Abstract
Insulin resistance from chronic kidney disease (CKD) stimulates muscle protein wasting but mechanisms causing this resistance are controversial. To help resolve this, we used microarray analyses to identify initiators of insulin resistance in the muscles of mice with CKD, glucose intolerance and insulin resistance. CKD raised mRNAs of inflammatory cytokines in muscles and there was a 5.2-fold increase in signal regulatory protein-α (SIRP-α), a transmembrane glycoprotein principally present in muscle membranes. By immunoprecipitation we found it interacts with the insulin receptor and insulin receptor substrate-1 (IRS-1). Treatment of myotubes with a mixture of inflammatory cytokines showed that SIRP-α expression was increased by a NF-κB-dependent pathway. Blockade of NF-κB using a small molecule chemical inhibitor or a dominant-negative IKKβ reduced cytokine-induced SIRP-α expression. The overexpression of SIRP-α in myotubes impaired insulin signaling and raised proteolysis while SIRP-α knockdown with siRNAs in skeletal muscle cells increased tyrosine phosphorylation of the insulin receptor and IRS-1 despite inclusion of cytokines. This led to increased p-Akt and suppression of protein degradation. Thus, SIRP-α is part of a novel mechanism for inflammation-mediated insulin resistance in muscle. In catabolic conditions with impaired insulin signaling, targeting SIRP-α may improve insulin sensitivity and prevent muscle atrophy.
Introduction
Insulin resistance complicates chronic kidney disease (CKD) even in patients with mild renal insufficiency. For example, Fliser et al. identified insulin resistance in patients with serum creatinine values as low as 1.0 mg/dL and inulin clearances as high as 119 ml/min/1.73 m2 (1). Because these subjects had other diseases besides diabetic nephropathy, it was concluded that CKD rather than specific kidney diseases cause insulin resistance. It is well known that insulin resistance extends to patients with advanced kidney failure (2;3). Studies of circulating blood cells or tissue samples from hemodialysis patients have led to the conclusion that the glucose intolerance is due to defects in intracellular signaling processes rather than insulin receptor binding (4). Evidence for a link between glucose intolerance in CKD and defects in intracellular signaling also occurs in several complications of CKD (e.g., metabolic acidosis, increased glucocorticoid production, excess angiotensin II and inflammation) (5-9). There is no general agreement about mechanism(s) causing insulin resistance in CKD (10;11).
Our interest in this topic arises because disorders with impaired insulin signaling are frequently associated with loss of muscle mass. The metabolic acidosis of CKD causes both impaired insulin signaling and stimulation of at least two proteases, caspase-3 and the ubiquitin-proteasome system which in turn causes loss of muscle protein (12;13). Activation of these proteases is complicated. For example, in mice with CKD, we found depressed activity of phosphatidylinositol 3-kinase (PI3K) in muscles plus an increase in Bax related to release of cytochrome C and activation of caspase-3 (6;7;14). Furthermore, decreased PI3K activity also reduces p–Akt in muscle leading to reduced phosphorylation of forkhead transcription factors (FoxO). FoxO's translocate to muscle nuclei, stimulating UPS proteolytic activity by increasing the expression of E3 ubiquitin ligases, Atrogin-1 and MuRF1.
We found another mechanism causing muscle wasting, suppression of muscle progenitor or satellite cells function (15). Following injury or loss of muscle mass, these cells differentiate into myofibrils and repair the injury or contribute to correcting loss of muscle mass but in CKD, satellite cell function is depressed by a process involving impaired IGF-1 signaling (15). Inflammation is also associated with insulin resistance and muscle wasting. In mice with CKD or in response to infusion of angiotensin II, circulating interleukin (IL-6) and tumor necrosis factor (TNF-α) increase and impair insulin/IGF-1 signaling in muscle (8;16). Thus, insulin resistance in CKD is pathophysiologically important because it stimulates muscle proteolysis producing muscle atrophy.
What mechanisms cause insulin resistance? Insulin resistance could arise from accumulation of unexcreted toxins such as indoxyl sulfate or urea but how these compounds impair insulin signaling is unclear (17-19). Alternatively, defective phosphorylation of intracellular mediators of insulin/IGF-1 action could cause defects in insulin signaling pathway (7;20-22). For example, changes in tyrosine phosphorylation could impair IGF-1-initiated signaling, decreasing phosphatidylinositol 3-kinase (PI3K) and p-Akt activities leading to muscle protein wasting (6;13;23).
We have uncovered a new mechanism for CKD-induced insulin resistance, upregulation of signal regulatory protein-alpha (SIRP-α). SIRP-α is a transmembrane glycoprotein which contains three extracellular immunoglobulin-like domains and a cytoplasmic region containing src homology-2 (SH-2) binding motifs. Following tyrosine phosphorylation of SIRP-α, a complex forms with SHP2 triggering tyrosine phosphatase activity (24;25). We examined how SIRP-α influences insulin-induced intracellular signaling responses and whether it exerts pathophysiologically important changes in muscle protein metabolism.
Results
CKD increases inflammatory cytokines causing insulin resistance
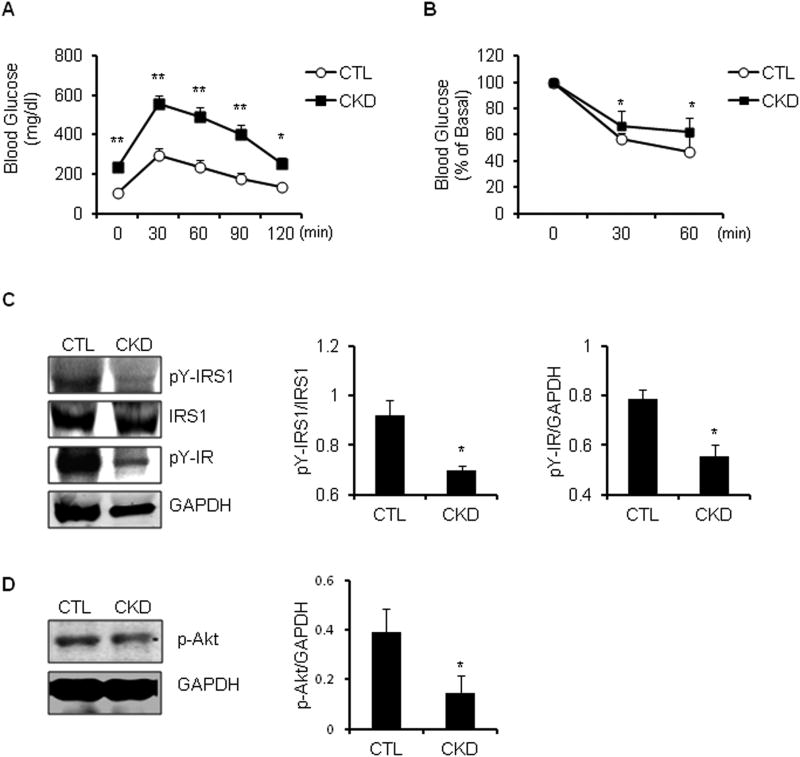
Our mouse model of CKD exhibits blood chemistries similar to those of patients with CKD, including an increase in BUN, serum creatinine and metabolic acidosis (Table 1). In glucose tolerance testing, the baseline blood glucose was higher and remained so for 2 hours after injecting either glucose or insulin. Thus, in comparison to results from pair-fed, sham-operated, control mice the mouse model of CKD develops glucose intolerance and insulin resistance (Figure 1A, B). Notably, tyrosine phosphorylation of the insulin receptor and IRS-1 were decreased in muscles of mice with CKD vs. responses in control mice (Figure 1C). These changes led to decreased phosphorylation of Akt (Figure 1D) and increased activity of caspase-3 and expression of the E3 ubiquitin ligases, atrogin-1 and MuRF1 (12;14).
