Summary
Binding of a pre-mRNA substrate triggers spliceosome activation while the release of the mRNA product triggers spliceosome disassembly. The mechanisms that underlie the regulation of these rearrangements remain unclear. We find evidence that the GTPase Snu114p mediates the regulation of spliceosome activation and disassembly. Specifically, both unwinding of U4/U6, required for spliceosome activation, and disassembly of the post-splicing U2/U6•U5•intron complex are repressed by Snu114p bound to GDP and derepressed by Snu114 bound to GTP or nonhydrolyzable GTP analogs. Further, similar to U4/U6 unwinding, spliceosome disassembly requires the DExD/H-box ATPase Brr2p. Together, our data define a common mechanism for regulating and executing spliceosome activation and disassembly. Although sequence similarity with EF-G suggests Snu114p functions as a molecular motor, our findings indicate that Snu114p functions as a classic regulatory G protein. We propose that Snu114p serves as a signal-dependent switch that transduces signals to Brr2p to control spliceosome dynamics.
Introduction
In nuclear pre-messenger RNA (pre-mRNA) splicing, two sequential transesterification reactions excise an intron and ligate flanking exons, yielding mRNA (Will and Lührmann, 2006). Conserved sequences in the intron define the sites of chemistry: the 5′ splice site, the branch site adenosine and the 3′ splice site. In the first reaction, the branch site adenosine attacks the 5′ splice site, yielding a free 5′ exon and a lariat intermediate. In the second reaction, the 5′ exon attacks the 3′ splice site, yielding ligated exons and an excised lariat intron. These reactions are catalyzed by the spliceosome, a conserved, dynamic machine composed of five small nuclear RNAs (snRNAs) – U1, U2, U4, U5, U6 – and over 100 proteins (Staley and Guthrie, 1998; Jurica and Moore, 2003). The snRNAs play key roles in substrate recognition and likely catalysis (Valadkhan, 2005). These roles require rearrangements of the snRNAs (Staley and Guthrie, 1998).
The most conserved snRNA, U6, undergoes major rearrangements during splicing (Staley and Guthrie, 1998). U6 binds the 5′ splice site and is implicated in catalysis (Valadkhan, 2005). Significantly, when associating with a pre-mRNA substrate, U6 is base-paired extensively with U4 (Brow and Guthrie, 1988). In a key step in the catalytic activation of the spliceosome, U4/U6 unwinds, allowing the formation of structures mutually exclusive with U4/U6, structures that include the U6 intramolecular stem-loop (ISL), which binds a metal that promotes 5′ splice site cleavage (Yean et al., 2000; Huppler et al., 2002), and U2/U6 helix I (Madhani and Guthrie, 1992), which, minimally, promotes exon ligation (Hilliker and Staley, 2004). During recycling of the spliceosome for subsequent rounds of splicing, such structures must unwind to allow reformation of base-paired U4/U6.
These and other RNA rearrangements in splicing each require a member of the DExD/H-box family of ATPases, which function in nearly all RNA-dependent processes (Cordin et al., 2006). The family is defined by eight conserved motifs that function in RNA binding, ATP binding and hydrolysis, and coupling RNA binding to ATP binding and hydrolysis. In vitro, some family members unwind generic, duplex RNA and/or displace proteins from single-stranded RNA. In vivo, DExD/H-box ATPases perform specific functions, but the mechanisms that regulate their activity and establish their specificity are understood poorly (Silverman et al., 2003).
U4/U6 unwinding requires the DExD/H-box ATPase Brr2p, a conserved splicing factor (Lauber et al., 1996; Noble and Guthrie, 1996; Xu et al., 1996; Kim and Rossi, 1999). In budding yeast, U4/U6 unwinding is not only catalyzed after assembly of a spliceosome on a pre-mRNA substrate but also before assembly within the U4/U6•U5 small nuclear ribonucleoprotein (snRNP) particle (Cheng and Abelson, 1987). This snRNP has served as a model system for understanding U4/U6 unwinding during spliceosome activation. U4/U6 unwinding in this snRNP requires ATP hydrolysis and Brr2p (Raghunathan and Guthrie, 1998). Because Brr2p unwinds RNA, including protein-free U4/U6 (Laggerbauer et al., 1998), Brr2p is thought to unwind U4/U6 directly during spliceosome activation. Brr2p contains two DExD/H-box ATPase domains, each followed by a Sec63 domain (Ponting, 2000), a domain of unknown function found in two additional DExD/H-box ATPases and Sec63p itself, a component of the translocon. Brr2p, a core component of the U5 and U4/U6•U5 snRNPs (Lauber et al., 1996; Gottschalk et al., 1999; Stevens and Abelson, 1999; Stevens et al., 2001), is a component of the spliceosome throughout the splicing cycle in budding yeast (Ohi et al., 2002; Stevens et al., 2002; Lebaron et al., 2005) and in mammals (Jurica and Moore, 2003), suggesting that Brr2p requires regulation.
U4/U6 unwinding also requires the conserved splicing factor Snu114p, a GTPase (Fabrizio et al., 1997). Mutations in SNU114 block U4/U6 unwinding during spliceosome activation (Bartels et al., 2002; Bartels et al., 2003; Brenner and Guthrie, 2006). Further, snu114 mutants enhance brr2 mutants, in addition to other mutants defective in spliceosome assembly (Brenner and Guthrie, 2005). Like Brr2p, Snu114p is an integral component of the U5 and U4/U6•U5 snRNPs (Fabrizio et al., 1997; Gottschalk et al., 1999; Stevens and Abelson, 1999; Stevens et al., 2001) and a component of the spliceosome throughout the splicing cycle in budding yeast (Ohi et al., 2002; Stevens et al., 2002; Lebaron et al., 2005) and in mammals (Jurica and Moore, 2003). Intriguingly, Snu114p shares high sequence identity with the translation elongation factor EF-G/EF-2 (Fabrizio et al., 1997), which drives translocation of tRNA and mRNA through the ribosome (Rodnina et al., 1997). This similarity inspired the hypothesis that Snu114p functions as a chemo-mechanical motor that hydrolyzes GTP to drive RNA rearrangements during splicing (Fabrizio et al., 1997).
In contrast to our understanding of U4/U6 unwinding in spliceosome activation, much less is known about spliceosome disassembly. At the stage of exon ligation, the conserved DExD/H-box ATPase Prp22p binds to the spliceosome (James et al., 2002) to promote exon ligation (Schwer and Gross, 1998), to proofread exon ligation (Mayas et al., 2006) and to release the ligated product (Schwer and Gross, 1998; Wagner et al., 1998). Subsequently, the conserved DExD/H-box ATPase Prp43p replaces Prp22p (James et al., 2002) and promotes release of the excised intron and disassembly of the spliceosome (Arenas and Abelson, 1997; Martin et al., 2002; Tsai et al., 2005). The requirements for spliceosome disassembly, however, have not been thoroughly explored.
