Abstract
The flavonoids and the terpene lactones are regarded as the two main active components of Ginkgo biloba that affect human health. In the work discussed in this paper, two analytical methods for the characterization of G. biloba authentic materials and commercial products, an LC–UV chromatographic fingerprinting method and a traditional flavonol quantification method, were compared. The traditional method was used to determine the total flavonol content (as glycosides) after acid hydrolysis. The fingerprinting method examined the chromatographic profiles of methanol–water extracts using chemometric methods. The traditional method showed that all the commercial products met the current voluntary standard of 24% flavonols by weight. The chromatographic fingerprinting method revealed significant variations in the commercial products with regard to the relative concentration of individual flavonols.
Keywords: Ginkgo biloba, HPLC, UV, Dietary supplement, Fingerprint
Introduction
The Ginkgo biloba tree is one of the oldest known trees on earth with fossil records dating back more than 200 million years. G. biloba leaves and berries have long been used in China as a traditional medicine for various ailments. Scientific studies of G. biloba began in the 1950s and have shown it to be a useful remedy for a wide range of disorders [1]. G. biloba has been reported to affect fundamental aspects of human physiology by improving blood flow to tissues, including the brain, and by enhancing cellular metabolism, both symptoms associated with old age [2]. A comprehensive summary of research on G. biloba has been published by the Office of Dietary of Supplements of the National Institute of Health (ODS, NIH) [3].
The pharmacological activity of extracts of G. biloba leaves (GBE) has been attributed to two major classes of compounds, terpene lactones and flavonols [4–7]. Terpene lactones are potent and selective antagonists of platelet activating factor. Flavonols are responsible for the free radical-scavenging and antioxidant activity of G. biloba preparations [8]. Quercetin, kaempferol, and isorhamnetin are aglycones of the flavonols present in GBE [8, 9]. Many flavonol glycosides have been identified in GBE [10].
GBEs are currently prepared to meet the unofficial industrial standard of EGb 761 established by W. Schwabe (Karlsruhe, Germany). Preparations meeting the EGb 761 standard contain 24% flavonol glycosides and 6% terpene lactones (a standard deviation of 20% is allowed for both). The recommended daily amount is 120–240 mg [11]. Chemistry, pharmacology, toxicology, and clinical studies conducted using EGb 761 were reviewed in 1998 [12].
High-performance liquid chromatography with UV detection (HPLC–UV) is currently the most popular system for flavonol determination. The total flavonol content is determined by converting the glycosides to aglycones by acid hydrolysis [4] and comparison with aglycone standards. Terpene lactones are poor chromophores with weak absorption in the range 200–220 nm. Light loss in this wavelength region is primarily because of refraction and analyses are subject to interferences from sample matrices and solvents. This makes UV detection an unsuitable option for a complex matrix [8], so other detection methods, for example evaporative light-scattering detection (ELSD), sonic spray ionization (SSI) mass spectrometry, atmospheric-pressure chemical ionization (APCI) mass spectrometry, and electrospray (ESI) mass spectrometry, have been used for detection [13–17].
Quantitation of total flavonols in G. biloba does not give any information about the composition of the individual flavonol glycosides, which leads to the possibility of adulteration [18, 19]. Commercial G. biloba products may fully meet label claims and still have dramatically different flavonol composition [19].
The growing demand for botanical dietary supplements requires new analytical approaches for assessment of the identity, potency, and purity of these materials. In 1991 chromatographic fingerprinting technology was accepted by the World Health Organization as a strategy for identification and evaluation of the quality of herbal medicines [20]. The reproducibility of such chromatograms depends on the instrumentation, experimental conditions, variations in HPLC columns and reagents, and the skills and backgrounds of personnel in different parts of the world, however. Thus, efficient and consistent comparison of chromatographic fingerprints is not a trivial undertaking.
In this study a method was developed that reduced the variability introduced by experimental conditions. Chromatographic fingerprints of fourteen commercial G. biloba dietary supplements (GDS), one American Herbal Pharmacopoeia (AHP)-verified G. biloba leaf sample, and three G. biloba standard reference materials (SRM 3246, leaf material, SRM 3247, GBE, and SRM 3248, tablet) from the National Institute of Standards and Technology (NIST) were analyzed. These results were compared with the total flavonol content obtained by use of traditional acid hydrolysis.
Materials and methods
Chemicals
The water and methanol used were of Optima grade (Fisher Scientific, Pittsburgh, PA, USA). The trifluoroacetic acid (TFA) used was of HPLC grade (Sigma–Aldrich, St Louis, MO, USA). Rutin, quercetin, kaempferol, and isorhamnetin were also purchased from Sigma; purities were all labeled as greater than 90%.
Commercial samples
Fourteen different GDS samples (designated samples 1 to 14) were purchased from local stores in the greater Washington DC area. All fourteen samples were purchased within the same week, were in the solid form (tablets and capsules), and were analyzed before their expiration dates. G. biloba standard reference materials SRM 3246 (dried leaves), SRM 3247 (GBE), and SRM 3248 (tablet) were obtained from NIST. A verified botanical reference standard BRS-GB (dried leaves) was purchased from the American Herbal Pharmacopoeia (Scotts Valley, CA, USA). The NIST SRMs are high-quality, authentic, and well homogenized samples and AHP verified G. biloba leaves are authentic material from a different source. None of the standards provides, or is intended to provide, any information about seasonal or processing variation.
Sample preparation
Ten tablets of each sample were weighed and ground to fine powder using a mortar and pestle. Ten capsules were opened and emptied, and the contents were mixed and weighed. The GBE content of each sample was calculated using the weight of the sample powder and the labeled amount of GBE in each tablet or capsule and expressed as a ratio (w/w). Each sample (200 mg) was extracted twice with H2O–MeOH (1:1, 10 mL) as described elsewhere [19]. The two extracts were combined and filtered through 0.45 μm Nylon syringe filters (Alltech Associates, Deerfield, IL, USA). Each extract was analyzed in triplicate.
Acid hydrolysis
Before quantitation, flavonol glycosides were converted to their aglycones by acid hydrolysis, using the method reported by Wagner [20] with slight modification (a volume of 1 mL instead of 10 mL). Hydrolysis was performed by adding 1 mL 5.5% HCl in MeOH to 1 mL of the sample extract in a closed flask. The solution was heated for 1 h at 100 °C. The hydrolyzed samples (500 μL) were immediately transferred to standard 2-mL HPLC vials and analyzed.
Instrumentation and the LC–UV method
HPLC was performed with an Agilent Technologies (Palo Alto, CA, USA) 1100 Series system consisting of a binary pump with vacuum degasser, a thermostatted column compartment, an autosampler, and a diode-array detector (DAD). Compounds were separated on a Supelco Discovery C18 HPLC column (250 mm×3.0 mm, 5 μm particles; Sigma–Aldrich).
The HPLC conditions were the same as described previously [19]. Briefly, mobile phase A consisted of 0.05% TFA in H2O and mobile phase B consisted of 0.05% TFA in MeOH. The initial ratio of A to B was 70:30; this was changed linearly to 30:70 in 26 min, to 5:95 after 28 min, held constant until 33 min, and then reset to 70:30 where it was held until 40 min when the next sample was injected. The wavelength for UV detection was 360 nm. These conditions afforded satisfactory separation of all the flavonol glycosides and flavonol aglycones investigated.
Calculation
The total flavonol content (as glycosides) was calculated from the measured quantities of flavonol aglycones by use of the method described by Hasler [10]. The factors for conversion from aglycone mass to glycoside mass are 2.51 for quercetin, 2.64 for kaempferol, and 2.39 for isorhamnetin.
The total flavonol content (as glycosides) was expressed as a percentage of the GBE to facilitate sample-to-sample comparison. AHP verified leaves and NIST SRM 3246 (leaves) and 3248 (tablet) were not included in the quantitative studies because the results cannot be converted to percentages of GBE. The results were calculated by using the formula:
where Cs is the flavonol glycoside concentration (μg mL−1) (calculated from the measured concentration of aglycones in hydrolyzed G. biloba samples), DF is the dilution factor (samples were diluted twofold during hydrolysis and fivefold before analysis for aglycone quantitation and injected directly for rutin quantification), V is the volume of sample extract solution (mL), Ws is the mass of sample extracted (∼200 mg), and GBE(w/w) is the GBE mass ratio calculated from the mass of the sample and the labeled amount of GBE contained in the sample.