Table 1. Serum markers evaluating kidney function.
| Control | CKD | |
|---|---|---|
| BUN (mg/dl) | 30.6 ± 2.4 | 103.7 ± 3.9 ** |
| Creatinine (mg/dl) | 0.35 ± 0.05 | 1.5 ± 0.22 ** |
| Bicarbonate (mM) | 20 ± 1 | 14.6 ± 1.6 * |
Serum BUN (Control: n=10 vs. CKD: n= 25), Creatinine (n=5), and Bicarbonate (n=3) were evaluated in CKD vs. sham-operated control mice. (*, p<0.05 and **, p<0.01, vs. CTL).
Figure 1. Insulin resistance is present in CKD mice.
A. Intraperitoneal glucose tolerance test was performed in CKD vs. control (CTL) mice after 16h fast (*, p<0.01 and **, p<0.001, vs. CTL, n=5) B. Intraperitoneal insulin tolerance test was performed in CKD vs. CTL mice after a 4h fast. (*, p<0.05 vs. CTL, n=3). C. Representative immunoblots from gastrocnemius muscle lysates of CKD vs. CTL mice after 6 h fast and stimulation with 10 U/kg of insulin, which was allowed to circulate for 5 minutes before muscles were collected (left panel). The band density of pY-IRS1 to IRS1 and pY-IR to GAPDH is shown in right panel (*, p<0.05 vs. CTL, n=3). D. Representative immunoblots of p-Akt protein from gastrocnemius muscle lysates from CKD vs. control mice (left panel). The band density of p-Akt to GAPDH is shown in right panel (*, p<0.05 vs. CTL; n=3)
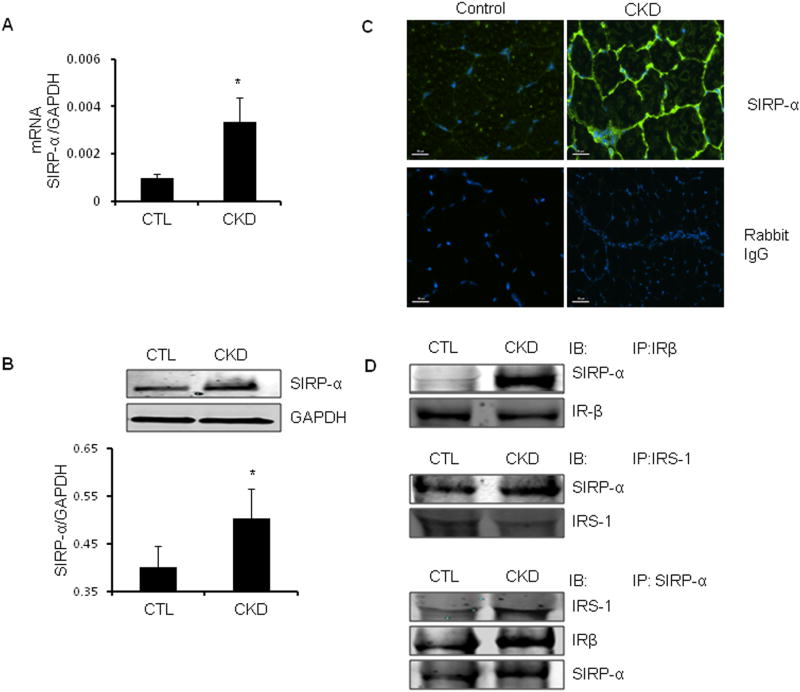
Using a microarray analysis of mRNAs, we examined candidate mediators of CKD-induced insulin resistance in gastrocnemius muscles from CKD and control mice. CKD was associated with upregulation of several inflammatory genes (Supplemental Table 1, 2). Besides these factors, there was a 5.2-fold increase of SIRP-α in muscles of mice with CKD, confirmed by RT-PCR (Figure 2A). Like SIRP-α mRNA, SIRP-α protein increased in muscles of CKD mice vs. control mice (Figure 2B). By immunostaining frozen sections of the mixed fiber, tibialis anterior (TA) muscles from CKD and control mice, we confirmed that SIRP-α is principally present in muscle membranes (Figure 2C).
Figure 2. In CKD muscle, SIRP-α expression is upregulated.
A. SIRP-α mRNA expression in gastrocnemius muscles of CKD vs. sham-operated control (CTL) mice were evaluated by RT-PCR (*, p<0.05 vs. CTL; n=3). B. Representative immunoblots of SIRP-α protein from gastrocnemius muscle lysates from CKD vs. control mice (upper panel). The band density of SIRP-α to GAPDH in the lower panel (*, p<0.05 vs. CTL; n=3). C. Cross-sections of the tibialis anterior muscle from CKD vs. control mice were immunostained with SIRP-α (green) or negative control (rabbit IgG). Scale =50μm. D. In gastrocnemius muscle lysates of CKD vs. CTL, IR-β (top panel) and IRS-1 (middle panel) immunoprecipitates were immunblotted with SIRP- α, or proteins were immunoprecipitated for SIRP-α and immunoblotted with IRS-1, IRβ, or SIRP- α (bottom panel).
SIRP-α associates with the insulin receptor and insulin receptor substrate-1
Insulin signaling depends on tyrosine phosphorylation of the insulin receptor and IRS-1 followed by sequential activation of phosphatidylinositol (PI3K) and p-Akt (26). Since SIRP-α is predominantly present in muscle membranes, we hypothesized that juxtaposition of SIRP-α and the insulin receptor or IRS-1 would cause an interaction between SIRP-α and the insulin receptor or IRS-1, critical initiators of insulin signaling. We immunoprecipitated the insulin receptor or IRS-1 from lysates of gastrocnemius muscles of CKD and control mice. Western blotting uncovered SIRP-α in the immunocomplexes from the imunoprecipitation of the insulin receptor or IRS-1 (Figure 2D, upper and middle panels). Alternatively, muscle lysates of CKD vs. control mice were immunoprecipitated with the SIRP-α antibody and IRS-1 or the insulin receptor were present in the immunocomplexes; the association was stronger in lysates from mice with CKD (Figure 2D, lower panel). Thus, SIRP-α interacts with the insulin receptor or IRS-1 in muscles and CKD seems to increase these interactions. Based on these in vivo results, next, we will examine the mechanisms stimulating SIRP-α expression and insulin signaling in cultured muscle cells to manipulate mediators of insulin or IGF-1 signaling in order to test mechanisms rigorously.
