codons are often decoded by distinct tRNA species), or the need to preserve a conserved overlapping open reading frame.
Analyzing nearly 1300 genome sequences from diverse isolates, Jagger et al. found that IAV gene segment 3 (there are eight segments), which encodes the PA subunit of the viral RNA polymerase, exhibits a highly conserved internal sequence that potentially encodes a 61-residue polypeptide in a +1 open reading frame [because of the triplet nature of the genetic code, each messenger RNA (mRNA) potentially encodes information in three reading frames]. Noting the lack of a proximal potential +1 canonical translation start codon or mRNA splice acceptor site to shift frames, the authors focused on a decanucleotide sequence. The near-absolute conservation of this sequence between IAV strains is striking, given the potential use of 15 synonymous codons for the three encoded amino acids and the presence of a CGU codon, which is rarely used by human or viral genes. Such rare codons are typically decoded more slowly because of the low concentration of their cognate tRNAs—a mechanism that can promote ribosomal frame-shifting (3) (see the figure). Because the abundance of individual tRNA species can vary widely among mammalian cell types (4), this mechanism can potentially modulate frameshifting in different cell types.
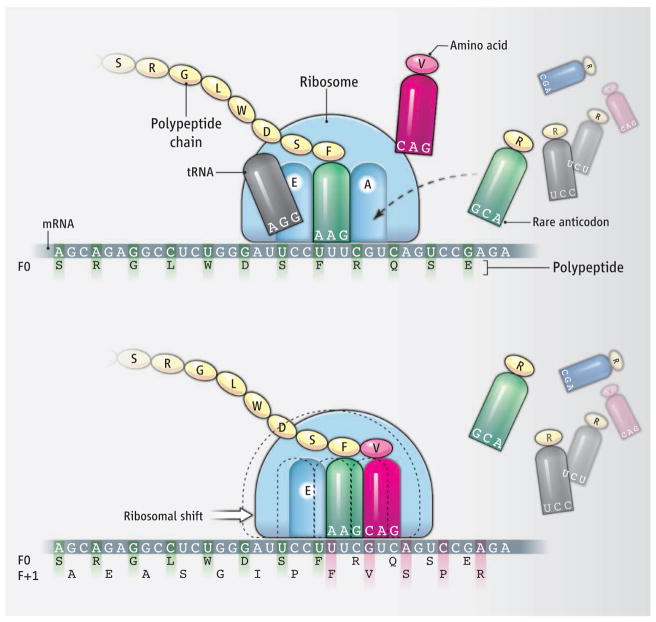
Figure. Ribosomal shift.
As the ribosome (60S subunit shown) transits PA mRNA in the standard reading frame (F0), the codon in the ribosomal A site is matched to the cognate aminoacyl tRNA, whose amino acid cargo is bonded to the elongating polypeptide. Deacylated tRNA is ejected from the E (exit) site. Ribosomal pausing as the slowly decoded arginine (R) codon CGU enters the A site and waits for its rare 3′GCA5′ anticodon allows the upstream phenylalanine (F) anticodon 3′AAG5′ to occasionally slip into the preferred +1 UUC codon, thus changing the reading frame to F+1 and altering the C-terminal peptide to generate the PA-X fusion protein.
Jagger et al. show that the translation of PA mRNA generates a protein that consists of the amino-terminal 191 amino acids of PA fused to 61 amino acids that result from +1 frameshifting. Expression of the PA-X protein from cDNA that encodes PA likely explains the observed decreased expression of a number of host cell proteins (5)—a phenomenon that had long been attributed to PA protease activity despite the absence of sequence or structural features that suggest protease function (6). Rather than PA itself, it is PA-X, using the PA endonuclease domain, which selectively degrades host mRNAs, abrogating their translation and reducing expression of short-lived cellular proteins.
PA-X exhibits remarkable sequence conservation across strains, although it does come in two forms, with many strains of the recently introduced swine-origin IAV (SOIV) possessing a stop codon that truncates PA-X by 20 residues. This situation is eerily similar to PB1-F2, the first IAV gene product shown to be generated from a non-spliced alterative reading frame (encoding 87 to 90 amino acids in PB1) (7). PB1-F2 exhibits similar length polymorphism [typically, SOIVs encode an 11-residue fragment, and recent classical H1N1 viruses express a 57-residue form (8)]. Further, like PB1-F2 (8), PA-X is a viral accessory protein (i.e., dispensable for replication, but rather modulates host immune response).
Jagger et al. show that eliminating PA-X expression in the highly pathogenic 1918 IAV (whose associated pandemic caused perhaps 50 million deaths worldwide) enhances viral pathogenicity while inducing many changes in host gene expression, as determined by global transcriptional profiling. This may reflect, at least in part, the direct and cascading effects of host mRNA degradation by PA-X, although it is probable that PA-X, like many viral accessory proteins, is highly multifunctional. Although IAV pathogenesis is complex, it seems that PA-X diminishes viral damage by reducing the expression of select proinflammatory cytokines.
Intriguingly, PA-X and PB1-F2 exert opposite effects on pathogenesis, because PB1-F2 enhances morbidity and mortality (albeit in a viral strain– and host-dependent manner) (9). Presumably, the two proteins work together to optimally modulate host immunity. It will be interesting to examine the effect of PA-X on secondary bacterial pneumonia, whose severity is enhanced by PB1-F2 in mouse-model infections (10).
How do PA-X and PB1-F2 contribute to viral transmission between hosts, given that transmission is the ultimate selective factor in viral evolution? IAV’s high mutation rate ensures that genes that do not confer increased fitness quickly succumb to loss-of-function mutations.
Viral pathogenesis and transmission are intimately related: Viruses have no interest in incapacitating their hosts if it limits viral transmission. Indeed, few viruses exhibit both high lethality and transmissibility (among human viruses, smallpox is the only known example). What is IAV aiming for? Perhaps to diminish immunological memory to preserve a susceptible host population. Perhaps to induce sufficient sneezing, coughing, rhinorrhea, etc., to maximize transmission, but not such severe disease that the host remains at home in bed with minimal chance to infect others.
It may be exceedingly difficult to accurately model the effects of these evolutionary constraints in animal models, which are unlikely to recapitulate subtle immunological, behavioral, or physical effects critical to human IAV transmission. Complicating matters is the constant flow of IAV between host species, which may preserve genes that confer a significant selective advantage in some species but not others, and de-optimize transmission for any one species. Identifying factors such as PA-X, however, is an essential step on the path to greater understanding.
Acknowledgments
Supported by the Division of Intramural Research, NIAID.
References
- 1.Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. PLoS Genet. 2011;7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagger BW, et al. Science. 2012;337:199. doi: 10.1126/science.1222231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belcourt MF, Farabaugh PJ. Cell. 1990;62:339. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmar KA, Goodenbour JM, Pan T. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz-Ezquerro JJ, de la Luna S, Ortín J, Nieto A. J Virol. 1995;69:2420. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boivin S, Cusack S, Ruigrok RWH, Hart DJ. J Biol Chem. 2010;285:28411. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, et al. Nat Med. 2001;7:1306. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 8.Krumbholz A, et al. Med Microbiol Immunol. 2011;200:69. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- 9.Zamarin D, Ortigoza MB, Palese P. J Virol. 2006;80:7976. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAuley JL, et al. Cell Host Microbe. 2007;2:240. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]