Abstract
Epidermal growth factor receptor (EGFR) gene mutation and copy number are useful predictive markers that guide the selection of non-small cell lung cancer (NSCLC) patients for EGFR-targeting therapy. This study aimed to investigate the correlation between EGFR gene mutation and copy number and clinicopathologic characteristics of Chinese patients with NSCLC. NSCLC specimens collected from 205 patients between November 2009 and January 2011 were selected to detect EGFR gene mutations with real-time polymerase chain reaction (RT-PCR) and to detect EGFR gene copy number with fluorescence in situ hybridization (FISH). EGFR mutations primarily occurred in females, non-smokers, and patients with adenocarinomas (all P < 0.001). Tissues from 128 (62%) patients were FISH-positive for EGFR, including 37 (18%) with gene amplification and 91 (44%) with high polysomy. EGFR gene mutation was correlated with FISH-positive status (R = 0.340, P < 0.001). Multivariate analysis showed that not smoking (OR = 5.910, 95% CI = 2.363–14.779, P < 0.001) and having adenocarcinoma (OR = 0.122, 95% CI = 0.026–0.581, P = 0.008) were favorable factors for EGFR gene mutation. These results show a high frequency of EGFR FISH positivity in NSCLC tissues from Chinese patients and a significant relevance between EGFR gene mutations and FISH-positive status. Among the FISH-positive samples, EGFR gene mutation occurred more frequently in samples with gene amplification compared to those with high polysomy, suggesting that EGFR mutation and gene amplification should be used as clinical decision parameters to predict response to EGFR-targeting therapy.
Keywords: Epidermal growth factor receptor, gene mutation, gene copy number, non-small cell lung cancer, correlation
Lung cancer is the leading cancer in men, comprising 17% of total new cancer cases and 23% of total cancer deaths[1]. The epidermal growth factor receptor (EGFR) family of proteins is well known to play an important role in lung carcinogenesis, tumor cell survival and proliferation[2]. Somatic gene mutations clustered within the tyrosine kinase domain of EGFR are common in non-small cell lung cancer (NSCLC) and occur most frequently in women, East Asians, non-smokers, and patients with adenocarcinomas[3]–[6]. Studies on agents targeting mutated EGFR sparked an interest in the predictive and prognostic significance of gene status[7]. The average response rate to therapy with anti-EGFR tyrosine kinase inhibitors (TKIs) was 75% for NSCLC patients with EGFR mutations[8]; however, a high response rate to anti-EGFR TKIs has also been observed in patients with increased EGFR gene copy number[9],[10]. Molecular analysis using fluorescence in situ hybridization (FISH) indicated that increased EGFR gene copy number with balanced polysomy occurs in approximately 10% to 40% of patients with NSCLC[11]. Some studies have shown that EGFR mutations are highly predictive of drug response, and others indicated that EGFR gene copy number may be equally or more predictive of improved survival than EGFR mutation status[12]. Further studies suggested that EGFR gene mutation and amplification can occur simultaneously, and both were proposed as potential biomarkers of anti-EGFR TKI responsiveness[13],[14].
Although the most useful biomarker for selecting candidates for anti-EGFR TKI therapy still remains controversia[15],[16], concurrent analyses of gene mutation and copy number have revealed the relevance and association of these factors with clinical outcome. This study aimed to analyze the correlation between EGFR gene mutations and gene copy number in Chinese patients with NSCLC and to further clarify the relationship between clinicopathologic features and gene mutations and copy number.
Materials and Methods
Tumor specimens
Tumor specimens were obtained from 205 consecutive patients who underwent surgery for pathologically proven NSCLC between November 2009 and January 2011 at Sun Yat-sen University Cancer Center. Detailed demographic and clinical information of these patients was available at the Sun Yat-sen University Cancer Center Surveillance System. No patient underwent either neoadjuvant chemoradiotherapy or target therapy. Tumors were staged according to the International Association of the Study for Lung Cancer (IASLC) TNM staging system[17]. The tumor blocks were fixed in 10% buffered formaldehyde and embedded in paraffin. The blocks were cut in 4-µm consecutive sections and stained with hematoxylin and eosin (HE). Slides rich in viable tumor cells were submitted for FISH analysis.