In differentiated muscle cells, cytokines stimulate SIRP-α expression and suppress p-Akt
In mice, circulating levels of IL-6, TNF-α, and interferon-gamma (IFN-γ) are increased by CKD (8;21) while in both CKD and dialysis patients, these cytokines are increased along with an increase in LPS (27;28). These results emphasizing the potential of inflammatory cytokines plus our finding that cytokines change proteasome proteolytic activity in muscle stimulated us to examine how cytokines might increase SIRP-α expression in muscle (29). First, cultured, differentiated C2C12 myotubes were exposed to IL-6, TNF-α, INF-γ or lipopolysaccharide (LPS) individually and the expression of SIRP-α was measured (data not shown). SIRP-α protein increased with the mixture of IL-6, TNF-α and INF-γ (Supplemental Figure 1), but not as much as with the cytokine combination of IL-6, TNF-α, INF-γ and LPS. These responses are consistent with our report that the cytokine combination stimulates NF-κB and proteolysis in muscle cells (29). To mimic in vivo conditions present in mice or patients with CKD, we exposed differentiated C2C12 myotubes to a mixture of the four cytokines and examined SIRP-α expression and function.
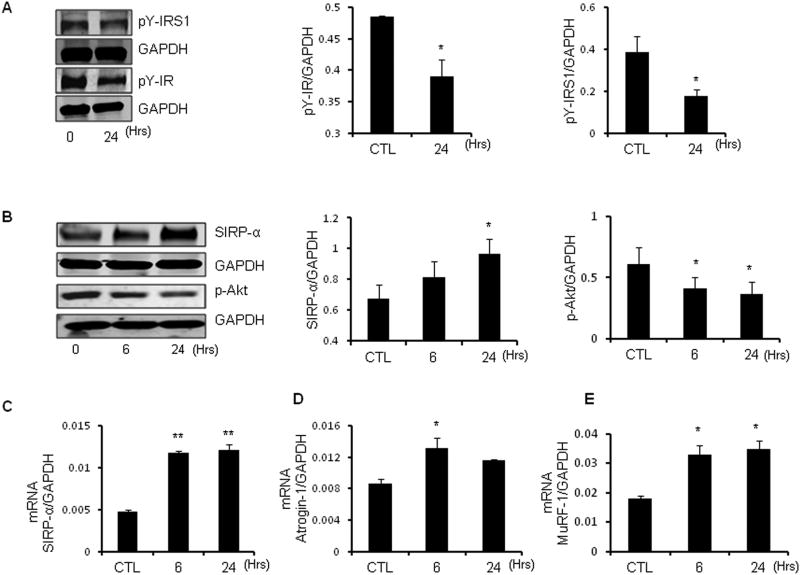
In myotubes, the cytokine mixture produced a time-dependent decrement in phosphorylated tyrosines of both IRS-1 and insulin receptor (Figure 3A). This was associated with an increase in SIRP-α protein and mRNA after 6 h, reaching a peak at 24 h (Figures 3B, 3C) and persisting for at least 72 h (data not shown). The increased SIRP-α and decrease in insulin receptor and IRS-1 tyrosine phosphorylation led to a reduced p-Akt (Figure 3B). This decrease in response to cytokines was similar in magnitude to the change in muscle levels of p-Akt present in CKD mice (Figure 1D). The increase in SIRP-α was also associated with increased expression of mRNAs encoding the E3 ubiquitin ligases, Atrogin-1and MuRF-1 (Figures 3D, 3E). This is relevant because these enzymes lead to increased muscle protein degradation (6;7).
Figure 3. Cytokines trigger SIRP-α expression, impair insulin signaling and activate E3 ubiquitin ligases in C2C12 myotubes.
C2C12 myotubes were serum starved, and then treated with a mixture of IL-6 (2ng/ml), TNF-α (2ng/ml), IFN-γ (2 ng/ml), LPS (10 ng/ml) for 6 or 24 hours and compared with control (CTL) myotubes without cytokine exposure. A. Representative immunoblots were probed with p-Tyrosine-IRS-1 (pY-IRS1), pY-insulin receptor (IR) and GAPDH (left panel). The quantitative analysis of protein expression relative to GAPDH is shown (right panel; *, p<0.05 vs. CTL, n=3 independent experiments). B. Representative immunoblots of SIRP-α and p-Akt shown (left panel). The quantitative analysis of protein expression to GAPDH is shown (right panel; *, p<0.05 vs. CTL, n=3 independent experiments). C-E. mRNA expression of SIRP-α (C), Atrogin-1(C) and MuRF-1(D) were measured by RT-PCR at 6 or 24 hours. (*, p<0.05 vs. CTL, n=3 repeats).
SIRP-α impairs insulin signaling in muscle cells
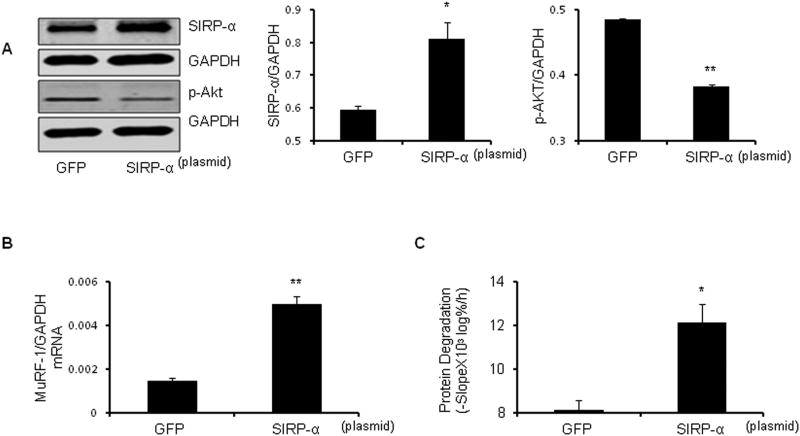
We examined whether the increase in CKD-stimulated SIRP-α (Figure 2A-C) affects p-Akt in muscle. C2C12 myoblasts were transfected with a plasmid that expresses SIRP-α or a GFP-expressing, control plasmid. After differentiating into myotubes, the cytokine mixture was added for 30 minutes to activate SIRP-α. Transfection with the SIRP-α plasmid significantly (p<0.001) decreased p-Akt (Figure 4A) vs. myotubes with GFP transfection. Moreover, raising SIRP-α increased expression of the E3 ubiquitin ligase, MuRF-1 and accelerated the protein degradation of myotubes (Figure 4B, 4C).