To investigate the requirements for spliceosome disassembly, we developed a biochemical assay for the disassembly of spliceosomes assembled in vivo, allowing tests of mutations that would otherwise inhibit spliceosome assembly in vitro. We found that both Brr2p and Snu114p promoted spliceosome disassembly. Further, Snu114p repressed spliceosome disassembly when bound to GDP and derepressed spliceosome disassembly when bound to GTP. Nonhydrolyzable GTP analogs similarly derepressed spliceosome disassembly, indicating that GTP hydrolysis was not required. The guanine nucleotide states of Snu114p similarly regulated U4/U6 unwinding. These findings indicate that U4/U6 unwinding and spliceosome disassembly are mediated by parallel mechanisms. Further, our data suggest that Snu114p, rather than acting as a motor, acts as a classic signaling G protein (Sprang, 1997) to regulate Brr2p activity and thereby to control spliceosome dynamics.
Results
ATP-dependent Disassembly of Spliceosomes Assembled In Vivo
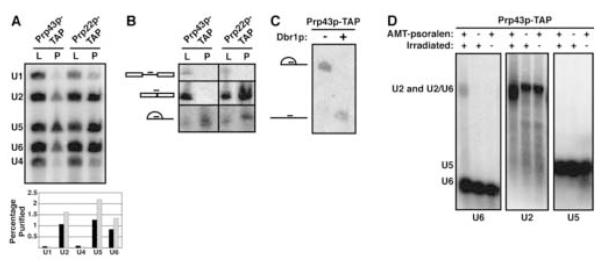
To examine the requirements for spliceosome disassembly in Saccharomyces cerevisiae, we purified a subpopulation of spliceosomes assembled in vivo that are poised for disassembly. Specifically, from yeast lysates we affinity-purified spliceosomes associated with the disassembly factor Prp43p, because Prp43p binds to the spliceosome only after the mRNA dissociates (James et al., 2002). The purified spliceosomes were enriched for U2, U5 and U6 (Figure 1A), as expected (Lebaron et al., 2005; Combs et al., 2006; Leeds et al., 2006). Further, by probing for the U3A snoRNA precursor and splicing products, the spliceosomes were enriched for excised, lariat intron but not for precursor or ligated product (Figure 1B,C), as expected (James et al., 2002); the lariat intermediate was undetectable. In contrast, spliceosomes affinity-purified by association with Prp22p were enriched for U2, U5, U6, the excised U3A intron and the ligated U3A snoRNA (Figure 1A,B), as expected (James et al., 2002). Although Prp43p interacts with many RNAs involved in ribosome biogenesis (Lebaron et al., 2005; Combs et al., 2006; Leeds et al., 2006), we did not observe association of Prp43p with U3A snoRNA, snR66 snoRNA or 27S pre-rRNA (Figure 1B; data not shown), due to pelleting of large particles during the high-speed spin used to clarify lysates. By crosslinking, U2 and U6 interacted in the Prp43p-associated spliceosomes (Figure 1D), suggesting that the two snRNAs were base-paired, as expected for an activated spliceosome (Will and Lührmann, 2006). Further, in the absence of ATP all of the Prp43p-associated RNAs, U2, U5, U6 and the excised U3A intron, comigrated in a single U2/U6•U5•intron complex (Figure 2A), indicative of a spliceosome poised for disassembly (Will and Lührmann, 2006). Thus, we succeeded in purifying a subpopulation of activated spliceosomes that had released the mRNA product but had not yet released the intron product or dissociated the snRNPs.
Figure 1.
Purification of in vivo Assembled Spliceosomes Poised for Disassembly (A) Prp43p- and Prp22p-associated spliceosomes, affinity-purified from yJPS797 or yJPS773 using a tandem affinity purification (TAP) tag (Rigaut et al., 1999), are enriched for U2, U5 and U6. RNA was extracted from lysate (L) or purified spliceosomes (P); 1% or 100%, respectively, was analyzed by primer extension (top). The percent yield for each snRNA is shown for Prp43p-(black) and Prp22p-(grey) associated spliceosomes (bottom). (B) Prp43p-associated spliceosomes are enriched specifically for excised intron, whereas Prp22p-associated spliceosomes are enriched for excised intron and ligated exons. RNA was extracted from lysate (L) or purified spliceosomes (P); 0.4% or 100%, respectively, was probed by Northern for the U3A pre-snoRNA, snoRNA and excised intron, indicated from top to bottom on the left. (C) In Prp43p-associated spliceosomes, the U3A intron is branched. RNA extracted from spliceosomes was incubated without (-) or with (+) debranchase (Dbr1p), as described (Khalid et al., 2005) for 30 min at 22°C, and probed by Northern for the branched and debranched introns, depicted from top to bottom on the left. (D) In Prp43p-associated spliceosomes, U2 crosslinks with U6. After UV-irradiating spliceosomes, RNA was extracted and probed sequentially by Northern for U6, U2 and U5, as indicated below the blots. The crosslinked species is indicated as U2/U6.
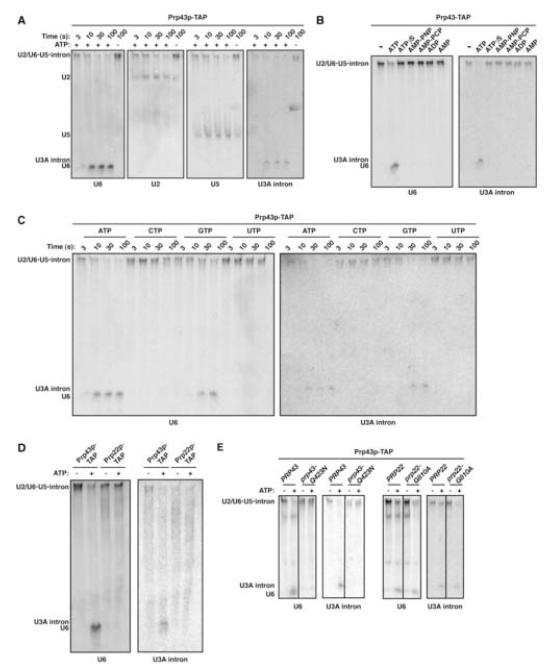
Figure 2.