Results and discussion
Quantification of the total flavonol content
Quantification of individual flavonol glycosides by LC–UV was not possible due to the lack of commercially available standards. Instead, the flavonol glycosides were converted to their aglycones (quercetin, kaempferol, and isorhamnetin) by acid hydrolysis, a well established procedure. Reference standards for the three aglycones are commercially available. Details of the quantification of total flavonols in G. biloba samples using this method have been discussed elsewhere [19]. The total flavonol content for all the samples ranged from 24.8 to 30.8% of their respective GBE (Table 1). All of the samples met or exceeded the voluntary standard of 24% flavonols. Although the traditional (acid hydrolysis) approach has been widely accepted for quality control of herbal preparations containing flavonol components [8, 16, 21, 22], it is known that the method are susceptible to adulteration [18, 19] and/or degradation [4, 22].
Table 1. Amounts (%) of rutin, free aglycones, and quercetin in G. biloba samplesa.
| Sample | PCA plot symbol | Amount of rutin (% total area) | Amount of free aglycone (% total area) | Amount of quercetin (% total area) | Quer/Aglycb |
|---|---|---|---|---|---|
| 1 | a | 4.47±0.03 | 74.76±0.50 | 66.54±0.81 | 0.89 |
| 2 | b | 13.28±0.21 | 27.76±0.99 | 24.75±0.96 | 0.89 |
| 3 | c | 14.47±0.05 | 7.56±0.16 | 3.71±0.19 | 0.49 |
| 4 | d | 24.54±1.21 | 29.19±0.37 | 15.63±0.28 | 0.53 |
| 5 | e | 25.18±0.27 | 10.16±0.26 | 5.08±0.26 | 0.50 |
| 6 | f | 27.23±0.69 | 28.72±0.21 | 12.16±0.28 | 0.42 |
| 7 | g | 32.60±0.11 | <5 | n/a | n/a |
| 8 | h | 42.08±0.28 | 7.00±0.13 | 2.86±0.09 | 0.41 |
| 9 | i | 42.68±0.10 | 5.19±0.17 | 2.26±0.05 | 0.44 |
| 10 | j | 46.96 ±0.07 | <5 | n/a | n/a |
| 11 | k | 60.01±0.24 | 14.25±0.39 | 7.78±0.46 | 0.55 |
| 12 | l | 74.35±0.01 | 7.93±0.04 | 3.95±0.03 | 0.50 |
| 13 | m | 77.84±0.11 | 9.34±0.18 | 7.21±0.21 | 0.77 |
| 14 | n | 80.82±0.23 | <5 | n/a | n/a |
| AHP Std | o | 25.44±0.08 | <5 | n/a | n/a |
| SRM 3246 | P | 20.09±0.14 | <5 | n/a | n/a |
| SRM 3247 | q | 21.24±0.19 | <5 | n/a | n/a |
| SRM 3248 | r | 20.90±0.28 | <5 | n/a | n/a |
Amounts of rutin, free aglycones, and quercetin were calculated by dividing the respective peak area(s) by the total peak areas of the twenty-one peaks (n=3).
Ratio of quercetin to free aglycones
Peak analysis of chromatographic fingerprints
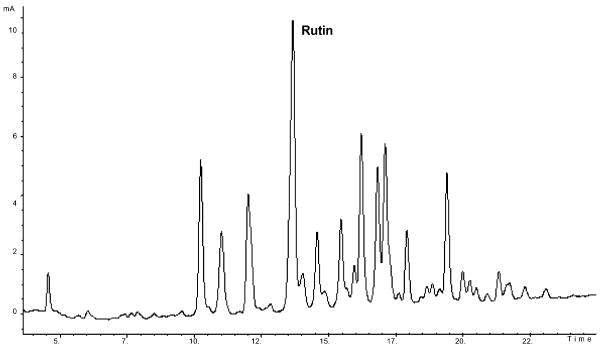
A typical chromatographic fingerprint obtained from NIST SRM 3247 is shown in Fig. 1. For the eighteen samples studied, a total of twenty-one major peaks was obtained (where a major peak was defined as any peak with at least 5% of the rutin peak area).
Fig. 1. LC–UV chromatogram obtained from NIST SRM 3247 (UV, 360 nm).
The peak areas for the same components varied substantially among samples and were occasionally less than 5% of the rutin peak area. Of the twenty-one peaks, four were identified using commercial standards (rutin at 13.75 min, quercetin at 19.21 min, kaempferol at 22.44 min, and isorhamnetin at 23.21 min). For most samples, rutin was the largest peak; the exceptions were samples 1 and 2 for which the quercetin peak was largest.
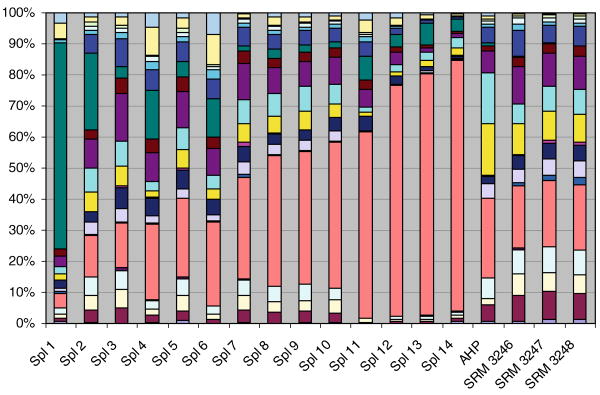
Samples were characterized by dividing individual peak areas by the sum of the total area of all the peaks. The peak areas for all the peaks are listed in Appendix 1. Table 1 shows peak-area percent for rutin, quercetin, and the sum of the free aglycones (found without hydrolysis). The ratio of quercetin to the free aglycones is also shown. The composition of the twenty-one peaks for each of the fourteen samples and the four standards is summarized graphically in Fig. 2.
Fig. 2. The percentages of the twenty one peaks in the LC–UV chromatograms of the eighteen G. biloba samples (from bottom upward, peak 1 to peak 21).
It is difficult to analyze the complex relationships among all the peaks by visual inspection. The patterns for rutin and quercetin are particularly striking, however. On the basis of these components the samples could be divided into four groups.
-
–
Samples in Group #1 (samples 1-3) contain less than 15% rutin. Noticeably, two of the three samples contain substantially higher percentages of quercetin compared with the total peak areas and the area of the free aglycones.
-
–
Samples in Group #2 (samples 4–7 and the standards) contained 20% to 33% rutin and their quercetin-to-aglycone ratios were approximately 0.5, when measurable. Most importantly, the samples in Group #2 have characteristics similar to the standards. Samples 4 and 6 contained higher percentages of the free aglycones, however, suggesting that decomposition of the flavonol glycosides may have been the source of the free aglycones.
-
–
Samples in Group #3 (samples 8–10) contained 40 to 50% rutin.
-
–
Samples in Group #4 (samples 11–14) contained more than 60% rutin, 2 to 4 times higher than in the standards.
PCA analysis of chromatographic fingerprints
PCA is a popular method that can be used to summarize multivariate data. The simplest approach to applying PCA to chromatographic data is to use the entire chromatogram as the input data set for each sample. Unfortunately, the analytical variability of the retention times and the detector response make this approach problematic. Sophisticated mathematical models have been used to address these problems [23–26]. Unfortunately, these models have limited applicability.
For our study, the analytical variability was reduced by using a template to identify the peaks of interest and normalizing the areas of each peak by the area of a preselected component—rutin. The chromatograms of all samples were compared with the chromatogram obtained from NIST SRM 3247 to ensure correct peak identification. Areas for each peak were then compiled, and if the area was less than 5% that of the rutin peak a value of zero was inserted in the data set (Appendix 2). All peak areas were then normalized by the area of the rutin peak. Using this approach the potential variations in retention time were eliminated. As long as the same HPLC column was used, the order of elution of the peaks did not change, and their identification was straightforward. Normalization of all peak areas by the area of the rutin peak minimized variations introduced by the detection system. The data set used for PCA analysis is included in Appendix 2.