Figure 4. Overexpression of SIRP-α impairs insulin signaling and increases protein degradation.
C2C12 myotubes were transfected with SIRP-α plasmid vs. plasmid expressing green fluorescent protein (GFP). Cells were allowed to differentiate into myotubes, serum starved, and treated with cytokines for 30 min. A. Representative western blots of SIRP-α and p-Akt (left panel) with measured band density of protein relative to GAPDH as illustrated (right panel; *, p<0.05 and **, p<0.01 vs. GFP; n=3 independent experiments). B. MuRF-1 mRNA expression was measured by RT-PCR (**, P<0.01 vs. GFP; n=3 independent experiments). C. Rate of protein degradation was measured in cells transfected with SIRP-α plasmid or GFP plasmid. (*, P<0.05 vs. GFP; n=3 independent experiments).
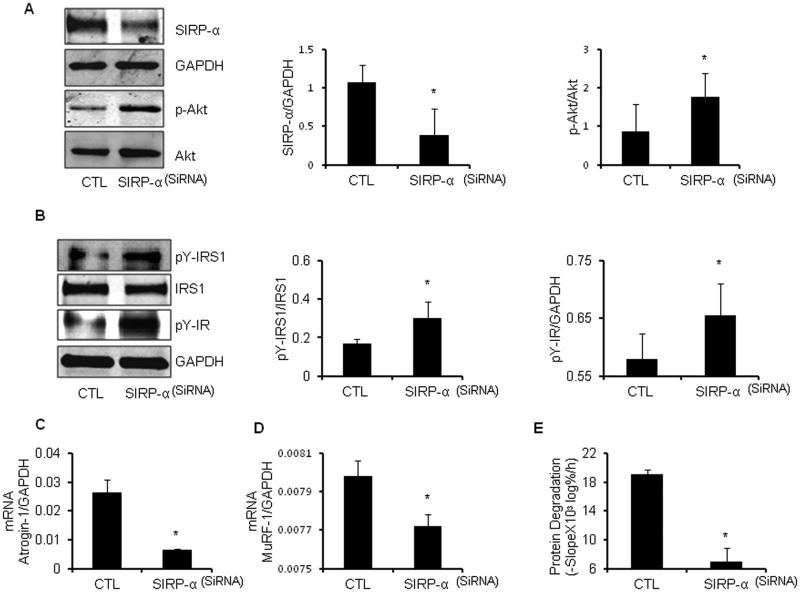
Silencing SIRP-α improves insulin signaling and protein metabolism in muscle cells despite cytokines
Since SIRP-α interferes with insulin-induced intracellular signaling, its suppression should improve insulin signaling. To test this proposal, we examined whether SIRP-α directly interacts with the insulin receptor or IRS-1. Myoblasts were treated with a SIRP-α SiRNA or a scrambled SiRNA and stimulated to differentiate into myotubes. We found that suppression of SIRP-α increased p-Akt even though inflammatory cytokines were present (Figure 5A). Secondly, we incubated myotubes in serum-free media plus the cytokine mixture for 6h, myotubes were exposed to cytokines plus 100 nM insulin for 5 min. In myotubes with silenced SIRP-α, insulin-induced tyrosine phosphorylation of the insulin receptor and IRS-1 were higher than in myotubes transfected with scrambled SiRNA despite cytokine exposure (Figure 5B). These results and the in vivo responses (Figure 2D) demonstrate that SIRP-α interacts with the insulin receptor and IRS-1, to regulate tyrosine phosphorylation of insulin signaling intermediates. Silencing SIRP-α in myotubes exposed to cytokines also decreased the mRNA expressions of Atrogin-1 and MuRF1 (Figure 5C, 5D). This is pathophysiologically relevant because atrogin-1 and MuRF1 lead to protein degradation by increasing muscle proteolysis (Figure 5E).
Figure 5. Suppression of SIRP-α improves insulin signaling, and blocks protein degradation despite the presence of cytokines.
A. C2C12 myoblasts were transfected with SIRP-α SiRNA (SIRP-α) vs. control scrambled SiRNA (CTL). After differentiating into myotubes, serum starved, and subsequently treated with cytokine mixture for 24 hours. Representative immunoblots of SIRP-α and p-Akt (left panel), and band density relative to GAPDH and Akt (respectively) is illustrated (right panel; *, p<0.05 vs. CTL; n=3 independent experiments). B. C2C12 myoblasts were transfected with SIRP-α SiRNA (SIRP-α) vs. control, scrambled SiRNA (CTL). Following differentiation, the myotubes were serum starved, and subsequently treated with cytokines for 6 hours. Subsequently, cells were washed with serum free media and then treated with 100 nm of insulin in fresh serum free media for 5 minutes. Representative immunoblots were probed with pY-IRS-1, pY-IR, and GAPDH (left panel). Quantitative analysis of proteins pY-IRS-1 to IRS1 or pY-IR to GAPDH is shown. (right panel; *, p<0.05 vs. CTL; n=3 repeats). C-E. With the same treatment as Figure 5A. E3 ubiquitin ligases, Atrogin-1(C) and MuRF-1(D), mRNA expression were measured based on RT-PCR (*, p<0.05 vs. CTL; n=3 repeats). The rate of protein degradation (E) was measured in cells transfected with SIRP-α SiRNA vs. CTL SiRNA (*, P<0.05 vs. CTL; n=3 independent experiments).