Prp43p-associated Spliceosomes Disassemble in an ATP- and Prp43p-Dependent Manner (A) A Prp43p-associated U2/U6•U5•intron complex disassembles rapidly in the presence of ATP. Complexes were purified from yJPS797 and incubated at 4°C in the presence (+) or absence (-) of ATP. At the indicated times, samples from the reactions were quenched and then analyzed by native gel followed by serial Northern probing. The migration of the U2/U6•U5•intron complex; the free U3A intron, and the free U6, U2 and U5 snRNAs are indicated on the left. (B) Disassembly of the U2/U6•U5•intron complex requires hydrolyzable ATP. Reactions were incubated at 4°C for 60 s and analyzed as in (A). (C) Disassembly of the U2/U6•U5•intron complex is promoted preferentially by ATP. Complexes were incubated at 4°C and analyzed as in (A). (D) ATP does not promote disassembly of Prp22p-associated spliceosomes. Prp43p-associated U2/U6•U5•intron complexes, purified from yJPS797, and Prp22p-associated U2/U6•U5•intron•mRNA complexes, purified from yJPS773, were incubated and analyzed as in (B). (E) Disassembly of Prp43p-associated spliceosomes is inhibited by the prp43-Q423N mutation but not by the prp22-G810A mutation. Spliceosomes were purified from wild-type PRP43 (yJPS797) or PRP22 (yJPS1049) and mutant prp43-Q423N (yJPS798) or prp22-G810A (yJPS1052) and then incubated and analyzed as in (B).
These purified, Prp43p-associated spliceosomes disassembled in an ATP-dependent manner. By native gel or glycerol gradient analysis, the purified U2/U6•U5•intron complex dissociated in the presence of ATP within 10 s at 4°C, yielding released excised intron and free U2, U6 and U5 (Figure 2A and data not shown; cf. Huang et al., 2002; Tsai et al., 2005). In the presence of AMP, ADP or the nonhydrolyzable ATP analogs ATPγS, AMP-PNP or AMP-PCP, the spliceosomes failed to release the excised U3A intron and to dissociate the snRNAs (Figure 2B; data not shown – throughout, the dissociation of U2 and U5 paralleled the dissociation of U6 and the intron), indicating that disassembly required ATP hydrolysis. In the presence of CTP or UTP, the spliceosomes similarly failed to release the excised intron and to dissociate the snRNAs and in the presence of GTP the spliceosomes disassembled ~10-fold slower than in the presence of ATP (Figure 2C; data not shown), indicating the requirement for an ATP-specific NTPase for both intron release and snRNA dissociation. Because dissociation of the complex did not require extract, disassembly was mediated exclusively by components of the purified complex (cf. Tsai et al., 2005).
We determined whether the dissociation of the U2/U6•U5•intron complex reflected genuine spliceosome disassembly. We tested for disassembly of the earlier, mRNA-containing, Prp22p-bound spliceosomes (Figure 1A,B). In contrast to the Prp43p-bound spliceosomes, the Prp22p-bound spliceosomes failed to dissociate the snRNAs or to release the excised intron (Figure 2D; data not shown). To test whether dissociation of the U2/U6•U5•intron complex required factors specific to spliceosome disassembly, we assayed for dissociation of Prp43p-bound spliceosomes purified from either the cold-sensitive mutant strain prp43-Q423N, which impedes release of the excised intron (Leeds et al., 2006), or from the cold-sensitive mutant strains prp22-G810A and prp22-H606A, which impede the earlier step of mRNA release (Schwer and Meszaros, 2000). Cold-sensitive mutants allowed us to purify spliceosomes from strains grown at the permissive temperature (30°C) and then to assay for disassembly defects at a cold temperature standard for our assay (4°C). Both intron release and snRNA dissociation were blocked by the prp43 mutation, as expected (Arenas and Abelson, 1997; Martin et al., 2002; Tsai et al., 2005), but not by the prp22 mutations (Figure 2E; data not shown), as expected (James et al., 2002). Thus, we have established an assay for genuine spliceosome disassembly.
Spliceosome Disassembly Requires the DExD/H-box ATPase Brr2p
The requirement for Prp43p in spliceosome disassembly (Figure 2E) did not likely account for the preference for ATP in disassembly (Figure 2C), because Prp43p and other members of the DEAH-box subclass of DExD/H-box ATPases do not require ATP specifically (Cordin et al., 2006 and references therein; Tanaka and Schwer, 2006). Because the DExD/H-box protein Brr2p is (i) an ATP-specific NTPase (Laggerbauer et al., 1998; Stevens et al., 2001), (ii) the only known integral ATPase of the budding yeast spliceosome (Ohi et al., 2002; Stevens et al., 2002; Lebaron et al., 2005) and (iii) associated with Prp43p particles (Lebaron et al., 2005), Brr2p was a strong candidate for the ATP-specific NTPase required for disassembly. To test for a requirement for Brr2p, we investigated whether the cold-sensitive brr2-1 mutation (Noble and Guthrie, 1996) impeded disassembly; this mutation, located in a putative RNA binding motif of the first DExD/H-box ATPase domain, impedes U4/U6 unwinding (Raghunathan and Guthrie, 1998). Spliceosomes purified from the brr2-1 mutant failed both to release the excised intron and to dissociate the snRNAs (Figure 3A; data not shown), indicating a requirement for the first DExD/H-box domain of Brr2p in spliceosome disassembly. To test for a role for the Sec63 domains of Brr2p (Ponting, 2000), we engineered single alanine substitutions at residues that are nearly invariant among all known Sec63 domains. One mutant, brr2-R1107A, in the first Sec63 domain, was cold-sensitive (Figure 3B). We purified Prp43p-associated spliceosomes from the brr2-R1107A mutant grown at the permissive temperature. These spliceosomes failed to release the excised intron and to dissociate the snRNAs (Figure 3A; data not shown). In vivo, after a 4 h shift to 15°C, the brr2-1 and brr2-R1107A mutants maintained the abundance of mature U3A snoRNA but accumulated intron excised from the U3A precursor (Figure 3C), indicating that the brr2 mutants impeded degradation of the excised lariat intron, consistent with a role for Brr2p in spliceosome disassembly. Thus, Brr2p is not only required for U4/U6 unwinding but also for spliceosome disassembly.
Figure 3.
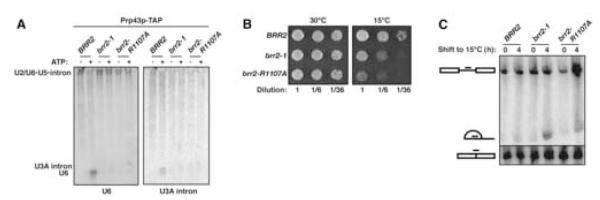
Spliceosome Disassembly Requires the DExD/H-box ATPase Brr2p (A) Disassembly of Prp43p-associated spliceosomes is inhibited by brr2 mutations. Spliceosomes were purified from the wild-type strain BRR2 (yJPS999) or the mutant strains brr2-1 (yJPS1000) and brr2-R1107A (yJPS1001). Spliceosomes were incubated and analyzed as in Figure 2B. (B) As for brr2-1, the mutant brr2-R1107A is cold sensitive. Cells were diluted from liquid culture and grown on rich media for 2 days at 30°C or 6 days at 15°C. (C) In brr2 mutants, the excised lariat intron from U3A accumulates. Wild-type BRR2 (yJPS999) or the mutants brr2-1 (yJPS1000) and brr2-R1107A (yJPS1001) were grown in rich liquid media at 30°C and shifted to 15°C for 4 h. RNA was analyzed by Northern as in Figure 1B.