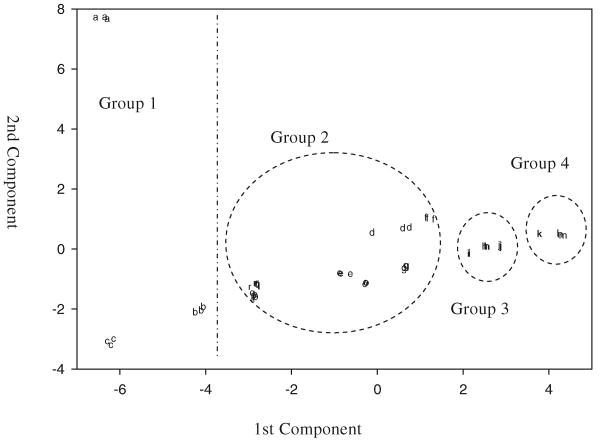
Figure 3 shows the score plot (principal component 2 as a function of principal component 1) for PCA analysis of the fourteen samples and four standards. With the exception of sample 1 (symbol a), there is little separation on the vertical axis. The primary dispersion is horizontal. This is supported by the eigenvalues that show that 80% of the variance is accounted for the first principal component. The samples can easily be organized into four groups that match those described in the previous section. Group #1 (samples 1–3, represented by symbols a–c) is on the left. These three samples contain the lowest percentages of rutin (<15%). Groups 2, 3, and 4 are found progressively to the right. The groupings are somewhat arbitrary, but the progression to the right corresponds to increasing rutin concentration.
Fig. 3. Two-dimensional PCA score plot of the chromatograms of the eighteen G. biloba samples.
Figure 4 shows the same plot as Fig. 3 but without samples 1–3. The second principal component now accounts for a larger fraction of the variance and the samples are more easily organized into five groups. Samples 11–14 (represented by symbols k–n) form a tight group (#4) on the right. The slight separation of sample 11 (symbol k) can be explained by its lower rutin percentage, 60% compared to 74.4%—80.8% of the other three samples. The rutin percentages for Group #3 (samples 8–10, represented by symbols h, i, and j) are between 40 and 50. Group #2, from Fig. 3, can be further divided into three subgroups. Subgroup 2a, on the left, comprises the three NIST standard reference materials. These results indicate that the extraction process (from 3246 to 3247) and the formulation process (from 3247 to 3248) did not change the characteristics of the G. biloba products substantially. The rutin concentrations of subgroup 2b (samples 4 and 6, represented by symbols d and f) are between 20% and 30%, but their free aglycone concentrations are close to 30%. Subgroup 2c comprises samples 5, 7, and the AHP-verified G. biloba leaves. Both the rutin free aglycone percentages of samples 5 and 7 are close to those of the standards.
Fig. 4. Two-dimensional PCA score plot of the chromatograms obtained from samples 4-14 and from the standards.
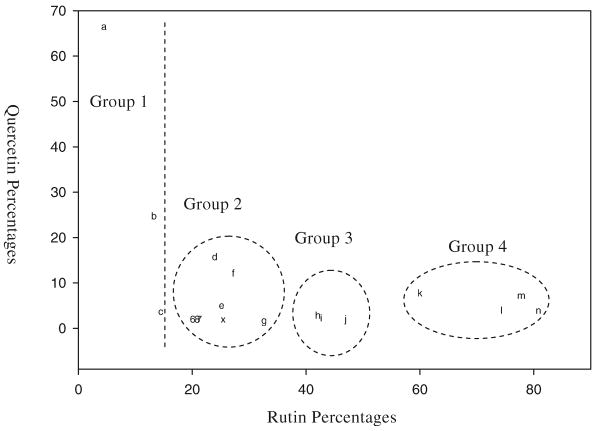
A criticism of PCA is that it can detect global differences between groups but cannot pinpoint the causes of the differences. The PCA of the chromatographic fingerprints shown in Fig. 3 suggest that the first component is strongly correlated with percentage rutin in the sample and the second component is correlated with percentage quercetin. A plot with percentage rutin on the X-axis and percentage quercetin on the Y-axis is shown in Fig. 5. Although group 2 was not separated as well from groups 1 and 3, it indeed has a similar pattern.
Fig. 5. Two-dimensional plot of the rutin and quercetin content of the eighteen samples.
The quantitative analyses in the preceding section and PCA in this section revealed there were substantial differences between rutin percentages, free aglycone percentages, quercetin percentages, and chromatographic fingerprints among the fourteen samples studied, even though the total flavonol contents were similar and met the standard set by EGb 761. These findings raise concern about the G. biloba content of dietary supplements sold in the USA.
Only four of the fourteen samples (samples 4, 5, 6, 7) have rutin percentages and chromatographic fingerprints similar to those of authentic G. biloba standards. Of these four samples, only two (samples 5 and 7) have free aglycone percentages similar to those of standards. The substantially larger amounts of free aglycones in samples 4 and 6 are probably because of either processing procedures (drying temperature too high, etc.) or storage conditions that led to hydrolysis of flavonol glycosides. Samples 1 and 2 contain substantially higher percentages of quercetin than the others, and samples 11 to 14 contain substantially higher percentages of rutin than the others. The AHP recently released a statement, “Code of Ethics and Business Conduct”, suggesting manufacturers adulterate GBE with rutin or quercetin to claim a higher total flavonol content, to meet the standard set by EGb 761 [18]. These six samples are certainly candidates for further investigation. The other samples can be regarded as borderline products. The question of whether the borderline samples are adulterated could be more easily resolved if a wider range of samples of authentic G. biloba over a period of years and from several locations were available.
Only two (samples 5 and 7) of the fourteen samples investigated could be recommended as authentic without degradation and six of fourteen (approx. 40%) are potentially adulterated. If only the traditional standard of total flavonols were used all fourteen GDS samples would be judged authentic. It is clear that the traditional standard of 24% total flavonols is not adequate to assess of the quality of GDS products.
A few groups have published results using chromatographic fingerprints for GBE analysis [27–29]. These studies concentrated on developing a fingerprint of a particular GBE, however, and/or on optimization of either the HPLC and/or the detector operating conditions for that particular fingerprint. These studies thus have limited application as quality-control tools for G. biloba supplements. Other studies have discussed the potential of using chromatographic fingerprints as a quality-control tool for herbal medicines [30, 31], but did not address specific productrelated problems in detail.
The NIST SRMs are large and well homogenized samples for which there is no concern about lot-to-lot reproducibility. By using the NIST SRMs as templates and minimizing instrumental variations, our approach to fingerprinting can be easily implemented. Compared with the traditional total flavonol quantitation method, the fingerprint method is easier, faster, less expensive, and less labor-intensive. Although it cannot replace the traditional method for determination of the total flavonol content of G. biloba products, it can certainly be used as a screening tool to eliminate samples that would not meet the criteria for authentic standards. The traditional method for quantification of total flavonols should be used as a second-stage quality-control tool only, because, alone, it cannot enable adequate assessment of the content of G. biloba supplements.
Table 2. Measured total flavonol and total rutin content of the G. biloba samples as percentages of GBEa.
| Symbol of PCA plot | Total flavonols (% GBE) | Total rutin (% GBE) | F/Rb | |
|---|---|---|---|---|
| 1 | a | 27.78±0.15 | 0.85±0.01 | 32.62 |
| 2 | b | 29.30±0.13 | 1.90±0.05 | 15.40 |
| 3 | c | 26.55±0.11 | 2.27±0.02 | 11.68 |
| 4 | d | 28.75±0.14 | 3.13±0.03 | 9.19 |
| 5 | e | 30.77±0.09 | 3.66±0.04 | 8.42 |
| 6 | f | 26.16±0.04 | 4.86±0.09 | 5.38 |
| 7 | g | 27.48±0.04 | 5.12±0.10 | 5.37 |
| 8 | h | 28.54±0.03 | 6.69±0.12 | 4.27 |
| 9 | i | 24.88±0.02 | 8.45±0.05 | 2.94 |
| 10 | j | 29.17±0.08 | 6.64±0.17 | 4.39 |
| 11 | k | 25.23±0.07 | 12.68±0.21 | 1.99 |
| 12 | l | 26.06±0.05 | 14.84±0.12 | 1.76 |
| 13 | m | 24.80±0.03 | 16.56±0.44 | 1.50 |
| 14 | n | 28.19±0.08 | 19.22±0.06 | 1.47 |
| AHP Std | o | n/a | n/a | n/a |
| SRM 3246 | p | n/a | n/a | n/a |
| SRM 3247 | q | 26.51±0.27 | 3.87±0.02 | 6.85 |
| SRM 3248 | r | n/a | n/a | n/a |
Total flavonol content and total rutin content were calculated as percentages on the basis of the labeled amount of GBE present in the samples. The GBE content of each sample was calculated from the mass of the tablets/capsules (powdered) and the labeled amount of GBE.