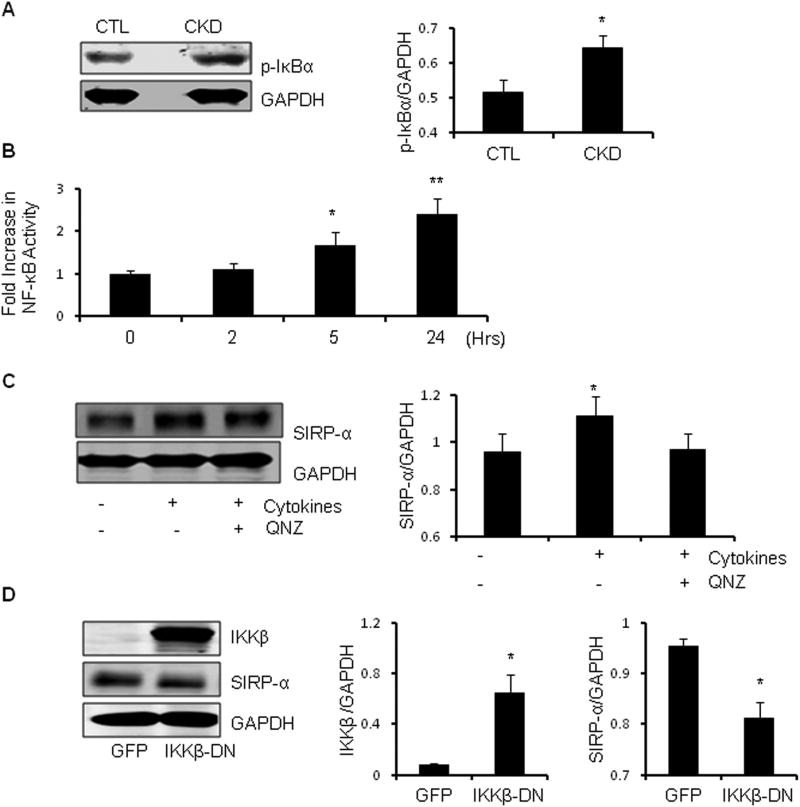
NF-κB activation stimulates SIRP-α expression in muscle
Since CKD raises circulating inflammatory cytokines, we hypothesized that they may affect SIRP-α expression and function through activation of NF-κB. There are several predicted NF-κB recognition sites in the SIRP-α promoter (http://genome.ucsc.edu) and by western blotting, we found a significant (p<0.05) increase in the phosphorylation of IκBα in muscles of CKD mice (Figure 6A). The latter would lead to IκBα degradation and translocation of NF-κB into the nucleus and expression of target genes (30). Secondly, we infected C2C12 myoblasts with a NF-κB promoter-luciferase adenovirus. When they differentiated into myotubes, we found that the cytokine mixture produced a significant increase (p<0.05) in NF-κB promoter activity at 5 or 24 h (Figure 6B). To confirm NF-κB involvement, we treated myotubes with QNZ, a NF-κB inhibitor and after 2 h added the cytokine mixture. The cytokines increased SIRP-α expression and its expression was suppressed by the NF-κB inhibitor (Figure 6C). To exclude non-specific responses, we infected myoblasts with an adenovirus expressing dominant-negative IKKβ (DNIKKβ). After differentiation, myotubes were exposed to cytokines. We found a suppressed levels of SIRP-α in myotubes infected with DNIKKβ vs. results infected with the GFP-expressing adenovirus (Figure 6D). Thus, NF-κB activation in muscle cells promotes SIRP-α expression.
Figure 6. NF-κBregulates SIRP-α expression.
A. Gastrocnemius muscle lysates were obtained from CKD vs. control (CTL) mice. Representative immunoblots of p-IκBα as illustrated (left panel). Band density was measured relative to GAPDH as shown (right panel; *, p<0.05 vs. CTL; n=3). B. C2C12 myoblasts were infected with a NF-κB promoter-luciferase construct. Following their differentiation, the myotubes were treated with the cytokine mixture in serum starved media. Activation of the NF-κB promoter at times listed were measured and the fold change over 0 h was quantified (*, p<0.05 and **, p<0.001 vs. 0 h; n=4 independent experiments). C. Serum starved myotubes were treated with cytokine mixture with or without the NF-κB inhibitor, QNZ. Representative immunoblots of SIRP-α expression (left panel) and band density relative to GAPDH is shown (right panel; *, p<0.05 vs. CTL; n=3 independent experiments). D. C2C12 myoblasts were infected with a DNIKKβ adenovirus or a control-adenovirus that expresses green fluorescent protein (GFP). After differentiation both cell groups were serum starved and treated with cytokines for 24h. Representative western blot analysis (left panel), with quantitative band density of IKKβ and SIRP-α relative to GAPDH as shown (right panel; *, p<0.05 vs. CTL; n=3 independent experiments).
Discussion
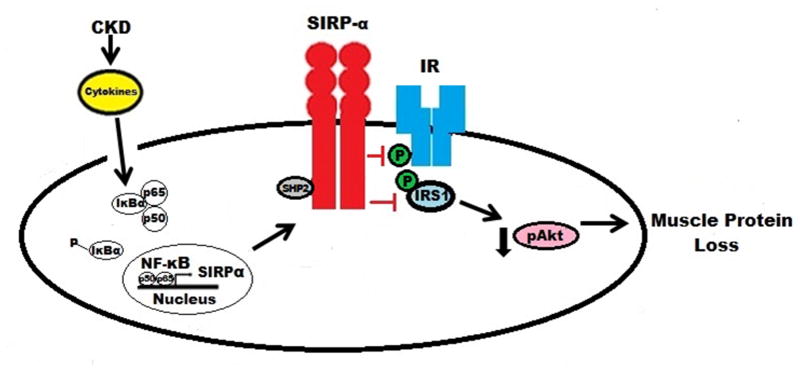
We have uncovered a novel mechanism for the CKD-induced insulin resistance that is associated with muscle protein wasting (6;13). The mechanism involves stimulation of the expression of the SIRP-α in muscle membranes. From this position, SIRP-α can interact with the insulin receptor and IRS-1, resulting in their decreased tyrosine dephosphorylation with decreased Akt phosphorylation. The latter stimulates the activities of caspase-3 and the ubiquitin-proteasome system with stimulation of protein degradation. Components of this mechanism are: 1) CKD increases circulating inflammatory cytokines stimulating NF-κB. 2) SIRP-α is increased by activation of NF-κB; and 3) SIRP-α interacts with the insulin receptor and IRS-1 to decrease their tyrosine phosphorylation, impairing intracellular insulin signaling (Figure 7). We speculate that the insulin resistance and muscle protein wasting occurring in other conditions associated with inflammation (e.g., diabetes, acidosis, excess angiotensin II, cancer cachexia and aging) could also result from this mechanism.
Figure 7. Proposed Scheme: CKD-induced stimulation of SIRP-α impairs insulin signaling and induces muscle wasting.
CKD-induced cytokines stimulate NF-κB activation, allowing for increased expression of SIRP-α proteins. Stimulation of SIRP-α triggers tyrosine de-phosphorylation of IRS-1 and insulin receptor, leading to a reduction in p-Akt and finally muscle protein loss.
