Summary
Circadian (ca. 24 hr) oscillations in expression of mammalian “clock genes” are found not only in the suprachiasmatic nucleus (SCN), the central circadian pacemaker, but also in peripheral tissues [1]. Under constant conditions in vitro, however, rhythms of peripheral tissue explants [2] or immortalized cells [3] damp partially or completely. It is unknown whether this reflects an inability of peripheral cells to sustain rhythms as SCN neurons can, or a loss of synchrony among cells. Using bioluminescence imaging of Rat-1 fibroblasts transfected with a Bmal1::luc plasmid, and primary fibroblasts dissociated from mPer2Luciferase-SV40 knockin mice, we monitored single cell circadian rhythms of clock gene expression for 1-2 weeks. We found that single fibroblasts can oscillate robustly and independently, with undiminished amplitude and diverse circadian periods. Cells were partially synchronized by medium changes at the start of an experiment, but due to different intrinsic periods their phases became randomly distributed after several days. Closely spaced cells in the same culture did not have similar phases, implying a lack of functional coupling among cells. Thus, like SCN neurons, single fibroblasts can function as independent circadian oscillators; however, lack of oscillator coupling in dissociated cell cultures leads to a loss of synchrony among individual cells and damping of the ensemble rhythm at the population level.
Results and Discussion
The suprachiasmatic nucleus (SCN) of the hypothalamus was long thought to be the exclusive driver of mammalian circadian rhythms. This conclusion was based on SCN lesion and transplant studies, as well as demonstrations that SCN tissue can generate rhythms of neuronal firing and other physiological outputs [4, 5]. When monitored at the single cell level on multielectrode arrays, dissociated SCN neurons exhibit independently phased firing rhythms that persist without damping [6, 7, 8]. Such studies left little doubt that SCN cells are competent, self-sustained circadian oscillators, or “clock cells”. In contrast, there was little evidence of physiological rhythms in isolated peripheral tissues of mammals, except for a study showing rhythmic melatonin secretion from cultured retina [9]. This stood in contrast to well-established autonomous rhythms of cultured pineal tissue in reptiles and birds [10, 11, 12].
When the first core genetic components of the mammalian circadian clock were identified, it was discovered that these “clock genes” are expressed rhythmically outside the SCN [13], and even outside the brain [1], in many peripheral tissues. Following the discovery of independent peripheral oscillators throughout Drosophila tissues using period::luciferase reporters [14] came the discovery that rhythmic clock gene expression can even be found in immortalized mammalian cell lines [3]. Further application of luciferase reporter genes has permitted gene expression rhythms to be followed longitudinally in explants of peripheral tissues from transgenic mPer1::luc rats [2], and in transfected fibroblasts [15, 16]. The peripheral cell or tissue rhythms, however, tend to damp out within a few cycles until re-started by serum shock or medium change, whereas rhythms of SCN explants are more robust, and can persist for more than 50 days [17]. This damping of peripheral circadian rhythms has led to the hypothesis that peripheral cells contain damped rather than self-sustained circadian oscillators [3, 16].
Several observations cast doubt on this hypothesis of damped peripheral circadian oscillators, however. First, some degree of damping is also observed in SCN explants from mPer1::luc rodents, despite firm evidence that SCN neurons are self-sustained oscillators. Second, with a more physiological “knockin” reporter, in which luciferase is fused to the endogenous mPER2 clock protein, circadian rhythmicity of peripheral tissue explants is still detectable after three weeks in isolation [18]. Thus, the difference in damping between SCN and other tissues appears to be quantitative rather than qualitative, and peripheral circadian clocks must have at least some capacity for self-sustained oscillation.