Ratio of total flavonols to rutin
Acknowledgments
This research was supported by the Office of Dietary Supplements at the National Institutes of Health under an Interagency Agreement. We thank Dr Lane C. Sander of NIST for providing the NIST Ginkgo Standard Reference Materials.
Appendix 1.
Peak areas for all peaks.
| Peak number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Retention time (min) | 4.564 | 10.305 | 11.097 | 12.078 | 12.87 | 13.753 | 14.075 | 14.657 | 15.547 | 16.041 | 16.308 | 16.913 | 17.188 | 18 | 19.207 | 19.487 | 20.094 | 21.452 | 21.846 | 22.436 | 23.21 |
| Sample 1 | 8.5 | 16.2 | 25.6 | 32.1 | 1.0 | 72.7 | 12.0 | 17.2 | 43.8 | 1.0 | 29.3 | 38.8 | 50.9 | 33.7 | 1104.6 | 0.1 | 17.6 | 1.0 | 1.0 | 78.7 | 55.7 |
| 9.0 | 16.5 | 25.6 | 31.4 | 1.0 | 72.8 | 12.3 | 17.1 | 44.6 | 1.0 | 29.5 | 39.0 | 50.9 | 33.5 | 1081.5 | 0.1 | 17.9 | 1.0 | 1.0 | 79.6 | 57.7 | |
| 8.6 | 16.6 | 25.8 | 32.4 | 1.0 | 72.8 | 11.1 | 15.7 | 44.6 | 1.0 | 29.6 | 40.7 | 56.6 | 43.2 | 1065.4 | 0.1 | 17.0 | 1.0 | 1.0 | 80.4 | 57.6 | |
| Sample 2 | 1.0 | 18.0 | 20.4 | 25.1 | 1.0 | 57.7 | 1.0 | 16.8 | 14.6 | 1.0 | 26.5 | 33.3 | 40.7 | 13.2 | 113.8 | 126.1 | 5.8 | 6.0 | 5.3 | 8.2 | 5.2 |
| 1.0 | 18.3 | 20.6 | 25.2 | 1.0 | 57.6 | 1.0 | 17.0 | 14.8 | 1.0 | 26.6 | 33.6 | 40.8 | 13.1 | 106.3 | 26.2 | 5.9 | 6.7 | 5.5 | 8.0 | 5.3 | |
| 1.0 | 18.1 | 20.9 | 25.3 | 1.0 | 58.6 | 1.0 | 17.5 | 14.4 | 1.0 | 26.6 | 33.7 | 40.9 | 13.3 | 104.1 | 26.2 | 5.8 | 6.6 | 5.1 | 7.7 | 5.2 | |
| Sample 3 | 1.0 | 61.9 | 79.4 | 77.0 | 14.0 | 185.6 | 1.0 | 60.2 | 83.7 | 11.9 | 78.9 | 100.7 | 196.0 | 63.3 | 45.0 | 116.0 | 27.1 | 17.7 | 15.7 | 32.5 | 17.5 |
| 1.0 | 61.7 | 78.9 | 76.5 | 14.7 | 184.9 | 1.0 | 52.9 | 86.1 | 10.8 | 79.6 | 101.1 | 196.5 | 63.9 | 47.9 | 116.4 | 24.3 | 16.5 | 14.7 | 33.2 | 17.2 | |
| 1.0 | 61.7 | 70.4 | 77.1 | 14.7 | 184.6 | 1.0 | 58.4 | 82.8 | 11.0 | 80.0 | 100.9 | 195.4 | 62.3 | 49.2 | 114.3 | 23.5 | 18.4 | 16.7 | 30.9 | 16.5 | |
| Sample 4 | 1.0 | 6.1 | 5.6 | 7.2 | 1.0 | 64.9 | 1.0 | 6.3 | 16.5 | 1.0 | 6.2 | 8.2 | 25.4 | 12.5 | 44.4 | 19.5 | 7.8 | 5.1 | 1.0 | 25.8 | 12.4 |
| 1.0 | 6.4 | 5.7 | 7.7 | 1.0 | 68.8 | 1.0 | 5.4 | 15.4 | 1.0 | 6.2 | 8.0 | 26.0 | 12.3 | 43.6 | 19.0 | 7.4 | 5.1 | 1.0 | 24.6 | 12.4 | |
| 11.3 | 38.1 | 62.2 | 64.1 | 10.7 | 309.1 | 1.0 | 38.6 | 74.1 | 12.1 | 71.4 | 85.8 | 146.9 | 61.3 | 66.4 | 73.7 | 19.1 | 19.2 | 13.9 | 42.2 | 20.5 | |
| Sample 5 | 11.2 | 38.2 | 62.4 | 64.2 | 8.8 | 310.5 | 1.0 | 35.6 | 74.6 | 11.4 | 70.6 | 85.2 | 142.8 | 58.0 | 62.2 | 79.4 | 20.4 | 18.8 | 13.2 | 43.4 | 19.8 |
| 10.8 | 38.5 | 62.4 | 64.1 | 10.8 | 311.8 | 1.0 | 36.6 | 69.7 | 10.9 | 70.8 | 85.0 | 142.7 | 57.8 | 59.3 | 79.5 | 20.3 | 18.9 | 13.2 | 42.4 | 19.5 | |
| 1.0 | 6.2 | 5.9 | 6.8 | 1.0 | 71.4 | 1.0 | 6.7 | 14.6 | 1.0 | 6.0 | 8.0 | 25.0 | 12.0 | 42.7 | 17.0 | 7.5 | 5.0 | 1.0 | 25.2 | 12.9 | |
| Sample 6 | 1.0 | 14.4 | 17.7 | 28.0 | 1.0 | 297.9 | 1.0 | 22.6 | 57.0 | 1.0 | 35.3 | 50.4 | 96.8 | 41.3 | 137.8 | 69.2 | 35.5 | 11.8 | 8.3 | 106.8 | 75.8 |
| 1.0 | 14.5 | 17.8 | 28.0 | 1.0 | 298.4 | 1.0 | 22.3 | 57.1 | 1.0 | 35.6 | 50.9 | 97.5 | 40.4 | 135.8 | 71.5 | 31.8 | 11.5 | 9.7 | 107.7 | 76.9 | |
| 1.0 | 14.5 | 17.7 | 28.0 | 1.0 | 311.4 | 1.0 | 22.9 | 54.7 | 1.0 | 34.2 | 49.6 | 94.6 | 40.2 | 131.7 | 68.9 | 33.1 | 11.9 | 8.9 | 108.9 | 75.6 | |
| Sample 7 | 1.0 | 16.6 | 19.9 | 21.2 | 1.0 | 136.0 | 5.0 | 16.3 | 21.3 | 5.0 | 25.0 | 32.9 | 48.5 | 16.3 | 7.8 | 24.3 | 6.0 | 5.8 | 1.0 | 6.5 | 1.0 |
| 1.0 | 16.4 | 20.0 | 21.2 | 1.0 | 136.4 | 5.0 | 16.4 | 20.8 | 5.0 | 25.0 | 33.1 | 48.8 | 16.3 | 7.8 | 24.1 | 5.9 | 6.2 | 1.0 | 6.7 | 1.0 | |
| 1.0 | 16.4 | 19.9 | 21.2 | 1.0 | 135.9 | 5.0 | 16.1 | 20.9 | 5.0 | 25.0 | 32.8 | 48.4 | 16.5 | 6.8 | 24.1 | 5.2 | 5.9 | 1.0 | 6.2 | 1.0 | |
| Sample 8 | 1.0 | 21.8 | 21.6 | 31.9 | 1.0 | 270.0 | 1.0 | 20.8 | 26.1 | 1.0 | 34.0 | 47.1 | 53.9 | 20.1 | 19.2 | 31.2 | 8.5 | 8.2 | 1.0 | 17.8 | 9.3 |
| 1.0 | 21.8 | 21.4 | 32.2 | 1.0 | 269.7 | 1.0 | 21.1 | 21.0 | 1.0 | 34.3 | 47.0 | 53.6 | 20.0 | 18.0 | 31.0 | 8.4 | 7.8 | 1.0 | 16.8 | 9.6 | |
| 1.0 | 21.6 | 21.3 | 31.9 | 1.0 | 267.7 | 1.0 | 19.8 | 21.7 | 1.0 | 34.1 | 46.4 | 53.1 | 20.0 | 17.7 | 31.1 | 8.3 | 7.6 | 1.0 | 16.3 | 9.8 | |
| Sample 9 | 1.0 | 15.2 | 13.5 | 21.9 | 1.0 | 174.9 | 1.0 | 12.9 | 13.2 | 1.0 | 24.3 | 32.8 | 33.2 | 12.3 | 9.0 | 18.5 | 5.2 | 5.9 | 1.0 | 7.8 | 5.3 |