CKD-induced expression of proinflammatory cytokines initiates this mechanism of insulin resistance. There are, however, other mechanisms by which cytokines interfere with insulin signaling. For example, excess Ang II leads to the expression of proinflammatory cytokine such as IL-6 in muscle (8). When IL-6 rises in conjunction with increased hepatic production of serum amyloid A, insulin resistance develops in relationship with suppression of IRS-1 in muscle. Alternatively, CKD induces inflammatory cytokines with activation of NF-κB which stimulates a 2- to 3-fold increase in the expression of myostatin leading to muscle protein wasting. In mice with CKD, we have shown that blocking myostatin suppresses NF-κB and improves insulin resistance as indicated by an increase in p-Akt (21). In preliminary experiments, we have found that myostatin inhibition can suppress SIRP-α expression (Thomas et al., in preparation) emphasizing how myostatin can change muscle metabolism. Another source of cytokines could be macrophages (CD68) in the damaged kidney or in inflamed tissues. Finally, inflammation has been linked to the development of insulin resistance in models of other catabolic conditions. For example, NF-κB is activated in tissues of obese and diabetic mouse models or in hepatoma cells treated with TNF-α (31;32). Indeed, the liver is a source of proinflammatory cytokines since Chen et al, found that giving growth hormone to endotoxin-treated mice increased hepatic production of cytokines (33). This is relevant because Cai et al. reported that NF-κB activation in a mice with muscle-specific, transgenic activation of IKKβ, there was severe muscle wasting (34). Thus, an increase in cytokines will stimulate NF-κB to cause muscle wasting. In muscles of CKD mice we have confirmed that inflammatory cytokines are upregulated and that NF-κB is activated (Supplemental Table 1; Figure 6A), supplying a new mechanism for the insulin resistance associated with NF-κB activation.
To determine if responses to cytokines we measured in mice with CKD constitute a cause and effect relationship, we exposed differentiated C2C12 myotubes to a mixture of cytokines present in mice or humans with CKD. There was activation of NF-κB (Figure 6B) and increased SIRP-α expression (Figure 3B) and these changes were blocked in C2C12 myotubes treated with a NF-κB inhibitor or with DNIKKβ which blocks NF-κB activation (Figure 6C, D). We conclude that the regulation of SIRP-α expression includes inflammation-induced activation of NF-κB (Figure 7).
We also evaluated whether activation of the SIRP-α blunts insulin signaling. First, we showed that insulin signaling was impaired because activation of SIRP-α in muscle decreased p-Akt (Figure 3B). Second, SIRP-α over expression in myotubes reduced p-Akt (Figure 4A). Conversely, silencing SIRP-α in myotubes improved insulin signaling as signified by increased p-Akt despite the presence of cytokines (Figure 5A).
Regarding SIRP-α, others report that the SIRP family of proteins influences growth factor-induced tyrosine phosphorylation in different cells: Kharitonenkov et al., concluded that SIRP-α exerts a negative regulatory effect on responses to growth factors or insulin in human epidermoid carcinoma cells, in mouse mammary tumor cells or in rat cells overexpressing the human insulin receptor (24;25). Maile et al. reported that a potential target of SIRP-α in smooth muscle cells is the IGF-1 receptor (35). Finally, Mitsuhashi et al. reported that SIRP-α expression is increased in denervated muscles of rats (36). Our results extend these reports because we determined that CKD not only increases expression of SIRP-α but also targets this protein to muscle membranes. This permits interactions between SIRP-α and components of insulin signaling (Figure 2D) or signaling by other growth factors that raise p-Akt. The interaction between SIRP-α and the insulin receptor or IRS-1 decreases tyrosine phosphorylation to suppress insulin signaling mechanisms. For example, Feinstein et al. reported there was a 65% reduction in insulin-induced tyrosine phosphorylation of the insulin receptor and IRS-1 in rat hepatoma cells treated with TNF-α (32).
We speculate that SIRP-α also could be the link by which other disorders cause insulin resistance. For example, we find that SIRP-α expression is stimulated in the tibialis muscles of db/db mice vs. db/m mice, another model of insulin resistance (unpublished data). Our results also emphasize that stimulation of SIRP-α has pathophysiological consequences: SIRP-α enhances protein degradation in muscle and when SIRP-α was silenced, cytokines no longer stimulated protein degradation in muscle cells (Figure 5E). In contrast, SIRP-α overexpression was shown to stimulate the ubiquitin-proteasome system to increase protein degradation in muscle cells (Figure 4C). In conclusion, our results suggest that targeting SIRP-α may prove to be a therapeutic modality, improving insulin signaling, and preventing the profound catabolic consequences of insulin resistance in CKD.
Materials and Methods
Reagents and Antibodies
Phosphatase inhibitor and protease inhibitor were obtained from Roche (Indianapolis, IN), protein A/G Plus beads from Santa Cruz Biotechology (Santa Cruz, CA), RNAeasy and Plasmid Maxi Kit were from Qiagen (Valencia, CA), the iScript cDNA Synthesis Kit was from Bio-Rad (Hercules, CA), Protein Block Serum Free and Antibody Diluent were from Dako (Glostrup, Denmark). Vectashield Mounting Media with DAPI was from Vectashield (Burlingame, CA), IL-6, IFN-γ, TNF-α, and LPS were from R&D Systems (Minneapolis, MN), QNZ, the NF-κB Inhibitor was from Enzo Life Sciences (Ann Arbor, MI) and the SIRP-α SiRNA and control SiRNA were from Santa Cruz Biotechnology. The Nucleofector kit and GFP plasmid were from Lonza (Allendale, NJ) and insulin was from Sigma Aldrich (St. Louis, MO). SIRP-α plasmid cDNA was from Open Biosystems (Lafayette, CO), C2C12 mouse myoblasts were from American Type Culture Collection (Manassas, VA) and DMEM and fetal bovine serum (FBS) were from Cellgro Mediatech (Manassas, VA). The antibodies against phospho-Akt (Ser473), total Akt and p-IκBα (Ser 32) were from Cell Signaling Technology (Beverly, MA), against SIRP-α was from Abcam (Cambridge, MA), against IKKβ, GAPDH, IRβ was from Santa Cruz Biotechnology, against phosphotyrosine (4G10) and IRS-1 was from Millipore (Temecula, CA) and the secondary antibody of anti-rabbit Alexa Fluor 488 was from Invitrogen (Eugene, OR).
Aortic blood obtained from anesthetized mice was used to measure BUN (5), serum creatinine using the QuantiChrom Creatinine Assay Kit (BioAssay Systems, Hayward, CA). Blood bicarbonate was estimated using an IRMA TruPoint blood analysis system with a CC Cartridge.
Western Blot and Immunoprecipitation
Gastrocnemius muscles were homogenized in RIPA buffer plus Phosphatase Inhibitor and Complete Mini Protease Inhibitor (1mg protein per 20μl RIPA) and cell lysates were evaluated by western blotting as described (8;21). Immunoprecipitation was performed by adding 2 μg of anti-IRS-1 or anti-insulin receptor or SIRP-α to 1 mg muscle lysates in PBS. Phosphatase and protease inhibitors were added and after shaking overnight at 4°C, 30 μL of Protein A/G Plus beads were added. The mixture was agitated in a rotational shaker for 2 h at 4°C and after centrifugation at 450 x g, the supernatant was removed. The beads were washed 5× with PBS and western blots were performed. The Odessey Infrared Imaging System (Li-Cor Lincoln, Nebraska) and enhanced chemiluminescence (ECL) were used to image results.