The reasons for the partial damping and the greater damping in peripheral tissues than in SCN remain obscure, however. One possibility is that only some cells damp, or that cells damp only partially, perhaps due to a defective molecular oscillator, or perhaps due to depletion of luciferase substrate or nutrients from the culture medium [18]. An alternative hypothesis, originally suggested by Balsalobre et al. [3], is that individual cellular oscillators, rather than damping out, merely drift out of phase, effectively canceling one another out, such that there is a decrease in amplitude of the ensemble rhythmic output from a culture. In this case, differences observed among SCN explants, peripheral tissue explants, and dissociated cells, might reflect differences in coupling among component cellular oscillators, each of which is autonomous and self-sustained. In order to test this cellular desynchrony hypothesis directly, we monitored circadian rhythms of clock gene expression with single cell resolution, using bioluminescence imaging of Rat-1 fibroblasts acutely transfected with an mBmal1::luc plasmid, and primary fibroblasts dissociated from mPer2Luciferase-SV40 (mPER2::LUC-SV40) knockin mice. We show that damping in Rat-1 and primary fibroblast cultures is explained by loss of synchrony among cells rather than damping of individual cell rhythms.
Individual Rat-1 Fibroblasts Are Persistent, Independent Circadian Oscillators
Because damped peripheral circadian oscillations were first observed in Rat-1 cells [3], and they continue to serve as a model system [15, 16], we first attempted single cell monitoring in these cells. Rat-1 cell cultures transfected with mBmal1::luc (n = 6) and left undisturbed in the luminometer expressed high levels of luminescence, with circadian rhythms that damped rapidly over several days, followed by a gradual rise to a peak level (>1000 counts/sec) at 2-3 wks post-transfection, and finally a gradual decline thereafter (Figure 1A). Medium changes were sufficient to re-initiate damped circadian rhythmicity at any point, even during the decline. In order to take into account possible effects of these long-term dynamics of expression, we scheduled single cell imaging experiments to begin when expression was rising at 5 days after transfection, and after expression had peaked at 24 days. Because we changed the medium before beginning single cell imaging, we expected to be able to observe the single cell correlates of both the long-term expression changes and the more rapid damping of circadian rhythms.
Figure 1.
Rat-1 Cell Rhythms
(A) Long-term luminometer recording of luminescence in a 35 mm culture dish of Rat-1 cells acutely transfected with the mBmal1::luc circadian reporter plasmid. Cells were transfected one day before the recording began, and treated with a serum shock (50% serum × 2 hrs) just prior to recording. Note the initial circadian oscillations which damp out quickly in the first few days, followed by a slow increase in luminescence to a broad peak at 2-3 weeks after transfection. Even when the luminescence begins to decline, changing medium restores circadian oscillations.
(B) Phases of individual Rat-1 cell luminescence rhythms were significantly clustered at the start of each experiment, but randomly distributed by the end. Each blue triangle indicates the phase of one cell, following the convention that 0˚ is the phase of the fitted peak of the luminescence rhythm. The radial line indicates the average start phase (127˚, or 15.7 circadian hrs before peak), and the arc indicates the 95% confidence interval.
(C) Plots of bioluminescence over the course of a 2 week experiment beginning 5 days after transfection. Medium was changed just prior to starting the experiment. Plots are of two representative Rat-1 cells (#22, 48), the sum of all 100 cells monitored in the experiment, and the entire microscope field. Note that the two individual cells begin with similar phases, peaking ~15 hrs after the start of the experiment, but that due to differences in period and phase instability the cells have drifted completely out of phase by the end of the experiment. The amplitudes of the individual cell rhythms are variable but do not damp significantly. Cell 22 actually increases its amplitude toward the end of the experiment. A movie of Cell 48 can be seen in Figure S1. As the cells become desynchronized, they begin to cancel out one another’s rhythms, and this is reflected in the rapidly damped oscillations seen in the arithmetic sum of luminescence from all 100 cells monitored, as well as in the luminescence from the entire microscope field. Finally, in accordance with the expected increase in plasmid expression at 5-10 days post-transfection (see A), there is an upward sloping baseline evident in the ensemble rhythms.