| 1.0 | 15.1 | 13.2 | 22.0 | 1.0 | 175.1 | 1.0 | 13.6 | 13.5 | 1.0 | 24.2 | 32.8 | 33.2 | 12.3 | 9.4 | 18.8 | 5.1 | 5.1 | 1.0 | 6.5 | 5.2 | |
| 1.0 | 15.1 | 13.0 | 21.9 | 1.0 | 174.5 | 1.0 | 13.2 | 13.3 | 1.0 | 24.1 | 32.7 | 33.0 | 12.4 | 9.4 | 18.6 | 5.2 | 5.5 | 1.0 | 6.0 | 5.3 | |
| Sample 10 | 1.0 | 19.0 | 25.5 | 20.2 | 1.0 | 276.0 | 1.0 | 21.3 | 23.8 | 1.0 | 26.2 | 36.0 | 52.3 | 16.4 | 12.3 | 26.5 | 6.8 | 6.5 | 1.0 | 9.8 | 5.2 |
| 1.0 | 19.0 | 25.6 | 20.6 | 1.0 | 276.0 | 1.0 | 20.7 | 24.1 | 1.0 | 26.1 | 35.8 | 52.0 | 16.4 | 11.6 | 26.7 | 7.4 | 5.7 | 1.0 | 9.4 | 5.1 | |
| 1.0 | 19.1 | 25.7 | 20.5 | 1.0 | 276.9 | 1.0 | 21.2 | 24.1 | 1.0 | 26.1 | 35.9 | 52.2 | 16.3 | 12.4 | 26.7 | 6.7 | 5.9 | 1.0 | 9.2 | 5.1 | |
| Sample 11 | 5.0 | 1.0 | 22.2 | 1.0 | 1.0 | 1004.4 | 1.0 | 1.0 | 80.0 | 1.0 | 19.3 | 27.8 | 95.3 | 49.9 | 137.5 | 78.8 | 24.4 | 12.7 | 10.3 | 66.5 | 40.2 |
| 5.0 | 1.0 | 22.5 | 1.0 | 1.0 | 1005.3 | 1.0 | 1.0 | 80.9 | 1.0 | 19.3 | 27.7 | 95.1 | 49.4 | 132.0 | 77.9 | 23.8 | 12.9 | 8.5 | 67.0 | 42.3 | |
| 5.0 | 1.0 | 22.2 | 1.0 | 1.0 | 1003.3 | 1.0 | 1.0 | 80.7 | 1.0 | 19.5 | 27.8 | 95.4 | 50.2 | 121.3 | 79.4 | 24.4 | 12.6 | 8.8 | 66.9 | 41.7 | |
| Sample 12 | 1.0 | 5.0 | 5.6 | 6.9 | 1.0 | 595.2 | 1.0 | 1.0 | 21.2 | 1.0 | 10.3 | 17.0 | 31.8 | 13.9 | 31.5 | 17.4 | 5.5 | 1.0 | 1.0 | 19.1 | 13.2 |
| 1.0 | 5.1 | 5.5 | 6.7 | 1.0 | 595.4 | 1.0 | 1.0 | 21.0 | 1.0 | 10.1 | 17.2 | 32.0 | 13.7 | 31.8 | 17.6 | 6.1 | 1.0 | 1.0 | 18.3 | 13.4 | |
| 1.0 | 5.1 | 6.1 | 6.1 | 1.0 | 594.8 | 1.0 | 1.0 | 21.2 | 1.0 | 10.1 | 17.0 | 31.9 | 13.7 | 31.4 | 17.3 | 6.4 | 1.0 | 1.0 | 19.0 | 12.8 | |
| Sample 13 | 1.0 | 5.6 | 5.8 | 8.4 | 1.0 | 674.5 | 1.0 | 5.2 | 11.4 | 1.0 | 16.2 | 24.3 | 12.3 | 6.5 | 64.6 | 7.4 | 1.0 | 1.0 | 1.0 | 11.2 | 6.9 |
| 1.0 | 5.4 | 6.7 | 8.8 | 1.0 | 675.2 | 1.0 | 6.6 | 11.1 | 1.0 | 16.3 | 24.2 | 12.4 | 6.6 | 62.4 | 7.0 | 1.0 | 1.0 | 1.0 | 11.1 | 7.3 | |
| 1.0 | 5.6 | 5.5 | 8.9 | 1.0 | 673.7 | 1.0 | 6.0 | 11.4 | 1.0 | 16.5 | 24.5 | 12.6 | 6.3 | 60.7 | 6.8 | 1.0 | 1.0 | 1.0 | 11.4 | 7.2 | |
| Sample 14 | 5.0 | 10.4 | 9.7 | 12.8 | 1.0 | 789.5 | 1.0 | 5.6 | 6.7 | 1.0 | 23.6 | 31.6 | 13.8 | 6.6 | 40.6 | 8.9 | 1.0 | 1.0 | 1.0 | 5.3 | 4.0 |
| 5.0 | 10.0 | 9.4 | 12.7 | 1.0 | 791.6 | 1.0 | 5.3 | 7.4 | 1.0 | 23.5 | 31.6 | 13.5 | 6.3 | 38.0 | 8.5 | 1.0 | 1.0 | 1.0 | 5.3 | 4.0 | |
| 5.0 | 10.1 | 8.9 | 12.8 | 1.0 | 791.4 | 1.0 | 6.0 | 7.4 | 1.0 | 23.6 | 31.5 | 13.5 | 6.3 | 37.7 | 7.8 | 1.0 | 1.0 | 1.0 | 5.3 | 4.0 | |
| AHP | 4.0 | 39.5 | 17.2 | 48.8 | 1.0 | 190.0 | 1.0 | 33.5 | 19.3 | 1.0 | 125.0 | 123.0 | 50.6 | 12.8 | 6.2 | 37.7 | 6.5 | 7.2 | 5.5 | 8.4 | 6.8 |
| 4.0 | 39.6 | 17.0 | 48.8 | 1.0 | 190.2 | 1.0 | 33.6 | 19.4 | 1.0 | 125.8 | 123.2 | 50.9 | 13.2 | 6.5 | 42.2 | 6.2 | 7.2 | 5.0 | 8.2 | 6.3 | |
| 4.0 | 39.7 | 17.1 | 48.8 | 1.0 | 190.0 | 1.0 | 33.8 | 19.4 | 1.0 | 123.6 | 123.6 | 51.2 | 13.8 | 6.4 | 38.1 | 6.6 | 7.7 | 5.3 | 7.8 | 6.3 | |
| SRM 3246 | 2.0 | 21.0 | 18.6 | 20.1 | 1.0 | 52.1 | 3.0 | 10.5 | 12.0 | 1.0 | 27.8 | 16.4 | 30.6 | 7.6 | 1.0 | 22.2 | 5.0 | 5.2 | 1.0 | 2.0 | 1.0 |
| 2.0 | 21.2 | 18.8 | 20.1 | 1.0 | 53.0 | 3.0 | 11.2 | 11.8 | 1.0 | 28.0 | 16.4 | 30.8 | 7.6 | 1.0 | 22.2 | 5.6 | 5.1 | 1.0 | 2.0 | 1.0 | |
| 2.0 | 21.0 | 18.8 | 20.0 | 1.0 | 51.4 | 3.0 | 9.9 | 11.6 | 1.0 | 22.5 | 16.4 | 30.5 | 7.4 | 1.0 | 22.0 | 5.0 | 5.5 | 1.0 | 2.0 | 1.0 | |
| SRM 3247 | 6.4 | 43.1 | 30.7 | 39.9 | 1.0 | 102.7 | 8.7 | 23.9 | 25.8 | 5.0 | 44.9 | 38.8 | 52.6 | 15.6 | 1.0 | 28.4 | 5.0 | 5.8 | 6.0 | 2.0 | 1.0 |
| 6.8 | 43.5 | 30.6 | 39.9 | 1.0 | 105.1 | 9.0 | 25.6 | 25.8 | 5.0 | 44.9 | 39.0 | 53.1 | 15.7 | 1.0 | 28.2 | 6.1 | 5.7 | 5.0 | 2.0 | 1.0 | |
| 6.9 | 43.7 | 30.6 | 39.8 | 1.0 | 105.9 | 9.2 | 26.6 | 25.4 | 5.0 | 44.8 | 39.2 | 53.0 | 15.5 | 1.0 | 28.2 | 5.1 | 5.6 | 5.0 | 2.0 | 1.0 | |
| SRM 3248 | 10.8 | 68.7 | 48.0 | 62.5 | 1.0 | 168.9 | 17.1 | 41.6 | 41.8 | 7.8 | 72.8 | 64.1 | 88.8 | 26.1 | 1.0 | 51.7 | 10.6 | 10.5 | 8.5 | 5.6 | 2.0 |
| 10.0 | 68.8 | 48.5 | 62.9 | 1.0 | 171.8 | 17.5 | 42.2 | 41.7 | 7.9 | 72.5 | 63.9 | 88.3 | 25.7 | 1.0 | 47.9 | 9.6 | 9.1 | 7.9 | 5.6 | 2.0 | |
| 10.0 | 68.7 | 48.9 | 62.9 | 1.0 | 173.1 | 19.3 | 42.3 | 41.4 | 7.8 | 72.7 | 63.7 | 88.6 | 26.0 | 1.0 | 47.8 | 10.3 | 9.9 | 8.6 | 5.7 | 2.0 |
Appendix 2.
Data set used for PCA analysis.