Spliceosome Disassembly Requires the EF-G-like GTPase Snu114p
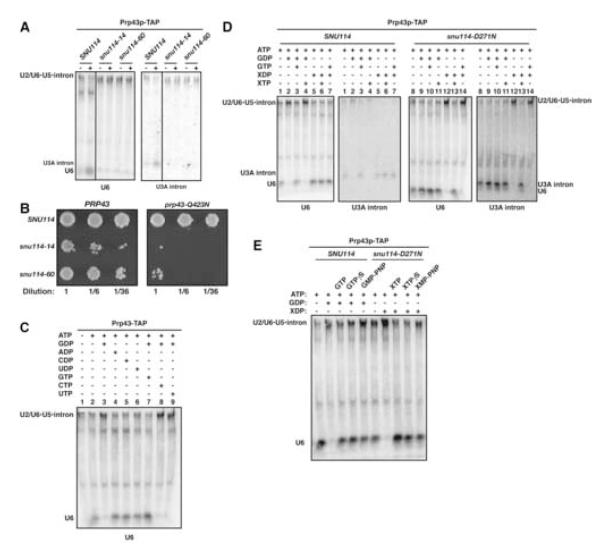
Because previous work suggested that Brr2p promotes spliceosome assembly in coordination with Snu114p (Bartels et al., 2002; Bartels et al., 2003; Brenner and Guthrie, 2005; Brenner and Guthrie, 2006), we evaluated the sensitivity of spliceosome disassembly to the cold-sensitive mutations snu114-14 and snu114-60 (Brenner and Guthrie, 2005). snu114-14 is mutated in the G” domain, which is specific to EF-G paralogs. In vivo, snu114-14 reduces the levels of U4/U6•U5 snRNP (Brenner and Guthrie, 2006). snu114-60 is truncated at the C-terminus and lacks a portion of a domain that undergoes significant rearrangements in EF-2 (Jorgensen et al., 2003). In vitro, snu114-60 blocks U4/U6 unwinding during spliceosome activation (Brenner and Guthrie, 2006). We grew the snu114-14 and -60 mutants at the permissive temperature and then purified Prp43p-associated spliceosomes. These purified spliceosomes failed to release the excised intron and to dissociate the snRNAs (Figure 4A; data not shown). Supporting these findings, prp43-Q423N was synthetically lethal with snu114-14 and synthetically sick with snu114-60 (Figure 4B; cf. Brenner and Guthrie, 2005). Thus, the GTPase Snu114p is not only required for spliceosome activation but also for spliceosome disassembly.
Figure 4.
Spliceosome Disassembly is Promoted and Regulated by the GTPase Snu114p (A) Disassembly of Prp43p-associated spliceosomes is inhibited by snu114 mutants. Spliceosomes were purified from the wild-type strain SNU114 (yJPS1053) or the mutant strains snu114-14 (yJPS1120) and snu114-60 (yJPS1056). Spliceosomes were incubated and analyzed as in Figure 2E. (B) prp43 and snu114 mutants genetically interact. yTB135 was cotransformed with plasmids encoding PRP43 (pJPS1346) or prp43-Q423N (pJPS1347) and SNU114 (pTB106), snu114-14 (pTB108) or snu114-60 (pTB113), grown in liquid media, diluted and grown on solid media containing 5-fluoroorotic acid for 5 days at 30°C. (C) Spliceosome disassembly is repressed specifically by GDP and derepressed specifically by GTP. Prp43p-associated spliceosomes, purified from a wild-type strain (yJPS797), were incubated at 4°C for 60 s and analyzed as in Figure 2B. (D) Spliceosome disassembly is regulated by Snu114p. Prp43p-associated spliceosomes were purified from a wild-type strain (yJPS1053) or a snu114-D271N mutant strain (yJPS1054) that appears to switch the specificity of Snu114p from guanine to xanthine (Bartels et al., 2003). Spliceosomes were incubated and analyzed as in Figure 2B. ATP, GDP and XDP concentrations were 2 mM; GTP and XTP concentrations were 0.5 mM. (E) Spliceosome disassembly does not require GTP hydrolysis. Prp43p-associated spliceosomes, purified from wild-type SNU114 (yJPS1053) or mutant snu114-D271N (yJPS1054), were incubated at 4°C for 60 s and analyzed as in Figure 2B.
Although Snu114p, in analogy to EF-G, has been proposed to function as a hydrolysis-dependent molecular motor (Fabrizio et al., 1997; Bartels et al., 2002; Bartels et al., 2003), the activities of G proteins are often modulated by guanine nucleotides (Sprang, 1997). To test for a role for guanine nucleotides in regulating spliceosome disassembly, we determined whether GTP or GDP modulated spliceosome disassembly. Both spliceosome disassembly and intron release were repressed by GDP but not ADP, CDP or UDP (Figure 4C, cf. lanes 3-6; data not shown). In the presence of ATP alone spliceosomes disassembled within 10 s (Figure 2A), but with the addition of GDP spliceosomes disassembled only after 600 s (data not shown). Although spliceosome disassembly and intron release did not require GTP, we found that GTP, but not CTP or UTP, alleviated the GDP-dependent repression of these processes (Figure 4C, cf. lanes 7-9); GTP alleviated repression even when GDP was in 4-fold excess. Thus, GDP represses spliceosome disassembly and GTP derepresses spliceosome disassembly.
Guanine nucleotides regulated spliceosome disassembly through Snu114p. We compared the disassembly of wild-type spliceosomes with the disassembly of spliceosomes purified from the temperature-sensitive mutant snu114-D271N, which has a mutation of an invariant aspartic acid to asparagine in the guanine binding pocket, a mutation that appears to switch the specificity of Snu114p from guanine nucleotides to xanthine nucleotides (Bartels et al., 2003), as in many other G proteins (Sprang, 1997). Although the disassembly of wild-type spliceosomes was repressed by GDP, disassembly was not repressed by xanthosine diphosphate (XDP; Figure 4D, cf. lanes 2,5). In contrast, the disassembly of snu114-D271N spliceosomes was repressed by XDP but not by GDP (Figure 4D, cf. lanes 9,12). Further, the GDP-mediated repression of disassembly in wild-type spliceosomes was alleviated by GTP but not by XTP (Figure 4D, cf. lanes 3,4). In contrast, the XDP-mediated repression of disassembly of snu114-D271N spliceosomes was alleviated by XTP but not by GTP (Figure 4D, cf. lanes 13,14). Thus, Snu114p is the GTPase that mediates the regulation of spliceosome disassembly by guanine nucleotides.