Real-time PCR analysis of EGFR mutations
We used an EGFR kit (GP Medical Technologies Ltd, Beijing, China) to detect a deletion in exon 19 (delE746-A750) and mutation in exon 21 (L858R) with real-time polymerase chain reaction (RT-PCR). RT-PCR was performed as follows: initial activation of DNA polymerase at 50°C for 2 min, denaturation at 95°C for 10 min, 40 cycles of amplification at 95°C for 15 s and at 62°C for 60 s. The cycle threshold (Ct) was used for results interpretation and was defined as the cycle at the highest peak of the second derivative curve, which represented the maximum point of the growth curve[18]. Positive results were defined as Ct ≤ 34 on the growth curve. The samples with positive results (34 < Ct ≤ 38) were subjected to repeated RT-PCR for result validation.
FISH analysis of EGFR gene copy number
FISH assays were performed using the EGFR FITC Red/CEP 7 Rhodamine Green probe (GP Medical Technologies Ltd, Beijing, China) according to the manufacturer's instructions. Tumor tissue sections were deparaffinized in 2× xylene washes at room temperature for 10 min, and dehydrated orderly in 100%, 85%, and 70% ethanol for 2 min each. After incubation in 30% saline sodium citrate (SSC) at 50°C for 20 to 30 min, sections were digested with proteinase K at 37°C and rinsed in 2× SSC for 30 min. The EGFR/CEP 7 probe set was applied to the selected area on each section. The sections were incubated at 90°C for 30 min for co-denaturation of chromosomal and probe DNAs, hybridized at 42°C for 16 h, and washed in SSC at room temperature thereafter. Chromatin was counterstained with DAPI (0.15 mg/mL in Vectashield Mounting Medium). The sections were observed under an epifluorescence microscope using single-band filters, and images of each section were merged by the Smart Capture software (Vysis, Downers Grove, IL, USA).
FISH analyses were defined according to the previously published criteria by Cappuzzo et al.[10] Six FISH strata were classified according to the frequency of tumor cells with specific number of copies of the EGFR gene and chromosome 7 centromere: 1) disomy (≤ 2 copies of EGFR per cell in > 90% of cells); 2) low trisomy (≤ 3 copies in 10%–40% of cells); 3) high trisomy (≤ 3 copies in > 40% of cells); 4) low polysomy (≥ 4 copies in 10%–40% of cells); 5) high polysomy (≥ 4 copies in > 40% of cells); 6) gene amplification (tight EGFR gene clusters and a ratio of EGFR gene to chromosome of ≥ 2, or ≥ 10 copies in ≥ 10% of cells). Gene amplification and high polysomy were defined as FISH-positive and all else were FISH-negative.
Statistical analyses
Chi-square (χ2) test or Fisher's exact test was performed to determine the associations between the presence of EGFR mutations or FISH phenotypes and patients' characteristics. Multivariate analyses (logistic regression models) were used to determine the influence factors for EGFR mutations. A P value < 0.05 was considered significant.
Results
Patient characteristics
Patients' clinical characteristics are presented in Table 1. Of the 205 patients, 138 were men and 67 were women, with a median age of 59 years (range, 29–80 years); 119 were smokers, and 86 were non-smokers. According to the WHO classification[19], 147 patients had adenocarcinoma (including bronchioloalveolar carcinoma), 52 had squamous cell carcinoma, 3 had adenosquamous carcinoma, 2 had sarcomatoid carcinoma, and 1 had large-cell carcinoma (lympho-epithelioma-like carcinoma); 85 presented with stage I disease (23 with IA, 62 with IB), 30 with stage II disease (5 with IIA, 25 with MB), 70 with stage III disease (51 with IIIA, 19 with NIB), and 20 with stage IV disease.