| Peak number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Retention time (min) | 4.564 | 10.305 | 11.097 | 12.078 | 12.87 | 13.753 | 14.075 | 14.657 | 15.547 | 16.041 | 16.306 | 16.913 | 17.188 | 18 | 19.207 | 19.487 | 20.094 | 21.452 | 21.846 | 22.436 | 23.213 |
| Sample 1 | 0.01 | 0.12 | 0.15 | 0.16 | 0.01 | 1.00 | 0.04 | 0.12 | 0.16 | 0.04 | 0.18 | 0.24 | 0.36 | 0.12 | 0.06 | 0.18 | 0.04 | 0.04 | 0.01 | 0.05 | 0.01 |
| 0.01 | 0.12 | 0.15 | 0.16 | 0.01 | 1.00 | 0.04 | 0.12 | 0.15 | 0.04 | 0.18 | 0.24 | 0.36 | 0.12 | 0.05 | 0.18 | 0.04 | 0.04 | 0.01 | 0.05 | 0.01 | |
| 0.01 | 0.12 | 0.15 | 0.16 | 0.01 | 1.00 | 0.04 | 0.12 | 0.15 | 0.04 | 0.18 | 0.24 | 0.36 | 0.12 | 0.06 | 0.18 | 0.04 | 0.05 | 0.01 | 0.05 | 0.01 | |
| Sample 2 | 0.01 | 0.09 | 0.07 | 0.13 | 0.01 | 1.00 | 0.01 | 0.08 | 0.08 | 0.01 | 0.14 | 0.19 | 0.19 | 0.07 | 0.05 | 0.11 | 0.03 | 0.03 | 0.01 | 0.03 | 0.03 |
| 0.01 | 0.09 | 0.08 | 0.13 | 0.01 | 1.00 | 0.01 | 0.08 | 0.08 | 0.01 | 0.14 | 0.19 | 0.19 | 0.07 | 0.05 | 0.11 | 0.03 | 0.03 | 0.01 | 0.04 | 0.03 | |
| 0.01 | 0.09 | 0.08 | 0.13 | 0.01 | 1.00 | 0.01 | 0.07 | 0.08 | 0.01 | 0.14 | 0.19 | 0.19 | 0.07 | 0.05 | 0.11 | 0.03 | 0.03 | 0.01 | 0.04 | 0.03 | |
| Sample 3 | 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.00 | 0.04 | 0.00 | 0.02 | 0.03 | 0.05 | 0.02 | 0.05 | 0.03 | 0.01 | 0.00 | 0.00 | 0.03 | 0.02 |
| 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.00 | 0.04 | 0.00 | 0.02 | 0.03 | 0.05 | 0.02 | 0.05 | 0.03 | 0.01 | 0.00 | 0.00 | 0.03 | 0.02 | |
| 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.00 | 0.04 | 0.00 | 0.02 | 0.03 | 0.05 | 0.02 | 0.05 | 0.03 | 0.01 | 0.00 | 0.00 | 0.03 | 0.02 | |
| Sample 4 | 0.00 | 0.08 | 0.08 | 0.12 | 0.00 | 1.00 | 0.00 | 0.07 | 0.08 | 0.00 | 0.13 | 0.17 | 0.20 | 0.07 | 0.07 | 0.12 | 0.03 | 0.03 | 0.00 | 0.06 | 0.04 |
| 0.00 | 0.08 | 0.08 | 0.12 | 0.00 | 1.00 | 0.00 | 0.08 | 0.08 | 0.00 | 0.13 | 0.17 | 0.20 | 0.07 | 0.07 | 0.11 | 0.03 | 0.03 | 0.00 | 0.06 | 0.04 | |
| 0.00 | 0.08 | 0.08 | 0.12 | 0.00 | 1.00 | 0.00 | 0.08 | 0.10 | 0.00 | 0.13 | 0.17 | 0.20 | 0.07 | 0.07 | 0.12 | 0.03 | 0.03 | 0.00 | 0.07 | 0.03 | |
| Sample 5 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.02 | 0.03 | 0.10 | 0.05 | 0.12 | 0.08 | 0.02 | 0.01 | 0.01 | 0.07 | 0.04 |
| 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.02 | 0.03 | 0.09 | 0.05 | 0.14 | 0.08 | 0.02 | 0.01 | 0.01 | 0.07 | 0.04 | |
| 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.08 | 0.00 | 0.02 | 0.03 | 0.09 | 0.05 | 0.13 | 0.08 | 0.02 | 0.01 | 0.01 | 0.07 | 0.04 | |
| Sample 6 | 0.01 | 0.09 | 0.08 | 0.10 | 0.01 | 1.00 | 0.01 | 0.09 | 0.20 | 0.01 | 0.08 | 0.11 | 0.35 | 0.17 | 0.60 | 0.24 | 0.11 | 0.07 | 0.01 | 0.35 | 0.18 |
| 0.02 | 0.09 | 0.09 | 0.11 | 0.02 | 1.00 | 0.02 | 0.10 | 0.25 | 0.02 | 0.10 | 0.13 | 0.39 | 0.19 | 0.68 | 0.30 | 0.12 | 0.08 | 0.02 | 0.40 | 0.19 | |
| 0.01 | 0.09 | 0.08 | 0.11 | 0.01 | 1.00 | 0.01 | 0.08 | 0.22 | 0.01 | 0.09 | 0.12 | 0.38 | 0.18 | 0.63 | 0.28 | 0.11 | 0.07 | 0.01 | 0.36 | 0.18 | |
| Sample 7 | 0.01 | 0.33 | 0.43 | 0.41 | 0.08 | 1.00 | 0.01 | 0.29 | 0.47 | 0.06 | 0.43 | 0.55 | 1.06 | 0.35 | 0.26 | 0.63 | 0.13 | 0.09 | 0.08 | 0.18 | 0.09 |
| 0.01 | 0.33 | 0.43 | 0.41 | 0.08 | 1.00 | 0.01 | 0.32 | 0.45 | 0.06 | 0.43 | 0.54 | 1.06 | 0.34 | 0.24 | 0.63 | 0.15 | 0.10 | 0.08 | 0.18 | 0.09 | |
| 0.01 | 0.33 | 0.38 | 0.42 | 0.08 | 1.00 | 0.01 | 0.32 | 0.45 | 0.06 | 0.43 | 0.55 | 1.06 | 0.34 | 0.27 | 0.62 | 0.13 | 0.10 | 0.09 | 0.17 | 0.09 | |
| Sample 8 | 0.00 | 0.07 | 0.09 | 0.07 | 0.00 | 1.00 | 0.00 | 0.08 | 0.09 | 0.00 | 0.09 | 0.13 | 0.19 | 0.06 | 0.04 | 0.10 | 0.03 | 0.02 | 0.00 | 0.03 | 0.02 |
| 0.00 | 0.07 | 0.09 | 0.07 | 0.00 | 1.00 | 0.00 | 0.08 | 0.09 | 0.00 | 0.09 | 0.13 | 0.19 | 0.06 | 0.04 | 0.10 | 0.02 | 0.02 | 0.00 | 0.03 | 0.02 | |
| 0.00 | 0.07 | 0.09 | 0.07 | 0.00 | 1.00 | 0.00 | 0.08 | 0.09 | 0.00 | 0.09 | 0.13 | 0.19 | 0.06 | 0.04 | 0.10 | 0.02 | 0.02 | 0.00 | 0.04 | 0.02 | |
| Sample 9 | 0.01 | 0.01 | 0.01 | 0.02 | 0.00 | 1.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.03 | 0.04 | 0.02 | 0.01 | 0.05 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 |
| 0.01 | 0.01 | 0.01 | 0.02 | 0.00 | 1.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.03 | 0.04 | 0.02 | 0.01 | 0.05 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | |
| 0.01 | 0.01 | 0.01 | 0.02 | 0.00 | 1.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.03 | 0.04 | 0.02 | 0.01 | 0.05 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | |
| Sample 10 | 0.02 | 0.31 | 0.35 | 0.44 | 0.02 | 1.00 | 0.02 | 0.29 | 0.25 | 0.02 | 0.46 | 0.58 | 0.71 | 0.23 | 1.97 | 0.45 | 0.10 | 0.10 | 0.09 | 0.14 | 0.09 |
| 0.02 | 0.32 | 0.36 | 0.44 | 0.02 | 1.00 | 0.02 | 0.30 | 0.26 | 0.02 | 0.46 | 0.58 | 0.71 | 0.23 | 1.85 | 0.45 | 0.10 | 0.12 | 0.09 | 0.14 | 0.09 | |
| 0.02 | 0.31 | 0.36 | 0.43 | 0.02 | 1.00 | 0.02 | 0.30 | 0.25 | 0.02 | 0.45 | 0.58 | 0.70 | 0.23 | 1.78 | 0.45 | 0.10 | 0.11 | 0.09 | 0.13 | 0.09 | |
| Sample 11 | 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.01 | 0.02 | 0.00 | 0.02 | 0.04 | 0.02 | 0.01 | 0.09 | 0.01 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 |
| 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.01 | 0.02 | 0.00 | 0.02 | 0.04 | 0.02 | 0.01 | 0.09 | 0.01 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 | |
| 0.00 | 0.01 | 0.01 | 0.01 | 0.00 | 1.00 | 0.00 | 0.01 | 0.02 | 0.00 | 0.02 | 0.04 | 0.02 | 0.01 | 0.10 | 0.01 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 | |
| Sample 12 | 0.03 | 0.12 | 0.20 | 0.21 | 0.03 | 1.00 | 0.00 | 0.12 | 0.22 | 0.03 | 0.23 | 0.27 | 0.46 | 0.19 | 0.19 | 0.25 | 0.06 | 0.06 | 0.04 | 0.14 | 0.06 |
| 0.04 | 0.12 | 0.20 | 0.21 | 0.03 | 1.00 | 0.00 | 0.11 | 0.24 | 0.04 | 0.23 | 0.27 | 0.46 | 0.19 | 0.20 | 0.26 | 0.07 | 0.06 | 0.04 | 0.14 | 0.06 | |
| 0.04 | 0.12 | 0.20 | 0.21 | 0.03 | 1.00 | 0.00 | 0.12 | 0.24 | 0.04 | 0.23 | 0.28 | 0.48 | 0.20 | 0.21 | 0.24 | 0.06 | 0.06 | 0.04 | 0.14 | 0.07 | |
| Sample 13 | 0.12 | 0.23 | 0.35 | 0.45 | 0.01 | 1.00 | 0.15 | 0.22 | 0.61 | 0.01 | 0.41 | 0.56 | 0.78 | 0.59 | 14.63 | 0.00 | 0.23 | 0.01 | 0.01 | 1.10 | 0.79 |
| 0.12 | 0.23 | 0.35 | 0.43 | 0.01 | 1.00 | 0.17 | 0.23 | 0.61 | 0.01 | 0.41 | 0.54 | 0.70 | 0.46 | 14.86 | 0.00 | 0.25 | 0.01 | 0.01 | 1.09 | 0.79 | |
| 0.12 | 0.22 | 0.35 | 0.44 | 0.01 | 1.00 | 0.17 | 0.24 | 0.60 | 0.01 | 0.40 | 0.53 | 0.70 | 0.46 | 15.19 | 0.00 | 0.24 | 0.01 | 0.01 | 1.08 | 0.77 | |
| Sample 14 | 0.00 | 0.05 | 0.06 | 0.09 | 0.00 | 1.00 | 0.00 | 0.07 | 0.18 | 0.00 | 0.11 | 0.16 | 0.30 | 0.13 | 0.42 | 0.22 | 0.11 | 0.04 | 0.03 | 0.35 | 0.24 |
| 0.00 | 0.05 | 0.06 | 0.09 | 0.00 | 1.00 | 0.00 | 0.07 | 0.19 | 0.00 | 0.12 | 0.17 | 0.33 | 0.14 | 0.46 | 0.24 | 0.11 | 0.04 | 0.03 | 0.36 | 0.26 | |
| 0.00 | 0.05 | 0.06 | 0.09 | 0.00 | 1.00 | 0.00 | 0.08 | 0.19 | 0.00 | 0.12 | 0.17 | 0.32 | 0.14 | 0.46 | 0.23 | 0.12 | 0.04 | 0.03 | 0.36 | 0.25 | |
| AHP | 0.02 | 0.21 | 0.09 | 0.26 | 0.01 | 1.00 | 0.01 | 0.18 | 0.10 | 0.01 | 0.66 | 0.65 | 0.27 | 0.07 | 0.03 | 0.20 | 0.03 | 0.04 | 0.03 | 0.04 | 0.04 |
| 0.02 | 0.21 | 0.09 | 0.26 | 0.01 | 1.00 | 0.01 | 0.18 | 0.10 | 0.01 | 0.66 | 0.65 | 0.27 | 0.07 | 0.03 | 0.22 | 0.03 | 0.04 | 0.03 | 0.04 | 0.03 | |
| 0.02 | 0.21 | 0.09 | 0.26 | 0.01 | 1.00 | 0.01 | 0.18 | 0.10 | 0.01 | 0.65 | 0.65 | 0.27 | 0.07 | 0.03 | 0.20 | 0.03 | 0.04 | 0.03 | 0.04 | 0.03 | |
| SRM 3246 | 0.04 | 0.40 | 0.36 | 0.39 | 0.02 | 1.00 | 0.06 | 0.20 | 0.23 | 0.02 | 0.53 | 0.31 | 0.59 | 0.15 | 0.02 | 0.43 | 0.10 | 0.10 | 0.02 | 0.04 | 0.02 |
| 0.04 | 0.40 | 0.35 | 0.38 | 0.02 | 1.00 | 0.06 | 0.21 | 0.22 | 0.02 | 0.53 | 0.31 | 0.58 | 0.14 | 0.02 | 0.42 | 0.11 | 0.10 | 0.02 | 0.04 | 0.02 | |
| 0.04 | 0.41 | 0.37 | 0.39 | 0.02 | 1.00 | 0.06 | 0.19 | 0.23 | 0.02 | 0.44 | 0.32 | 0.59 | 0.14 | 0.02 | 0.43 | 0.10 | 0.11 | 0.02 | 0.04 | 0.02 | |
| SRM 3247 | 0.06 | 0.42 | 0.30 | 0.39 | 0.01 | 1.00 | 0.08 | 0.23 | 0.25 | 0.05 | 0.44 | 0.38 | 0.51 | 0.15 | 0.01 | 0.28 | 0.05 | 0.06 | 0.06 | 0.02 | 0.01 |
| 0.06 | 0.41 | 0.29 | 0.38 | 0.01 | 1.00 | 0.09 | 0.24 | 0.25 | 0.05 | 0.43 | 0.37 | 0.51 | 0.15 | 0.01 | 0.27 | 0.06 | 0.05 | 0.05 | 0.02 | 0.01 | |
| 0.07 | 0.41 | 0.29 | 0.38 | 0.01 | 1.00 | 0.09 | 0.25 | 0.24 | 0.05 | 0.42 | 0.37 | 0.50 | 0.15 | 0.01 | 0.27 | 0.05 | 0.05 | 0.05 | 0.02 | 0.01 | |
| SRM 3248 | 0.06 | 0.41 | 0.28 | 0.37 | 0.01 | 1.00 | 0.10 | 0.25 | 0.25 | 0.05 | 0.43 | 0.38 | 0.53 | 0.15 | 0.07 | 0.31 | 0.06 | 0.06 | 0.05 | 0.03 | 0.01 |
| SRM 3248 | 0.06 | 0.40 | 0.28 | 0.37 | 0.01 | 1.00 | 0.10 | 0.25 | 0.24 | 0.05 | 0.42 | 0.37 | 0.51 | 0.15 | 0.06 | 0.28 | 0.06 | 0.05 | 0.05 | 0.03 | 0.01 |