Real time-PCR
Gastrocnemius muscles were obtained from CKD and sham-operated, pair-fed control mice. RNA was obtained using RNAeasy; cDNAs were synthesized using iScript cDNA Synthesis Kit . RT-PCR was performed with CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The primer sequences for mouse Atrogin-1, MuRF-1 and GAPDH are reported (8). The mouse SIRP-α primer sequences were: forward 5′-CTCTGTGGACGCCTGTAA-3′, reverse 5′-GATGCTGCTGCTGTTGTT-3′. RT-PCR primers related to Supplemental Table 2 are in Supplemental Table 3.
Chronic kidney disease model
Animal experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Anesthetized C57/BL6 mice underwent subtotal nephrectomy in two stages as described (5;21). From anesthetized mice, tibialis anterior and gastrocnemius muscles were obtained.
Glucose and insulin tolerance tests
For glucose tolerance, mice were fasted 16 h before glucose in tail vein blood was measured using Truetrack Glucometer (Nipro Diagnostics, Fort Lauderdale, FL). Subsequently, 1.5 mg glucose/g mouse was injected intraperitoneally and blood glucose was measured at 30, 60, 90 and 120 minutes after the glucose injection. For insulin tolerance, mice were fasted for 4 h before blood glucose was measured. Subsequently, insulin (0.375 mU/g mouse) was injected intraperitoneally and blood glucose was measured 30 and 60 min later.
Immunohistochemical staining
Frozen serial transverse cryosections (8 μm) from the midbelly of TA muscles from control and CKD mice were mounted on glass slides (15). The slides were incubated with an anti-SIRP-α antibody (diluted to 1:100 with Dako Antibody Diluent) and then with a secondary antibody, anti-Rabbit Alexa Fluor 488 (diluted to 1:400 in PBS). Vectashield Mounting Media with DAPI was applied and images examined by Nikon 80i microscope (Melville, NY).
Cell culture
C2C12 cells (ATCC, Manassas, VA) were cultured and differentiated as described (37). Myotubes were treated with a cytokine mixture of IL-6 (2 ng/ml), TNF-α (2 ng/ml), IFN-γ (2 ng/ml), LPS (10 ng/ml). Net protein degradation in myotubes was measured as the release of free [L-14C]phenylalanine from cell proteins prelabled with L-[U14C]phenylalanine without the use of protein synthesis inhibitor (29).
NF-κB activity
C2C12 myoblasts were infected with an adenovirus expressing NF-κB-luciferase. After differentiation into myotubes, the media was replaced and cytokines were added. After harvesting, cellular luciferase activity was assayed according to Promega (Madison, WI). NF-κB activity in myotubes was inhibited using 500 nM QNZ added 2 h before adding the cytokine mixture. We infected C2C12 myoblasts with an adenovirus expressing DNIKKβ or GFP as described (38). Differentiated myotubes were placed in serum-free media before cytokines were added for 24 h.
Silencing SIRP-α and overexpression of SIRP-α
C2C12 myoblasts were electroporated with either SiRNAs or plasmid cDNAs using the Amaxa Nucleofector technology and protocol (Lonza, Allendale, NJ). Myoblasts were transfected with 2 μg of plasmid SIRP-α or plasmid encoding GFP then differentiated into myotube, myotubes were placed in serum-free media and treated with cytokines for 30 min. Alternatively, The myoblasts were transfected with 0.4 μg of SIRP-α siRNA or Control (scrambled) SiRNA. The transfected cells were allowed to differentiate into myotubes, and placed in serum free medium before being treated with cytokines for 6 or 24 h.
Statistics
Values are presented as means ±SEM, and results were analyzed using Student's t test when results from 2 experimental groups were compared or using ANOVA when data from 3 or more groups were studied. For ANOVA analyses, pairwise comparisons were made by the Student-Newman-Keuls test. Statistical significance was set at p<0.05.
Supplementary Material
Fold change of mRNA of CKD mice compared to control mice was evaluated by microarray analysis and confirmatory RT-PCR. (n=3).
Supplemental Table 2. RT-PCR Primers.
C2C12 myotubes were serum starved, and then treated with a mixture of IL-6 (2 ng/ml), TNF-α (2 ng/ml), and IFN-γ (2 ng/ml) for 24 hours and compared with control (CTL) myotubes without cytokine exposure. Representative immunoblots of SIRP-α and GAPDH (left panel) with quantitative band density relative to GAPDH is shown (right panel; *, p<0.05 vs. CTL; n=3 independent experiments).
Acknowledgments
This study was sponsored by American Diabetes Association (1-11-BS-194 to L.Z.), Satellite Health (to L. Z.), NIH T32 DK-07656, R37DK37175 (W.E.M.) and support from Dr. and Mrs. Harold Selzman. We would like to thank John Lin, Zhaoyong Hu, and Yanlan Dong for excellent technical assistance.
Footnotes
Disclosures: None.