Table 1. Relationships between clinicopathologic characteristics and EGFR gene mutation and copy number in patients with non-small cell lung cancer (NSCLC).
| Variable | Total |
EGFR gene mutation |
FISH-positive phenotype |
||||
| Cases | % | P | Cases | % | P | ||
| Gender | 0.112 | ||||||
| Male | 138 | 26 | 19 | <0.001 | 81 | 59 | |
| Female | 67 | 40 | 60 | 47 | 70 | ||
| Age (years) | 0.624 | ||||||
| ≥ 59 | 111 | 33 | 35 | 0.412 | 71 | 64 | |
| <59 | 94 | 33 | 30 | 57 | 61 | ||
| Smoking status | 0.209 | ||||||
| Ever | 119 | 17 | 14 | <0.001 | 70 | 59 | |
| Never | 86 | 49 | 57 | 58 | 67 | ||
| Histological type | <0.001 | 0.095 | |||||
| ADC | 147 | 63 | 43 | 97 | 66 | ||
| Non-ADC | 58 | 3 | 5 | 31 | 54 | ||
| Differentiation | 0.063 | 0.080 | |||||
| Grade 1 | 17 | 5 | 29 | 9 | 53 | ||
| Grade 2 | 116 | 46 | 40 | 80 | 69 | ||
| Grade 3 | 63 | 13 | 21 | 36 | 57 | ||
| Unknown | 9 | 2 | 22 | 3 | 33 | ||
| TNM stage | 0.038 | 0.127 | |||||
| IA | 23 | 11 | 48 | 15 | 65 | ||
| IB | 62 | 13 | 21 | 31 | 50 | ||
| IIA | 5 | 0 | 0 | 2 | 40 | ||
| IIB | 25 | 6 | 24 | 16 | 64 | ||
| IIIA | 51 | 20 | 39 | 38 | 75 | ||
| IIIB | 19 | 6 | 32 | 11 | 58 | ||
| IV | 20 | 10 | 50 | 15 | 75 | ||
EGFR, epidermal growth factor receptor; FISH, fluorescence in situ hybridization; ADC, adenocarcinoma.
EGFR gene mutations
In the 205 specimens of NSCLC, we detected EGFR mutations in 66 (32%) samples, including exon 19 deletion mutations in 34 (17%) and exon 21 point mutations in 32 (15%) samples (Figure 1). Of the 66 patients with mutations, 40 were women and 26 were men; 17 were smokers and 49 were non-smokers; 63 had adenocarcinomas and 3 had non-adenocarcinomas. There were significantly higher mutation rates in women, non-smokers, and patients with adenocarcinomas (all P < 0.001); no relationships were observed between EGFR mutation rates and age (P = 0.412) and histological differentiation (P = 0.063) (Table 1).
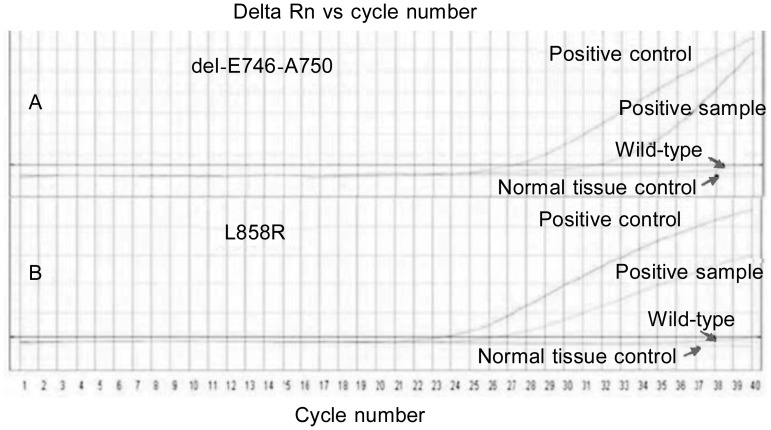
Figure 1. Representative real-time polymerase chain reaction (RT-PCR) curves for mutations in EGFR exons 19 and 21.