Spliceosome disassembly did not require GTP hydrolysis. The GDP-mediated repression of spliceosome disassembly was derepressed not only by GTP but also by the nonhydrolyzable GTP analogs GTPγS, GMP-PNP and GMP-PCP (Figure 4E; data not shown). Similarly, disassembly of snu114-D271N spliceosomes, repressed by XDP, was derepressed not only by XTP but also by XTPγS and XMP-PNP (Figure 4E). Thus, Snu114p promotes spliceosome disassembly not by hydrolyzing GTP but rather by simply binding GTP, suggesting that Snu114p acts as a classic regulatory G protein.
U4/U6 Unwinding is also Regulated by the GTPase Snu114p
Because Snu114p has also been implicated in U4/U6 unwinding during spliceosome activation (Bartels et al., 2002; Bartels et al., 2003; Brenner and Guthrie, 2006), we tested for a regulatory role for Snu114p in U4/U6 unwinding using U4/U6•U5 snRNPs (see Introduction). Although previously characterized U4/U6•U5 snRNPs from budding yeast lack Prp28p (Gottschalk et al., 1999; Stevens and Abelson, 1999), we were able to affinity purify U4/U6•U5 snRNPs using TAP-tagged Prp28p (Figure 5A), indicating that Prp28p is a component of at least a subpopulation of U4/U6•U5 snRNPs in budding yeast, as in mammals (Teigelkamp et al., 1997). ATP-dependent U4/U6 unwinding in these snRNPs was inhibited specifically by GDP (Figure 5B, cf. lanes 3-6). This inhibition of U4/U6 unwinding was derepressed specifically by GTP (Figure 5B, cf. lanes 7-9). To determine if this regulation was mediated by Snu114p, we assayed U4/U6 unwinding in U4/U6•U5 snRNPs purified from the snu114-D271N mutant (Bartels et al., 2003). U4/U6 unwinding in these snRNPs was inhibited by XDP but not by GDP (Figure 5C, cf. lanes 11,14), and this inhibition was derepressed by XTP but not by GTP (Figure 5C, cf. lanes 15,16). These data suggest that the guanine nucleotide state of Snu114p regulates spliceosome activation, in addition to spliceosome disassembly.
Figure 5.
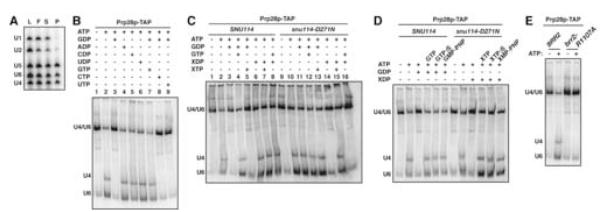
U4/U6 Unwinding is Regulated by Snu114p (A) Purification of U4/U6•U5 snRNPs associated with Prp28p. U4/U6•U5 snRNPs were affinity purified from lysates of yJPS1004 after glycerol gradient fractionation. By primer extension, RNA was analyzed from lysate (L) and pooled glycerol gradient fractions (F) and after immunoprecipitation from the supernatant (S) and the purified snRNP (P). (B) U4/U6 unwinding is repressed specifically by GDP and derepressed specifically by GTP. Prp28p-associated U4/U6•U5 snRNPs were incubated at 4°C and quenched after 60 s. RNA was extracted at 4°C, resolved on a native RNA gel and analyzed by Northern, probing simultaneously for U4 and U6. The migration of base paired U4/U6 and free U4 and U6 are indicated to the left. (C) U4/U6 unwinding is regulated by Snu114p. Prp28p-associated U4/U6•U5 snRNPs were purified from wild-type SNU114 (yJPS1108) or mutant snu114-D271N (yJPS1111). Nucleotide concentrations were as in Figure 4D. snRNPs were incubated and analyzed as in (B). (D) U4/U6 unwinding does not require GTP hydrolysis. Prp28p-associated U4/U6•U5 snRNPs purified from wild-type SNU114 (yJPS1108) or mutant snu114-D271N (yJPS1111) were incubated and analyzed as in (B). (E) U4/U6 unwinding requires the first Sec63 domain of Brr2p. Prp28p-associated U4/U6•U5 snRNPs were purified from a wild-type BRR2 (yJPS1115) or mutant brr2-R1107A (yJPS1117) strain and incubated and analyzed as in (B).
Similar to spliceosome disassembly, U4/U6 unwinding did not require GTP hydrolysis. Wild-type U4/U6•U5 snRNPs, repressed by GDP, unwound U4/U6 in the presence of the nonhydrolyzable GTP analogs GTPγS, GMP-PNP, and GMP-PCP (Figure 5D; data not shown). Additionally, snu114-D271N mutant U4/U6•U5 snRNPs, repressed by XDP, unwound U4/U6 in the presence of the nonhydrolyzable XTP analogs XTPγS and XMP-PNP (Figure 5D, cf. Bartels et al., 2003). Thus, Snu114p promotes U4/U6 unwinding by binding rather than by hydrolyzing GTP, further supporting a role for Snu114p as a classic regulatory G protein.
To determine if the first Sec63 domain (Ponting, 2000) of Brr2p is required for U4/U6 unwinding, we purified Prp28p-associated U4/U6•U5 snRNPs from the brr2-R1107A mutant. U4/U6 unwinding was inhibited by this mutation (Figure 5E). Thus, U4/U6 unwinding requires both the first DExD/H-box domain (data not shown; Raghunathan and Guthrie, 1998) and the first Sec63 domain of Brr2p, as for spliceosome disassembly (Figure 3A).
Discussion
To investigate the requirements for spliceosome disassembly, we purified spliceosomes assembled in vivo that were poised for disassembly (Figure 1). We found that disassembly not only requires ATP and the DEAH-box ATPase Prp43p (Figure 2A,E), as expected (Arenas and Abelson, 1997; Martin et al., 2002; Tsai et al., 2005), but also the DExD/H-box ATPase Brr2p and the GTPase Snu114p (Figures 3A, 4A), two spliceosome activation factors that promote U4/U6 unwinding (Laggerbauer et al., 1998; Raghunathan and Guthrie, 1998; Kim and Rossi, 1999; Bartels et al., 2002; Bartels et al., 2003; Brenner and Guthrie, 2006). Supporting these in vitro observations, brr2 mutants accumulate excised intron in vivo (Figure 3C) and snu114 mutants interact genetically with a prp43 mutant (Figure 4B). Further, we found that GDP-bound Snu114p represses both spliceosome disassembly and U4/U6 unwinding and GTP-bound Snu114p derepresses these rearrangements, without hydrolyzing GTP (Figures 4D,E and 5C,D). Our data suggest that the spliceosome activates and disassembles by a common pathway and that the GTPase Snu114p regulates this pathway.