| 0.06 | 0.40 | 0.28 | 0.36 | 0.01 | 1.00 | 0.11 | 0.24 | 0.24 | 0.05 | 0.42 | 0.37 | 0.51 | 0.15 | 0.07 | 0.28 | 0.06 | 0.06 | 0.05 | 0.03 | 0.01 |
References
- 1.Michel PF. Presse Med. 1986;15:1450–1454. [PubMed] [Google Scholar]
- 2.Gaby AR. Alt Med Rev. 1996;1:236–242. [Google Scholar]
- 3.Strømgaard K, Vogensen SB, Nakanishi K. Encyclopedia of dietary supplements. Marcel Dekker; New York: 2005. pp. 249–257. [Google Scholar]
- 4.Dubber MJ, Kanfer I. J Pharm Sci. 2004;7:3030–3039. [PubMed] [Google Scholar]
- 5.Wu Y, Wu Z, Butko P, Christen Y, Lambert MP, Klein WL, Link CD, Luo Y. J Neurosci. 2006;26:13102–13113. doi: 10.1523/JNEUROSCI.3448-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto Y, Adachi Y, Fujii Y, Kamei C. Brain Res. 2007;1128:70–78. doi: 10.1016/j.brainres.2006.08.102. [DOI] [PubMed] [Google Scholar]
- 7.Mdzinarishvili A, Kiewert C, Kumar V, Hillert M, Klein J. Neuroscience. 2007;144:217–222. doi: 10.1016/j.neuroscience.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 8.van Beek TA. J Chromatogr A. 2002;967:21–55. doi: 10.1016/s0021-9673(02)00172-3. [DOI] [PubMed] [Google Scholar]
- 9.Watson DG, Oliveria EJ. J Chromatogr B. 1999;723:203–210. doi: 10.1016/s0378-4347(98)00509-x. [DOI] [PubMed] [Google Scholar]
- 10.Hasler A, Sticher O. J Chromatogr. 1992;605:41–48. [Google Scholar]
- 11.Herbal Medicine Expanded Commission E Monographs. American Botanical Council; 6200 Manor Rd, Austin, TX: 2000. [Google Scholar]
- 12.DeFeudis FV. G biloba extract (EGb 761): from chemistry to the clinic. Ullstein Medical; Wiesbaden, Germany: 1998. [Google Scholar]
- 13.Ding C, Chen E, Zhou W, Lindsay RC. Anal Chem. 2004;76:4332–4336. doi: 10.1021/ac049809a. [DOI] [PubMed] [Google Scholar]
- 14.Ding C, Chen E, Lindsay RC. Anal Chem. 2005;77:2966–2970. doi: 10.1021/ac048510p. [DOI] [PubMed] [Google Scholar]
- 15.Jensen AG, Ndjoko K, Wolfender JL, Hostettmann K, Camponovo F, Soldati F. Phytochem Anal. 2002;13:31–38. doi: 10.1002/pca.614. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Fitzloff JF. J Pharm Biomed Anal. 2002;30:67–75. doi: 10.1016/s0731-7085(02)00201-7. [DOI] [PubMed] [Google Scholar]
- 17.Mauri P, Migliazza B, Pietta P. J Mass Spectrom. 1999;34:1361–1367. doi: 10.1002/(SICI)1096-9888(199912)34:12<1361::AID-JMS895>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.AHPA. Code of ethics and business conduct. American Herbal Products Association; 2006. [Google Scholar]
- 19.Ozcan M, Mcauley B, Chen P. J Food Drug Anal. 2007;15:55–62. [Google Scholar]
- 20.Wagner H, Bladt S, Hartmann U, Daily A, Berkulin W. Deutisch Apothek Zeit. 1989;129:2421–2424. [Google Scholar]
- 21.Li W, Fitzloff JF. J Liq Chromatogr Related Technol. 2002;25:2501–2514. [Google Scholar]
- 22.Sticher O, Meier B, Hasler A. The analysis of G biloba flavonoids. In: van Beek TA, editor. G biloba. Harwood; Amsterdam: 2000. pp. 179–202. [Google Scholar]
- 23.Gong F, Liang YZ, Xie PS, Chau FT. J Chromatogr A. 2003;1002:25–40. doi: 10.1016/s0021-9673(03)00648-4. [DOI] [PubMed] [Google Scholar]
- 24.Gong F, Liang YZ, Fung YS, Chau FT. J Chromatogr A. 2004;1029:173–183. doi: 10.1016/j.chroma.2003.12.049. [DOI] [PubMed] [Google Scholar]
- 25.Zeng ZD, Liang YZ, Wang YL, Li XR, Laing LM, Xu QS, Zhao CX, Li BY, Chau FT. J Chromatogr A. 2006;1107:273–285. doi: 10.1016/j.chroma.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Zeng ZD, Liang YZ, Chau FT, Wang YL. Anal Bioanal Chem. 2006;1107:392–400. doi: 10.1007/s00216-006-0405-6. [DOI] [PubMed] [Google Scholar]
- 27.van Nederkassel AM, Vijverman V, Massart DL, Vander Heyden Y. J Chromatogr A. 2005;1085:230–239. doi: 10.1016/j.chroma.2005.05.110. [DOI] [PubMed] [Google Scholar]
- 28.Ji YB, Xu QS, Hu YZ, Heyden YV. J Chromatogr A. 2005;1066:97–104. doi: 10.1016/j.chroma.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Ji YB, Aleart G, Xu CJ, Hu YZ, Heyden YV. J Chromatogr A. 2006;1128:273–281. doi: 10.1016/j.chroma.2006.06.053. [DOI] [PubMed] [Google Scholar]
- 30.Xie P, Chen S, Liang YZ, Wang X, Tian R, Upton R. J Chromatogr A. 2006;1112:171–180. doi: 10.1016/j.chroma.2005.12.091. [DOI] [PubMed] [Google Scholar]
- 31.Liang YZ, Xie P, Chan K. J Chromatogr B. 2004;812:53–70. doi: 10.1016/j.jchromb.2004.08.041. [DOI] [PubMed] [Google Scholar]