A total of 188 Rat-1 cells were monitored in two separate cultures, and all of these were rhythmic for at least 3 days (period 18-32 hrs, p<0.05). For 144 cells monitored and rhythmic over the entire experiment (Figure S1 for movie), the average period was 23.14 ± 1.98 hrs (± SD), but ranged widely between 19.1 and 30.3 hrs. Some cells could be followed for only a portion of the experiment because of cell movement, change in brightness, or occasional cell death. Other cells could not be fitted with a single sine wave for the entire experiment (9-15 days) because of phase or period instability. Therefore, start and end phases were estimated based on the first and last 3 days of the experiment, rather than on a single fitted curve. For 105 cells rhythmic during the first 3 days, start phases were significantly clustered (p<0.0001) at a mean vector of 125˚(15.7 circadian hrs before peak luminescence). On the other hand, for 111 cells rhythmic during the last 3 days, end phases were randomly distributed (p=0.9; Figure 1B). Thus, individual Rat-1 cells were all rhythmic (though somewhat unstable), and were partially synchronized by the medium change at the start of the experiment, but expressed diverse and variable circadian periods, such that their phases were randomly distributed by the end of the experiment.
Of 144 cells monitored and rhythmic for the entire experiment, there was no significant change in amplitude overall. This was quantified by the amplitude factor (AF), which specifies the factor by which fitted amplitude changes per day, and in this case was not significantly different from 1.0 (mean = 0.98, i.e. a decrease of 2% per day, SE = 0.01, p>0.05). However, amplitude changes were not consistent between experiments: in the experiment beginning 5 days post-transfection and lasting 15 days, overall field brightness increased by 44%, and 54 of 66 cells (82%) increased in amplitude (mean AF = 1.07, SE = 0.01, p<0.001); whereas, in a second experiment beginning 24 days after transfection and lasting 9 days, overall field brightness decreased by 31%, and 57 of 78 cells (73%) decreased in amplitude (mean AF = 0.93, SE = 0.01, p<0.001). These upward and downward trends of single cell amplitude observed at different times post-transfection are consistent with the broad peak of luminescence observed in the luminometer at 2-3 wks post-transfection, even in the absence of circadian oscillations at the culture level (Figure 1). They likely reflect the same slow dynamics of reporter plasmid copy number, unrelated to circadian clock function. Moreover, damping of the ensemble rhythm of the cultures is clearly present even soon after transfection, when most individual cells are increasing in amplitude (Figure 1C). Therefore, the rapid ensemble damping is much better explained by loss of synchrony among cells than by damping of individual cells.
Individual Primary Fibroblasts Are Persistent, Independent Circadian Oscillators
We next studied single cell circadian oscillations in a more physiological cell type and reporter system. To circumvent irregularities that might be associated with genetic modifications endemic to cell lines, we used primary fibroblasts. We also wanted to avoid the confounding influence of plasmid dynamics associated with acute transfection, and to use a reporter more faithful to endogenous clock gene dynamics. Thus, we turned to primary fibroblasts dissociated from mPER2::LUC-SV40 knockin mice. Cultures of these cells were also rhythmic when monitored in the luminometer, but with lower luminescence intensity of ~100-200 counts/sec. Unlike for Rat-1 cells, average daily luminescence intensity remained stable for months when cultures were healthy. Damped ensemble circadian rhythms were evident after each medium change, with amplitude factors ~0.8. Unlike tissue explants [18], the dissociated fibroblasts were usually completely arrhythmic by 3 weeks after a medium change. Single cell studies were performed on 3 cultures that had expressed such patterns in the luminometer for 2-4 months.