The positive control curve shows the mutation. Two ascending curves denoted “positive sample” represent exon 19 delE746-A750 mutation (A) and exon 21 L858R mutation (B). Smooth curves represent wild-type EGFR.
EGFR gene copy number
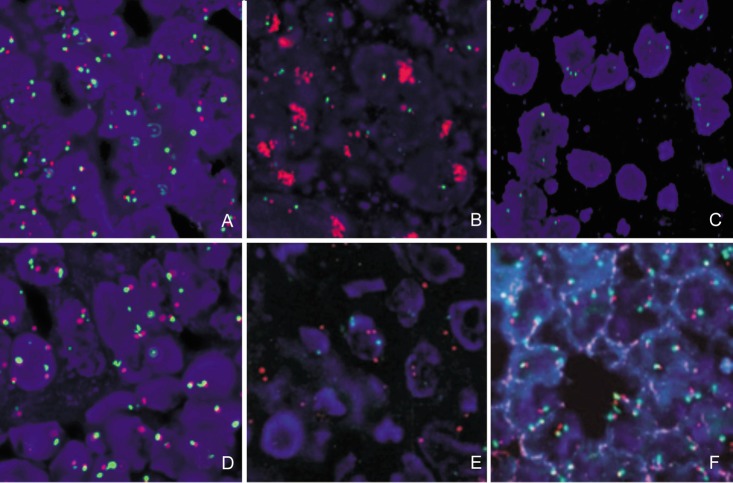
FISH analysis for EGFR gene copy number was performed on 205 specimens. FISH was positive in 128 (62%) of these 205 specimens, including 91 (44%) cases with high polysomy (Figure 2A) and 37 (18%) cases with amplification (Figure 2B). We also detected the disomy (Figure 2C) in 36 cases, low polysomy (Figure 2D) in 35 cases, low trisomy (Figure 2E) in 4 cases, and high trisomy (Figure 2F) in 2 cases. No relationship was found between EGFR gene copy number and patients' characteristics (Table 1). In the 2 cases of sarcomatoid carcinoma, one had amplification and the other had polysomy. In the 3 cases of adenosquamous carcinoma, 2 were detected with amplification and 1 with disomy.
Figure 2. EGFR gene copy number analyzed with fluorescence in situ hybridization (FISH).
Chromosome 7 centromere was labeled with FITC (green) and EGFR gene was labeled with rhodamine (red). A, EGFR high polysomy; B, EGFR amplification; C, EGFR disomy; D, EGFR low polysomy; E, EGFR low trisomy; F, EGFR high trisomy.
Relationship between EGFR gene mutations and gene copy number
Comparison between gene mutations and copy number showed that 57 (86%) of the 66 samples with EGFR mutation were also FISH-positive (24 with amplification and 33 with high polysomy), suggesting a relationship between EGFR mutation and the FISH-positive phenotype (P < 0.001) (Table 2). In the NSCLC patients with EGFR mutation, stratified analysis showed that the FISH-positive phenotype occurred more frequently than the FISH-negative phenotype (Table 3). In these two groups, we found that the FISH-positive phenotype primarily occurred in cases with EGFR mutation and adenocarcinoma (P < 0.001) or tumor differentiation of grade >1 (P < 0.001) (Table 3).
Table 2. Relationship between EGFR mutation and the FISH-positive phenotype in patients with NSCLC.
| EGFR mutation | Total | FISH phenotype [cases (%)] |
R | P | |
| Positive | Negative | ||||
| Presence | 66 | 57 (86) | 9 (14) | 0.34 | <0.001 |
| Absence | 139 | 71 (51) | 68 (49) | ||
Abbreviations as in Table 1.