Intron Release and Spliceosome Disassembly Share Their Requirements
Our findings demonstrate that dissociation of the excised intron, U2, U5 and U6 all require Prp43p, Brr2p and Snu114p. Both intron release and spliceosome disassembly are blocked by prp43 (Figure 2E, cf. Tsai et al., 2005), brr2 (Figure 3A) and snu114 mutants (Figure 4A). Additionally, efficient intron release and spliceosome disassembly both require ATP specifically (Figure 2C), consistent with a requirement in both processes for Brr2p, an ATP-specific DExD/H-box ATPase (Laggerbauer et al., 1998; Stevens et al., 2001). Further, both intron release and spliceosome disassembly are repressed by GDP-bound Snu114p and derepressed by GTP-bound Snu114p (Figures 4D,E and 5C,D). Thus, our data do not distinguish intron release from spliceosome disassembly, indicating either that these rearrangements occur downstream of all three factors or that the rearrangements are interdependent. Further, our data do not strictly order the functions of Prp43p, Brr2p and Snu114p, which could function sequentially or coordinately to effect intron release and spliceosome disassembly.
Dual Roles for Brr2p and Snu114p Suggest Parallel Pathways
Brr2p plays a dual role in promoting both U4/U6 unwinding (Figure 5E; Laggerbauer et al., 1998; Raghunathan and Guthrie, 1998; Kim and Rossi, 1999) and spliceosome disassembly (Figure 3). This dual role suggests that Brr2p promotes these processes by a common mechanism. Whereas Brr2p likely promotes U4/U6•U5 snRNP disassembly and spliceosome activation by unwinding U4/U6, Brr2p may promote spliceosome disassembly by unwinding U2/U6, a component of Prp43p-associated spliceosomes (Figure 1D). A role for Brr2p in unwinding U2/U6 is consistent with the observation that the ATPase activity of Brr2p is specifically stimulated by annealed U2/U6 (Xu et al., 1996). Because both U4/U6 unwinding and U2/U6 unwinding result in displacement of an snRNA from U6, we propose that Brr2p generally translocates along U6, displacing U4 during spliceosome activation and U2 during spliceosome disassembly (cf. Mayas et al., 2006).
Similar to Brr2p, Snu114p plays a dual role in promoting both U4/U6 unwinding (Figure 5; Bartels et al., 2002; Bartels et al., 2003; Brenner and Guthrie, 2006) and spliceosome disassembly (Figure 4). The dual role of Brr2p and Snu114p provides a rationale for the integral nature of these proteins in the spliceosome. The parallel roles for Snu114p and Brr2p as well as the synergistic interactions between snu114 and brr2 mutants (Brenner and Guthrie, 2005) suggests that Snu114p and Brr2p cooperate in U4/U6 unwinding and spliceosome disassembly. As a DExD/H-box ATPase, Brr2p likely promotes the RNA rearrangements of spliceosome activation and disassembly directly. Snu114p, as a regulator of these processes (Figures 4 and 5), likely functions upstream (cf. Bartels et al., 2002) either to establish the accessibility of Brr2p substrates or to activate Brr2p itself, perhaps through the Sec63 domain that is required for Brr2p activity (Figures 3, 5E).
Snu114p may regulate Brr2p directly, because the human orthologs of these factors interact directly by yeast two-hybrid analysis (Liu et al., 2006). Alternatively, Snu114p may regulate Brr2p indirectly through Prp8p. Like Snu114p and Brr2p, Prp8p is an integral U5 snRNP component that remains associated with the spliceosome throughout the splicing cycle (Grainger and Beggs, 2005 and references therein). Further, the human orthologs of Snu114p, Brr2p and Prp8p copurify, along with U5-40kD, as an RNA-free heterotetrameric complex (Achsel et al., 1998). Moreover, Prp8p interacts directly with both Snu114p and Brr2p (Achsel et al., 1998; van Nues and Beggs, 2001). In addition, prp8 mutants interact genetically with both snu114 (Brenner and Guthrie, 2005) and brr2 mutants (van Nues and Beggs, 2001; Kuhn et al., 2002). In particular, prp8 mutants suppress brr2 mutants (Kuhn et al., 2002), leading to the hypothesis that Prp8p controls Brr2p (see also Achsel et al., 1998), although these data do not rule out a function for Prp8p upstream of Snu114p (see below). Whether Brr2p is regulated directly or indirectly, given the central role of Brr2p in rearranging the spliceosome, Snu114p, by controlling Brr2p, may serve as a central regulator of the spliceosome.
Snu114p Promotes Brr2p-dependent Rearrangements without Hydrolyzing GTP
As a close paralog of the GTPase EF-G/EF-2, which hydrolyzes GTP to promote translocation of mRNA and tRNA through the ribosome (Rodnina et al., 1997), Snu114p has been proposed to act similarly as a motor that hydrolyzes GTP to translocate RNA within the spliceosome (Fabrizio et al., 1997). In support of this model, previous work implicated a role for GTP hydrolysis in promoting U4/U6 unwinding. Specifically, in spliceosomes reversibly stalled by the Snu114p-D271N variant, U4/U6 unwinding is stimulated by XTP but not by the nonhydrolyzable analog XMP-PNP (cited as data not shown in Bartels et al., 2003). However, we have found no evidence of a requirement for GTP hydrolysis or XTP hydrolysis in promoting either U4/U6 unwinding or spliceosome disassembly (Figures 4E and 5D; data not shown). Thus, our data support a role for GTP hydrolysis downstream of U4/U6 unwinding and spliceosome disassembly (see below). Further, instead of supporting a molecular motor function for Snu114p, our data suggest that Snu114p functions as a classic regulatory G protein.
Snu114p-Mediated Regulation of Brr2p-dependent Spliceosome Dynamics
Given the fundamental role of Brr2p in snRNA rearrangements, Brr2p likely requires downregulation at certain stages of the splicing cycle, such as the catalytic stages. Unlike the other spliceosomal DExD/H-box ATPases in budding yeast, which are controlled through regulated binding to the spliceosome (Silverman et al., 2003), Brr2p, as an integral spliceosome component, requires a regulatory mechanism that accommodates persistent binding to the spliceosome. Genetic studies have implicated Prp8p in regulating Brr2p activity (Kuhn et al., 2002), but the mechanism of regulation is unclear. As an integral GTPase of the spliceosome, Snu114p provides an ideal mechanism for continuous regulation of Brr2p.