Primary fibroblasts from the mPER2::LUC-SV40 knockin mice were nearly 100-fold dimmer than the Rat-1 cells (mean 2.23 vs. 199 photons/min/cell), but all had prominent circadian rhythms (n = 178, p<0.01; Figure 2; see also Figure S2 for a movie), with periods of 25.65 ± 1.40 hrs (mean ± SD, range 22.4-29.7 hrs; Figure 3A). Despite the much lower levels of luminescence, the circadian rhythms of these cells were more robust than those of the Rat-1 cells, with very stable periods and phases, and all could be fitted nicely with a single sine wave curve. The robustness of the rhythms evident in the raw data (Figure 2B) is reflected in the Q(P) value of the chi-square periodogram, which averaged 296.2 ± 49.3 (mean ± SD, n = 100) for the fibroblasts vs. 156.2 ± 56.6 (mean ± SD, n = 154) for the Rat-1 cells (excluding cells with incomplete data or < 30 min resolution, t-test, p<0.0001). Phases at the start of the experiment were significantly clustered (n = 178, p<0.0001) at a mean vector of 295˚ (4.3 circadian hrs before peak). This is approximately in antiphase to the Rat-1 cells with the mBmal1::luc reporter, consistent with the known antiphase relationship between mBmal1 and mPer2 expression in fibroblasts [19]. On the other hand, end phases were randomly distributed (n = 178, p=0.2; Figure 3B). Thus, compared to the Rat-1 cells, individual primary fibroblasts were much more robustly rhythmic, were partially synchronized to approximately the opposite phase by the medium change at the start of an experiment, but like the Rat-1 cells expressed a diversity of circadian periods, such that their phases were also randomly distributed by the end of the experiment (Figures 3C, 4).
Figure 2.
Primary Fibroblast Rhythms
(A) Bioluminescence images of primary fibroblasts dissociated from tails of mPER2::LUC-SV40 knockin mice, showing circadian rhythms of luminescence. Because the cells emitted only a few photons per minute, detection of single cell luminescence required 30 min exposures and binning of pixels 8 × 8 to reduce read noise per pixel. Cells 1-4 peak near 0 and 24 h elapsed time, whereas cells 5-8 peak about 15 h later. See the movie in Figure S2 for a dynamic view of single cell luminescence rhythms in a larger field of view.
(B) Representative circadian bioluminescence rhythms from individual primary fibroblasts, over the course of an experiment lasting more than 8 days. Compared to the Rat-1 cells, the primary fibroblast rhythms are much more robust, with greater phase and period stability, and more regular amplitude. No damping of single cell rhythms was evident.
Figure 3.
Desynchrony of Primary Fibroblasts
(A) Individual primary fibroblasts expressed diverse circadian periods. Above is a histogram of circadian period values for luminescence rhythms of 178 primary fibroblasts cultured from mPER2::LUC-SV40 knockin mice. Periods averaged 25.65 hrs, but ranged widely from 22.4 to 29.7 hrs. Below is a raster plot showing two cells with clearly different periods. In the raster plot, time of day is plotted left to right, and successive days down the page, such that vertically adjacent points are 24 hrs apart. Each row is extended to 48 hrs, duplicating data in the next row, so that patterns crossing midnight can be appreciated. One cell with a period > 24 hrs is plotted in red, another cell with a period < 24 hrs is plotted in blue, and thick bars designate times when the luminescence for a cell was above the mean for each row. Due to different circadian periods, the cells’ phase relationship changes over time.
(B) Phases of fibroblast luminescence rhythms were significantly clustered at the start of each experiment, but randomly distributed by the end. Each blue triangle represents the phase of one cell, following the convention that 0˚ is the phase of the fitted peak of the luminescence rhythm. The radial line indicates the average start phase (295˚, or 4.3 circadian hrs before peak), and the arc indicates the 95% confidence interval.
(C) Damping population rhythms emerge from undamped single cell rhythms, as illustrated by plots of bioluminescence over the course of an 11 day experiment. Medium was changed just prior to starting the experiment. Plots are of two representative fibroblasts (#6,10), the sum of all 25 cells monitored, and the entire microscope field. Damped luminescence rhythmicity previously recorded from the same culture dish in the luminometer is also plotted for comparison. Note first that the two individual cells begin with similar phases, peaking ~4-6 hrs after the start of the experiment, but that due to differences in period, the cells have drifted completely out of phase by the end. As cells become desynchronized, they begin to cancel out one another’s rhythms, and this is reflected in the rapidly damped oscillations seen in the arithmetic sum of luminescence from all 25 cells monitored, as well as in the luminescence from the entire microscope field, and finally also in the luminometer recording from the entire dish.
Figure 4.