Table 3. Analysis of factors that influence EGFR FISH phenotype in patients with NSCLC.
| Variable | EGFR mutation | FISH phenotype [cases (%)] |
P | |
| Negative | Positive | |||
| Gender | ||||
| Male | - | 54 (48) | 58 (52) | 0.001 |
| + | 3 (11) | 23 (89) | ||
| Female | - | 14 (52) | 13 (48) | 0.001 |
| + | 6 (15) | 34 (85) | ||
| Age (years) | ||||
| ≥ 59 | - | 34 (44) | 44 (56) | 0.011 |
| + | 6 (18) | 27 (82) | ||
| <59 | - | 34 (56) | 27 (44) | <0.001 |
| + | 3 (9) | 30 (91) | ||
| Smoking status | ||||
| Ever | - | 46 (45) | 56 (55) | 0.036 |
| + | 3 (18) | 14 (82) | ||
| Never | - | 22 (60) | 15 (40) | <0.001 |
| + | 6 (12) | 43 (88) | ||
| Histological type | ||||
| ADC | - | 41 (49) | 43 (51) | <0.001 |
| + | 9 (14) | 54 (86) | ||
| Non-ADC | - | 27 (49) | 28 (51) | 0.241 |
| + | 0 (0) | 3 (100) | ||
| Differentiation | ||||
| Grade 1 | - | 7 (58) | 5 (42) | 0.294 |
| + | 1 (20) | 4 (80) | ||
| Grade >1 | - | 61 (48) | 66 (52) | <0.001 |
| + | 8 (13) | 53 (87) | ||
| Stage | ||||
| Early (IA+IB) | - | 35 (57) | 26 (43) | 0.001 |
| + | 4 (17) | 20 (83) | ||
| Medium (IIA+IIB+IIIA) | 11 A) | 22 (40) | 33 (60) | 0.010 |
| + | 3 (11) | 23 (89) | ||
| Advanced (IIIB+IV) | 11 (48) | 12 (52) | 0.037 | |
| + | 2 (12) | 14 (88) | ||
Abbreviations as in Table 1.
Further analysis of the FISH-positive patterns revealed that amplification was more highly associated with EGFR mutation than high polysomy. In contrast, FISH-negativity was more associated with wild-type EGFR (P = 0.003) (Table 4). In patients with EGFR gene mutations, stratified analyses for influencing factors on FISH-positive phenotype showed that the frequency of amplification was related to EGFR mutation in patients who were female (P = 0.004) or younger than 59 (P = 0.001), who had never smoked (P = 0.023), or who had adenocarcinoma (P = 0.019), grade > 1 (P = 0.017), or early stage disease (P = 0.029) (Table 5).
Table 4. Relationship between EGFR mutations and FISH phenotype in patients with NSCLC.
| FISH phenotype | Total |
EGFR mutation [cases (%)] |
R | P | |
| Presence | Absence | ||||
| High polysomy | 91 | 33 (36) | 58 (64) | 0.375 | <0.001 |
| Amplification | 37 | 24 (65) | 13 (35) | ||
| FISH negativity | 77 | 9 (12) | 68 (88) | ||
Abbreviations as in Table 1.
Table 5. Stratified analysis of factors influencing FISH-positive phenotype in patients with NSCLC.
| Variable | EGFR mutation | FISH-positive phenotype [cases (%)] |
P | |
| High polysomy | Amplification | |||
| Gender | ||||
| Male | - | 45 (77) | 13 (59) | 0.127 |
| + | 14 (23) | 9 (41) | ||
| Female | - | 13 (41) | 0 (0) | 0.004 |
| + | 19 (59) | 15 (100) | ||
| Age (years) | ||||
| ≥ 59 | - | 34 (64) | 10 (56) | 0.516 |
| + | 19 (36) | 8 (44) | ||
| <59 | - | 24 (63) | 3 (16) | 0.001 |
| + | 14 (37) | 16 (84) | ||
| Smoking status | ||||
| Ever | - | 44 (86) | 12 (63) | 0.045 |
| + | 7 (14) | 7 (37) | ||
| Never | - | 14 (35) | 1 (6) | 0.023 |
| + | 26 (65) | 17 (94) | ||
| Histological type | ||||
| ADC | - | 35 (52) | 8 (27) | 0.019 |
| + | 32 (48) | 22 (73) | ||
| Non-ADC | - | 23 (96) | 5 (71) | 0.120 |
| + | 1 (4) | 2 (29) | ||
| Differentiation | ||||
| Grade 1 | - | 5 (83) | 0 (0) | 0.048 |
| + | 1 (17) | 3 (100) | ||
| Grade >1 | - | 53 (62) | 13 (38) | 0.017 |
| + | 32 (38) | 21 (62) | ||
| Stage | ||||
| Early (IA+IB) | - | 24 (65) | 2 (22) | 0.029 |
| + | 13 (35) | 7 (78) | ||
| Medium (IIA+IIB+IIIA) | - | 25 (64) | 8 (47) | 0.233 |
| + | 14 (36) | 9 (53) | ||
| Advanced (IIIB+IV) | - | 9 (60) | 3 (27) | 0.098 |
| + | 6 (40) | 8 (73) | ||
Abbreviations as in Table 1.