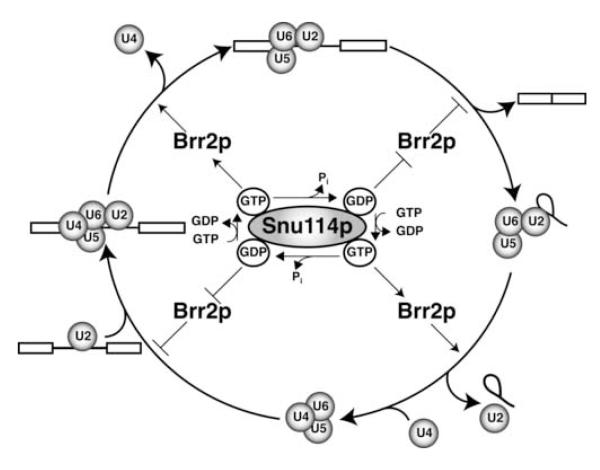
Our data suggest that changes of the guanine nucleotide state of Snu114p serve to regulate Brr2p and thereby control spliceosome dynamics (Figure 6). We propose that GTP-bound Snu114p promotes U4/U6 unwinding allowing activation of the spliceosome for catalysis. Further, subsequent to spliceosome activation, we suggest that a signal activates GTP hydrolysis, switching Snu114p from the GTP-bound state to the GDP-bound state, which would serve to downregulate Brr2p and to allow stabilization of the catalytic core of the spliceosome during the chemical steps of splicing. Finally, we propose that once the splicing reaction is complete a signal triggers exchange of GDP for GTP, switching Snu114p from the GDP-bound state to the GTP-bound state, which would activate intron release and spliceosome disassembly. Disassembly may stimulate GTP hydrolysis, switching Snu114p from the GTP-bound to the GDP-bound state, thereby downregulating Brr2p until a stage in spliceosome activation when exchange of GDP for GTP would be triggered to activate U4/U6 unwinding; such control could limit the apparently futile unwinding of U4/U6 in free U4/U6•U5 snRNPs (Cheng and Abelson, 1987; Raghunathan and Guthrie, 1998). Alternatively, Snu114p may remain in the GTP state promoting U4/U6 unwinding until the spliceosome reactivates in the next round of splicing. As a signal transducer, Snu114p may also sense errors in splicing and consequently control Brr2p to promote fidelity (e.g., Mayas et al., 2006). In addition, Snu114p may sense states of the cell, such as heat shock (Bond, 2006), and downregulate splicing globally.
Figure 6.
A Role for Snu114p in Regulating Brr2p-dependent Spliceosome Dynamics A simplified splicing cycle is shown, depicting the stages of spliceosome binding and activation (left) and splicing and spliceosome disassembly (right). A model for the roles of Snu114p and Brr2p is illustrated. In the model, Snu114p acts as switch that turns Brr2p “on” and “off”. In the GTP state, Snu114p turns Brr2p “on” to trigger spliceosome activation and disassembly (upper left and lower right, respectively). In the GDP state, Snu114p turns Brr2p “off” to repress inappropriate RNA rearrangements (lower left, upper right). GTP hydrolysis and exchange of GDP for GTP serves to toggle the switch, signaling transitions in the splicing cycle that require up- or down-regulation of Brr2p. See Discussion for details.
Although neither GTP nor GDP detectably enhance or inhibit splicing reactions in vitro (data not shown), these observations can be reconciled with our model. First, our model posits that the GTP-state of Snu114p promotes U4/U6 unwinding and spliceosome disassembly, but our data suggest that GTP is not necessary, in the absence of GDP, to achieve the GTP state; indeed, G protein effectors can promote the GTP state of cognate G proteins in the absence of GTP (Sprang, 1997). Second, our model posits that the GDP-state of Snu114p would be required to stabilize the spliceosome, but GDP may not be essential to achieve this state. Third, our model predicts that GDP would repress splicing, but in vitro at a temperature permissive for splicing (20°C), 5′ splice site cleavage and exon ligation are slow relative to U4/U6 unwinding and spliceosome disassembly – even when these rearrangements are delayed by GDP (data not shown). Fourth, our model suggests that GTP would inhibit certain stages of splicing, such as the catalytic stages, but Snu114p may fail to bind GTP at these stages or may repress stages that are not rate limiting in vitro. In vivo, where splicing likely occurs much faster than in vitro (Ares et al., 1999; Wetterberg et al., 2001), splicing may be more sensitive to the guanine nucleotide state of Snu114p. Indeed, a single amino acid mutation in Snu114p that is expected to abolish GTP binding is lethal in vivo (Fabrizio et al., 1997).
Implications: Regulators of Snu114p
Transitions between the guanine nucleotide states of Snu114p may require regulation by a GTPase-activating protein (GAP), a guanine nucleotide exchange factor (GEF) and/or a guanine nucleotide dissociation inhibitor (GDI; Sprang, 1997). Two candidates for such factors include the NineTeen Complex (NTC) and Prp8p. After U4/U6 unwinding, the NTC binds to and stabilizes the spliceosome (Chan et al., 2003). Further, concurrent with spliceosome disassembly, the NTC dissociates (Tsai et al., 2005). Because Snu114p is ubiquitylated (Peng et al., 2003) and the NTC includes the E3 ubiquitin ligase Prp19p (Ohi et al., 2003), Prp19p may regulate the guanine nucleotide state of Snu114p through reversible ubiquitylation. Consistent with this possibility, snu114 mutants enhance mutations in PRP19 as well as SAD1 (Brenner and Guthrie, 2005), which encodes a gene with similarity to ubiquityl C-terminal hydrolases.
Alternatively, since Prp8p interacts with all intronic consensus sites (Grainger and Beggs, 2005), Prp8p could signal to Snu114p the status of the splicing substrate and the stage of the splicing cycle (cf. Achsel et al., 1998). Further, Prp8p could send such a signal to Snu114p directly, given the physical interaction between Prp8p and Snu114p (Achsel et al., 1998; van Nues and Beggs, 2001; Liu et al., 2006). In addition, Prp8 contains a Jab1/MPN domain that binds ubiquitin (Bellare et al., 2006) and thereby could provide a link to Prp19p and Sad1p. Because both Prp8p and Snu114p interact with U5 (Dix et al., 1998) and U5 interacts with the substrate (Newman et al., 1995), U5 could also signal the status of the substrate to Snu114p. Indeed, crosslinking of purified Snu114p to GTP is significantly enhanced in the presence of poly(U) RNA (Bartels et al., 2003), suggesting that interactions between U5 or another RNA may serve to regulate Snu114p. Intriguingly, the U5 stem loop shares similarities with the tRNA anticodon loop, suggesting parallels between Snu114p and its paralog EF-G/EF-2 (Staley and Guthrie, 1998). Through the action of a GAP, a GEF and/or a GDI, Snu114p may not only transduce signals from the splicing substrate but also signals from the spliceosome and/or the cell to regulate Brr2p-dependent spliceosome dynamics.