Reference List
- 1.Fliser D, Pacini G, Engelleiter R, et al. Insulin resistance and hyperinsulinemia are already present in patients with incipient renal disease. Kidney Int. 1998;53:1343–1347. doi: 10.1046/j.1523-1755.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi S, Maesato K, Moriya H, et al. Insulin resistance in patients with chronic kidney disease. Am J Kid Dis. 2005;45:275–280. doi: 10.1053/j.ajkd.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA, Alvestrand A, Smith D, Hendler R. Insulin resistance in uremia. J Clin Invest. 1981;67:563–568. doi: 10.1172/JCI110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith D, DeFronzo RA. Insulin resistance in uremia mediated by postbinding defects. Kidney Int. 1982;22:54–62. doi: 10.1038/ki.1982.132. [DOI] [PubMed] [Google Scholar]
- 5.May RC, Kelly RA, Mitch WE. Mechanisms for defects in muscle protein metabolism in rats with chronic uremia: The influence of metabolic acidosis. J Clin Invest. 1987;79:1099–1103. doi: 10.1172/JCI112924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JL, Price SR, Zheng B, et al. Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atroply. J Am Soc Nephrol. 2006;17:1388–1394. doi: 10.1681/ASN.2004100842. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, Wang H, Lee IH, et al. Endogenous glucocorticoids and impaired insulin signaling are both required to stimulate muscle wasting under pathophysiological conditions in mice. J Clin Invest. 2009;119:7650–7659. doi: 10.1172/JCI38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Du J, Hu Z, et al. IL-6 and serum amyloid A synergy mediates angiotensin II-induced muscle wasting. J Am Soc Nephrol. 2009;20:604–612. doi: 10.1681/ASN.2008060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai D, Yuan M, Frantz DF, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman JE, Dohm GL, Elton CW, et al. Muscle insulin resistance in uremic humans: glucose transport, glucose transporters, and insulin receptors. Am J Physiol. 1991;261:E87–E94. doi: 10.1152/ajpendo.1991.261.1.E87. [DOI] [PubMed] [Google Scholar]
- 11.Cecchin F, Ittoop O, Sinha MK, Caro JF. Insulin resistance in uremia: Insulin receptor kinase activity in liver and muscle from chronic uremic rats. Am J Physiol. 1988;254:E394–E401. doi: 10.1152/ajpendo.1988.254.4.E394. [DOI] [PubMed] [Google Scholar]
- 12.Du J, Wang X, Meireles CL, et al. Activation of caspase 3 is an initial step triggering muscle proteolysis in catabolic conditions. J Clin Invest. 2004;113:115–123. doi: 10.1172/JCI200418330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey JL, Wang X, England BK, et al. The acidosis of chronic renal failure activates muscle proteolysis in rats by augmenting transcription of genes encoding proteins of the ATP-dependent, ubiquitin-proteasome pathway. J Clin Invest. 1996;97:1447–1453. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SW, Dai G, Hu Z, et al. Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol. 2004;15:1537–1545. doi: 10.1097/01.asn.0000127211.86206.e1. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Wang XH, Wang H, et al. Satellite cell dysfunction and impaired IGF-1 signaling contribute to muscle atrophy in chronic kidney disease. J Am Soc Nephrol. 2010;21:419–427. doi: 10.1681/ASN.2009060571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoelson S, Leo J, Goldfine A. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niwa T, Tsukushi S, Ise M, et al. Indoxyl sulfate and progression of renal failure-effects of a low-protein diet and oral sorbent on indoxyl sulfate production in uremic rats and undialyzed uremic patients. Min Elect Metab. 1997;23:179–184. [PubMed] [Google Scholar]
- 18.D'Apolito M, Du X, Zong H, et al. Urea-induced ROS generation causes insulin resistance in mice with chronic renal failure. J Clin Invest. 2010;120:203–213. doi: 10.1172/JCI37672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigalleau V, Blanchetier V, Combe C, et al. A low-protein diet improves insulin sensitivity of endogenous glucose production in predialytic uremic patients. Am J Clin Nutr. 1997;65:1512–1516. doi: 10.1093/ajcn/65.5.1512. [DOI] [PubMed] [Google Scholar]
- 20.Hotamisligil GS, Peraldi P, Budavari A, et al. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-a- and obesity-induced insulin resistance. Sci. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Rajan V, Lin E, et al. Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 2011;25:1653–1663. doi: 10.1096/fj.10-176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giorgino F, Pedrini MT, Matera L, Smith RJ. Specific increase in p85a expression in response to dexamethasone is associated with inhibition of insulin-like growth factor-1 stimulated phophatidylinositol 3-kinase activity in cultured muscle cells. J Biol Chem. 1997;272:7455–7463. doi: 10.1074/jbc.272.11.7455. [DOI] [PubMed] [Google Scholar]
- 23.Ding H, Gao XL, Hirschberg R, et al. Impaired actions of insulin-like growth factor-1 on protein synthesis and degradation in skeletal muscle of rats with chronic renal failure: Evidence for a postreceptor defect. J Clin Invest. 1996;97:1064–1075. doi: 10.1172/JCI118499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kharitonenkov A, Chen Z, Sures I, et al. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 25.Fujioka Y, Matozaki T, Noguchi T, et al. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandi A, Kitamura Y, Kahn CR, Accili D. Mouse models of insulin resistance. Physiol Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 27.Szeto CC, Kwan BC, Chow KM, et al. Endotoxemia is related to systemic inflammation and atherosclerosis in peritoneal dialysis patients. Clin J Am Soc Nephrol. 2008;3:431–436. doi: 10.2215/CJN.03600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIntyre CW, Harrison LE, Eldehni MT, et al. Circulating endotoxemia: a novel factor in systemic inflammation and cardiovascular disease in chronic kidney disease. Clin J Am Soc Nephrol. 2011;6:133–141. doi: 10.2215/CJN.04610510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du J, Mitch WE, Wang X, Price SR. Glucocorticoids induce proteasome C3 subunit expression in L6 muscle cells by opposing the suppression of its transcription by NF-κB. J Biol Chem. 2000;275:19661–19666. doi: 10.1074/jbc.M907258199. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 31.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Sci. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 32.Feinstein R, Kanety H, Papa MZ, et al. Tumor necrosis factor-a suppresses insulin-induced tyrosine phosphorylation of insulin receptor and its substrates. J Biol Chem. 1993;268:26055–26058. [PubMed] [Google Scholar]
- 33.Chen Y, Sun D, Krishnamurthy VM, Rabkin R. Endotoxin attenuates growth hormone-induced hepatic insulin-like growth factor I expression by inhibiting JAK2/STAT5 signal transduction and STAT5b DNA binding. Am J Physiol Endocrinol Metab. 2007;292:E1856–E1862. doi: 10.1152/ajpendo.00581.2006. [DOI] [PubMed] [Google Scholar]
- 34.Cai D, Frantz JD, Tawa NE, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Maile LA, CLEMMONS DR. Regulation of insulin-like growth factor I receptor dephosphorylation by SHPS-1 and the tyrosine phosphatase SHP-2. J Biol Chem. 2002;277:8955–8960. doi: 10.1074/jbc.M109258200. [DOI] [PubMed] [Google Scholar]
- 36.Mitsuhashi H, Yoshikawa A, Sasagawa N, et al. Denervation enhances the expression of SHPS-1 in rat skeletal muscle. J Biochem. 2005;137:495–502. doi: 10.1093/jb/mvi058. [DOI] [PubMed] [Google Scholar]
- 37.Tzanno-Martins C, Azevedo LS, Orii N, et al. The role of experimental chronic renal failure and aluminium intoxication in cellular immune response. Nephrol Dial Transplant. 1996;11:474–480. [PubMed] [Google Scholar]
- 38.Zhang L, Cheng J, Ma Y, et al. Dual pathways for nuclear factor kappaB activation by angiotensin II in vascular smooth muscle: phosphorylation of p65 by IkappaB kinase and ribosomal kinase. Circ Res. 2005;97:975–982. doi: 10.1161/01.RES.0000190589.52286.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fold change of mRNA of CKD mice compared to control mice was evaluated by microarray analysis and confirmatory RT-PCR. (n=3).
Supplemental Table 2. RT-PCR Primers.
C2C12 myotubes were serum starved, and then treated with a mixture of IL-6 (2 ng/ml), TNF-α (2 ng/ml), and IFN-γ (2 ng/ml) for 24 hours and compared with control (CTL) myotubes without cytokine exposure. Representative immunoblots of SIRP-α and GAPDH (left panel) with quantitative band density relative to GAPDH is shown (right panel; *, p<0.05 vs. CTL; n=3 independent experiments).