Multivariate analyses showed that never smoking (OR = 5.910, 95% CI = 2.36–14.78, P < 0.001) and adenocarcinoma (OR = 0.122, 95% CI = 0.026–0.581, P = 0.008) were related to EGFR mutations.
Discussion
Anti-EGFR TKIs are well known and are used for monotherapy in NSCLC. EGFR mutations were reported as predictive factors associated with anti-EGFR TKI sensitivity and response to EGFR-targeting therapies[6],[9],[20]–[22]. Many studies reported an association between EGFR mutations and some clinical features in NSCLC. The rates of EGFR mutation in those studies were 44%–55% in patients with adenocarcinoma, 51%–68% in non-smokers, 42%–62% in females, and 30%–50% in Asian ethnicity, but in contrast, the mutation rates were 10% in smokers, 14% in males, and 8% in adenocarcinoma patients from Western countries[20],[23]–[25]. In our current study of 205 specimens from Chinese patients with NSCLC, exon 19 deletion mutation and exon 21 point mutation were detected in 66 (32.2%) specimens by RT-PCR and occurred more frequently in adenocarcimoma (43%), females (60%), and non-smokers (57%), reflecting findings similar to those in the literature.
Although EGFR mutation was suggested to be the best predictor for TKI sensitivity, it should not be the only patient selection criterion for EGFR-targeting therapy; indeed, in studies with anti-EGFR TKIs, the survival benefit conferred by using EGFR mutation as a predictor is not only confined to individuals with tumor shrinkage, but also includes those with tumor stability or even disease progression[16]. Large randomized studies showed that EGFR gene copy number, as determined by FISH analysis, is probably an alternative for NSCLC patient selection and may show a survival benefit by predicting sensitivity to TKIs. In previous studies, high gene copy number or gene amplification tested by FISH was identified in 22%–45% of NSCLC patients[10],[26],[27]. Either EGFR FISH or mutation analyses could identify NSCLC with better outcome and response to TKIs[9],[10],[26],[28]. In our study, we identified 128 (62%) FISH-positive cases from 205 specimens, including 37 cases with amplification and 91 cases with high polysomy, which was a higher frequency than that seen in other studies. However, as seen in other studies, EGFR gene copy number was not correlated with age, gender, smoking status, stage, histological type, and differentiation[28].
Different trials have demonstrated a significant association between the EGFR FISH-positive phenotype and EGFR mutations[10],[26],[28],[29]. Our data have also demonstrated a positive correlation between the FISH-positive phenotype and gene mutations (R = 0.34, P < 0.001). Further analysis showed that histological type and grade could influence the FISH patterns when EGFR mutation status is taken into consideration.