Primary Fibroblast Rhythms Sorted by Start Phase Luminescence rhythms of all 75 primary fibroblasts from one experiment are represented in this plot. Each horizontal raster line represents a single cell, with elapsed time plotted left to right. Luminescence intensity data from all cells were normalized for amplitude, and then color-coded: higher than average values are red, and lower than average values are green. The cells are sorted in order of start phase, so that the emergence of desynchrony can be more easily appreciated.
The lack of significant phase clustering by the end of the experiment suggests an absence of functional coupling among individual cellular oscillators. They seem to drift freely out of phase over the course of the experiment without influencing one another’s rhythms. As a more rigorous test of this hypothesis, we examined all possible pairs of fibroblasts within a culture (n = 6078), and asked whether cells that were closer together tended to have more similar phases or periods. We found no correlation between the spatial distance and the difference in start or end phase, even when limiting the analysis to cells within 500 µm of each other (n = 444). We did find a small positive correlation between distance and period difference (r = 0.09, r2 = 0.01, p<0.0001), which could reflect a small effect on circadian period of subtle but stable local variations in a cell’s microenvironment within the dish. Thus, individual fibroblasts are independent single cell circadian oscillators.
Primary fibroblasts generally maintained robust rhythms throughout the entire experiment (7-11 days), and overall there was a small but significant increase in fitted amplitude (mean AF = 1.03, SE = 0.009, p<0.001), indicating that cells tended to gain amplitude at a rate of ~3% per day. On the other hand, the difference between mean fitted amplitude at the start of the experiment (1.87 photons/min) and at the end of the experiment (2.08 photons/min) was modest, and did not reach statistical significance (repeated measures t-test, p=0.14). Thus, individual fibroblasts are independent, non-damping single cell circadian oscillators (Figure 5).
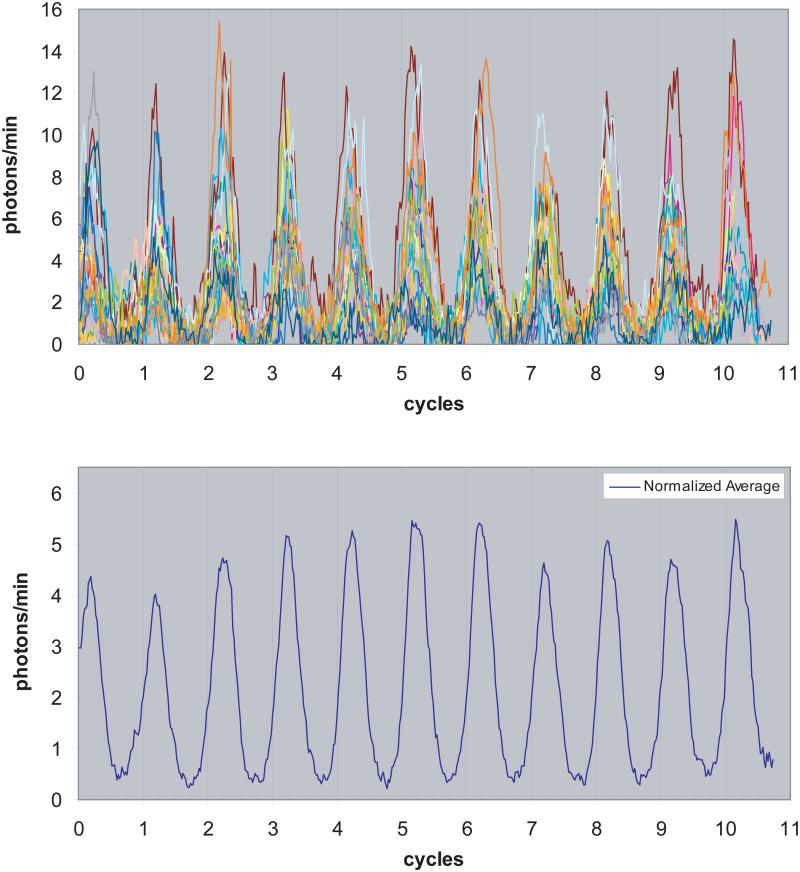
Figure 5.