Experimental Procedures
Oligonucleotides, plasmids and strains
Affinity Purification
Purification of spliceosomes or snRNPs was performed essentially as described (Rigaut et al., 1999). Ten liters of yeast were grown in rich media at 30°C to an optical density of 0.5 to 0.9 at 600 nm, harvested, washed and lysed by bead beating. After centrifugation at 140,000 × g for 1 hr, the soluble lysate was incubated with 1 mL immunoglobulin G-Sepharose beads (Amersham) for 5 hr at 4°C, after which the beads were washed three times with 50 mL buffer D (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 50 mM KCl, 20% [v/v] glycerol). The immunoprecipitated spliceosomes were eluted from the beads with TEV protease in elution buffer (Rigaut et al., 1999) for 2 hrs at 4°C followed by dialysis against buffer D. Before affinity purification of the Prp28p-associated snRNPs, cell lysates were fractionated on a 10% to 40% glycerol gradient by centrifugation for 18 hr at 140,000 × g in a Beckman SW-28 rotor. Fractions enriched for U4, U5 and U6 snRNAs were pooled and then affinity purifications were performed as above.
RNA Analysis
RNA was extracted using phenol/chloroform/isoamyl alcohol (25:24:1) at 65°C, unless noted otherwise. Primer extensions were performed as described (Staley and Guthrie, 1999). For Northern analysis, RNA was separated on a 6% denaturing polyacrylamide gel and transferred semidry to HybondN (Amersham), unless noted otherwise. Membranes were probed with 32P-labeled oligonucleotides or random-primed probes (NEBlot Kit from NEB) in Rapid-Hyb buffer (Amersham) and washed twice with 900 mM NaCl, 90 mM sodium citrate, 0.1% (w/v) sodium dodecyl sulfate. Primer extension gels and Northern blots were exposed to a phosphor screen and developed with a Storm PhosphorImager (Molecular Dynamics) and quantitated with ImageQuant (Molecular Dynamics).
Crosslinking
4′-amino-methyl-4,5′,8-trimethylpsoralen (AMT-psoralen) photochemical crosslinking was performed essentially as described (Wassarman, 1993). 300 μL of TEV eluate was mixed with AMT-psoralen (Sigma), to a final concentration of 40 μg/mL, added to a well of a 12-well plate on ice, covered with a glass plate and irradiated for 20 min at 365 nm with a hand held light source (Fisher) 8 cm from the sample. Samples were treated with 2 mg/mL proteinase K (Ambion) for 30 min at 50°C. RNA was extracted and treated with 0.9 M glyoxal, 10 mM sodium phosphate [pH 7.0], 50% (v/v) dimethyl sulfoxide for 60 min at 50°C, resolved on a 1.5% (w/v) agarose gel in 10 mM sodium phosphate [pH 7.0] and transferred by capillary action to HybondN and analyzed by Northern.
Spliceosome Disassembly and U4/U6 Unwinding Assays
For disassembly assays, 12.5 μL reactions were assembled with 40% TEV eluate and standard splicing buffer (3% [w/v] PEG8000, 2.5 mM MgCl2, 60 mM KPO4 [pH 7.0], 2 mM ATP) and incubated at 4°C for various times, quenched with 3.5 μL of 1.4 mg/ml heparin, 25 mM EDTA and separated on a 4.5% [w/v] polyacrylamide (80:1 acrylamide:bisacrylamide) nondenaturing gel run at 160 V for 4 hr at 4°C in TBE (50 mM Tris-borate [pH 8.3], 1 mM EDTA) and analyzed by Northern. Nonhydrolyzable analogs, GTP, GDP, XTP and XDP were purified by fast protein liquid chromatography using a NH4HCO3 gradient. Note: NTPs were added with equimolar concentrations of MgCl2. NTP and NDP concentrations were 2 mM unless otherwise noted. In the figures shown, NDPs were not added with MgCl2, but addition of NDPs with equimolar MgCl2 yielded equivalent results (data not shown). 25 μL U4/U6 unwinding reactions were assembled similarly but quenched with 500 μL NET-50 (Raghunathan and Guthrie, 1998). RNA was extracted at 4°C and separated on a 9% [w/v] polyacrylamide (80:1 acrylamide:bisacrylamide) nondenaturing gel run at 450 V for 2 hr at 4°C in TBE and analyzed by Northern.
Supplementary Material
Acknowledgements
We thank H. Maita and A. Andress for plasmid construction; B. Schwer for Dbr1p; P. Fabrizio for technical advice; T. Brenner and C. Guthrie for strains and sharing results before publication; R. Seifert for XDP; J. Piccirilli, L. Rothman-Denes, E. Sontheimer, D. Black and members of his lab, and members of the Staley lab for critical reading of the manuscript; and C. Jordan, V. Shaw and M. Norman for technical assistance. This research was supported by grants to J.P.S from the National Institutes of Health (GM62264) and the David and Lucille Packard Foundation (19057).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M, Jr., Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Klatt C, Lührmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Urlaub H, Lührmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C-terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner TJ, Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA. 2006;12:862–871. doi: 10.1261/rna.2319806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng S-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Cheng S-C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M, Jr., Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23) EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Biol. 2003;10:379–385. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res. 2005;33:6349–6360. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. USA. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J, Fabrizio P, Teigelkamp S, Lane WS, Hartmann E, Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rauhut R, Vornlocher HP, Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem. J. 2000;351(Pt 2):527–535. [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman E, Edwalds-Gilbert G, Lin R-J. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6•U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Mundt C, Achsel T, Will CL, Lührmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- Tsai R-T, Fu R-H, Yeh F-L, Tseng C-K, Lin Y-C, Huang Y-H, Cheng S-C. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA. Psoralen crosslinking of small RNAs in vitro. Mol. Biol. Rep. 1993;17:143–151. doi: 10.1007/BF00996222. [DOI] [PubMed] [Google Scholar]
- Wetterberg I, Zhao J, Masich S, Wieslander L, Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosome Structure and Function. In: Gestland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Xu D, Nouraini S, Field D, Tang SJ, Friesen JD. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin R-J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.