Primary Fibroblast Rhythms Normalized by Period and Phase Luminescence rhythms of all 25 primary fibroblasts from one experiment were normalized by period and phase, by plotting luminescence as a function of circadian cycles for each cell, in order to reveal any overall trends in amplitude or waveform. Individual cell rhythms are plotted in the upper panel, a different color for each cell. The mean is plotted below, revealing a reasonably stable mean amplitude and waveform over the course of the 11 day experiment, and no damping.
Discussion
These experiments demonstrate that individual fibroblast cells are capable of functioning as independently phased circadian oscillators that are self-sustained for many days in vitro. In fact, their circadian function appears very similar to SCN neurons assayed on multielectrode arrays for rhythms of neuronal firing [6], except that they are evidently sensitive to a wider range of resetting stimuli, including serum shock, glucocorticoids, cold pulses, and even medium changes [19, 20, 21, 22]. This supports the notion that fibroblasts may serve as a valid model for core circadian clock function that is largely generalizable to SCN [19, 23].
Our data also resolve a long-standing question about the source of damping previously observed in circadian rhythms of peripheral cells and tissues. For both Rat-1 fibroblasts and dissociated primary fibroblasts, damping in vitro reflects gradual desynchrony of many independent cellular oscillators with diverse circadian periods, and restoration of amplitude by chemical or environmental stimuli reflects temporary partial re-synchronization of these component oscillators. Izumo et al. [16] recently questioned whether desynchrony could explain damping in Rat-1 cell cultures, because population rhythms broadened only slightly as they damped, but this was only based on a qualitative assessment. The extent to which desynchrony explains damping for other peripheral cell populations or tissues is an open question; we cannot rule out the possibility that other cell types may be damped oscillators or arrhythmic. Nevertheless, our finding in fibroblasts has the fundamental implication that damping or arrhythmicity at a tissue or culture level can no longer be taken at face value as evidence for defective circadian function at a cellular level, because we now know that single peripheral cells as well as SCN neurons can become desynchronized and oscillate independently. It is now clear that hypotheses involving circadian rhythm damping or arrhythmicity must be tested at single cell resolution.
It should be noted that damping of peripheral circadian oscillators is not a phenomenon limited to mammals. It has also been observed widely in Drosophila [14, 24], and recent studies similar to ours indicate that damping in cultures of cyanobacteria [25] or zebrafish cells [26] is also due to loss of synchrony among single cell oscillators.
A final implication of our results is that single cell fibroblast circadian oscillators in dissociated culture are not functionally coupled. Again, we cannot rule out the possibility of coupling in other peripheral cell or tissue types, or even among fibroblasts themselves in vivo. Indeed, it seems likely that the different rhythm damping properties exhibited by SCN and other tissues observed in previous studies may primarily reflect differences in the extent to which component cellular oscillators are functionally coupled in a particular experimental preparation. Thus, the persistent residual rhythmicity recently observed in peripheral tissue explants [18] probably reflects preservation of weak coupling among cellular circadian oscillators in those tissues, whereas the relatively less damped persistent oscillations of SCN explants [17] reflects stronger coupling among SCN neurons.
Supplementary Material
Figure S1: Time Lapse Bioluminescence Movie of Rat-1 Cells Luminescence images of Rat-1 fibroblasts are shown throughout the course of a two week experiment. Original 29.8 min exposures were collected at 30 min intervals, with no binning. Cosmic ray artifacts were removed, and a background image was subtracted (see Supplemental Experimental Procedures). Intensity has been coded in pseudocolor to represent a wider dynamic range. Elapsed days are shown by a digital counter. One pixel is 3.3 μm wide. Note the two bright cells, which show somewhat irregular, out of phase circadian oscillations throughout the experiment. The cell on the right is #48; its rhythm is plotted in Figure 1.
Figure S2: Time Lapse Bioluminescence Movie of Primary Fibroblasts Luminescence images of primary fibroblasts from mPER2-LUC-SV40 knockin mice are shown throughout the course of an 8.6 day experiment. Original 29.9 min exposures were collected at 30 min intervals, with 8 × 8 binning of pixels to reduce read noise per pixel. Cosmic ray artifacts were removed, and a background image was subtracted (see Supplemental Experimental Procedures). Elapsed days are shown by a digital counter. One pixel is 26.4 μm wide. Note the many regularly oscillating cells, some of which move considerable distances over the course of the experiment.
Acknowledgments
We thank Kathy Spencer for expert microscopy support, Camilo Orozco for computer programming, Trey Sato for supplying the mBmal1::luc plasmid, Rie Yasuda for culturing primary fibroblasts, Hien Tran for earlier work on transfection of Rat-1 cells, and Tom Schultz for thoughtful comments on the manuscript. Supported in part by K08 MH067657 (DKW) and MH51573 (SAK). JST is an Investigator in the Howard Hughes Medical Institute.
References
- 1.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 3.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi JS, Turek FW, Moore RY, editors. Handbook of Behavioral Neurobiology. Kluwer Academic/Plenum Publishing; New York: 2001. [Google Scholar]
- 5.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Gen. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 8.Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nature Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 9.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 10.Menaker M, Wisner S. Temperature-compensated circadian clock in the pineal of Anolis. Proc Natl Acad Sci USA. 1983;80:6119–6121. doi: 10.1073/pnas.80.19.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson LM, Takahashi JS. Circadian clock in cell culture: I. Oscillation of melatonin release from dissociated chick pineal cells in flow-through microcarrier culture. J Neurosci. 1988;8:12–21. doi: 10.1523/JNEUROSCI.08-01-00012.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett RK, Takahashi JS. Lability of circadian pacemaker amplitude in chick pineal cells: a temperature-dependent process. J Biol Rhythms. 1997;12:309–318. doi: 10.1177/074873049701200403. [DOI] [PubMed] [Google Scholar]
- 13.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:10031011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 14.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 15.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 16.Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci USA. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 20.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 21.Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J Biol Chem. 2002;277:44244–44251. doi: 10.1074/jbc.M206233200. [DOI] [PubMed] [Google Scholar]
- 22.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 23.Rosbash M. Why the rat-1 fibroblast should replace the SCN as the in vitro model of choice. Cell. 1998;93:917–919. doi: 10.1016/s0092-8674(00)81197-6. [DOI] [PubMed] [Google Scholar]
- 24.Hardin PE. Analysis of period mRNA cycling in Drosophila head and body tissues indicates that body oscillators behave differently from head oscillators. Mol Cell Biol. 1994;14:7211–7218. doi: 10.1128/mcb.14.11.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 26.Carr A-J, Foulkes NS, Whitmore D. Does light start or synchronise circadian clocks in culture? (#226) Soc Res Biol Rhythms Abstr. 2004;9:138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Time Lapse Bioluminescence Movie of Rat-1 Cells Luminescence images of Rat-1 fibroblasts are shown throughout the course of a two week experiment. Original 29.8 min exposures were collected at 30 min intervals, with no binning. Cosmic ray artifacts were removed, and a background image was subtracted (see Supplemental Experimental Procedures). Intensity has been coded in pseudocolor to represent a wider dynamic range. Elapsed days are shown by a digital counter. One pixel is 3.3 μm wide. Note the two bright cells, which show somewhat irregular, out of phase circadian oscillations throughout the experiment. The cell on the right is #48; its rhythm is plotted in Figure 1.
Figure S2: Time Lapse Bioluminescence Movie of Primary Fibroblasts Luminescence images of primary fibroblasts from mPER2-LUC-SV40 knockin mice are shown throughout the course of an 8.6 day experiment. Original 29.9 min exposures were collected at 30 min intervals, with 8 × 8 binning of pixels to reduce read noise per pixel. Cosmic ray artifacts were removed, and a background image was subtracted (see Supplemental Experimental Procedures). Elapsed days are shown by a digital counter. One pixel is 26.4 μm wide. Note the many regularly oscillating cells, some of which move considerable distances over the course of the experiment.