Some authors consider high polysomy as significant gene alteration in carcinogenesis, though the biological mechanism was not clear. Moreover, EGFR amplification was closely associated with EGFR mutation[9],[22],[30],[31]. Our study also demonstrated the relationship between EGFR mutation and amplification, though high polysomy tended to relate with wild-type EGFR. Stratified analyses demonstrated that amplification was associated with EGFR mutations in patients who are female or young, who have never smoked, or who have adenocarcinoma, grade >1 (non-well differentiation), or early stage disease. Some articles inferred that EGFR mutant alleles were amplified selectively, and EGFR amplification happened during invasive growth of lung adenocarcinoma with EGFR mutations. Moreover, EGFR copy number gain was a late event compared to gene mutation in NSCLC pathogenesis, implying that EGFR mutation resulted in a high EGFR copy number[26],[32]–[34]. However, other authors thought that EGFR mutation and EGFR copy number gain are two independent events in NSCLC pathogenesis[35]. In our study, except for a number of observations already published in previous studies, we also found that early stage is associated with amplification in patients with EGFR mutations, which suggests that amplification may occur earlier in carcinogenesis than EGFR mutation.
Specific characteristics such as female gender, nonsmoking status, and adenocarcinoma histology are well known to relate with EGFR mutation in East Asians with NSCLC[6],[20]. However, multivariate analyses in our study only showed that non-smoking status and adenocarcinoma histology were related to EGFR gene mutation in Chinese patients with NSCLC. Thus, gender may not be a candidate selective factor for EGFR-targeting therapy in Chinese patients, but we need more evidence to prove that in further studies.
Conclusions
EGFR mutations are more common in Chinese NSCLC patients with adenocarcinoma histology and non-smoking status. Chinese NSCLCs are frequently EGFR FISH-positive. Among the FISH-positive patterns, gene amplification is related with EGFR gene mutation. Clinicopathologic characteristics including female gender, young age, non-smoking status, adenocarcinoma histology, high grade, and early stage are related to Chinese NSCLC patients with EGFR mutations and EGFR gene amplification simultaneously. Therefore, EGFR mutation and gene amplification should be used as clinical decision parameters to predict response to anti-EGFR TKI therapy.
Acknowledgments
This research was supported by grants from the Ministry of Science and Technology of China (No. 2006AA02A401).
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Dahabreh IJ, Linardou H, Siannis F, et al. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2010;16:291–303. doi: 10.1158/1078-0432.CCR-09-1660. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (the IDEAL 1 Trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 5.Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Jiang XL, Zhang CC, et al. Correlation of genes associated with drug response to prognosis of large cell lung carcinoma. Chin J Cancer. 2011;30:497–504. doi: 10.5732/cjc.010.10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 9.Tsao MS, Sakurada A, Cutz JC, et al. Erlotinib in lung cancer-molecular and clinical predictors of outcome. N Engl J Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 10.Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- 11.Dacic S, Flanagan M, Cieply K, et al. Significance of EGFR protein expression and gene amplification in non-small cell lung carcinoma. Am J Clin Pathol. 2006;125:860–865. doi: 10.1309/H5UW-6CPC-WWC9-2241. [DOI] [PubMed] [Google Scholar]
- 12.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 13.Chang JW, Liu HP, Hsieh MH, et al. Increased epidermal growth factor receptor (EGFR) gene copy number is strongly associated with EGFR mutations and adenocarcinoma in non-small cell lung cancers: a chromogenic in situ hybridization study of 182 patients. Lung Cancer. 2008;61:328–339. doi: 10.1016/j.lungcan.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Helfrich BA, Raben D, Varella-Garcia M, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- 15.Dziadziuszko R, Hirsch FR, Varella-Garcia M, et al. Selecting lung cancer patients for treatment with epidermal growth factor receptor tyrosine kinase inhibitors by immunohistochemistry and fluorescence in situ hybridization—why, when, and how? Clin Cancer Res. 2006;12:4409s–4415s. doi: 10.1158/1078-0432.CCR-06-0087. [DOI] [PubMed] [Google Scholar]
- 16.Cappuzzo F. EGFR fish versus mutation: different tests, different end-points. Lung Cancer. 2008;60:160–165. doi: 10.1016/j.lungcan.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 18.Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12:3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- 19.Morgensztern D, Govindan R. Baltimore, MD: Lippincott William & Wilkins; 2008. Lung cancer: a clinical guide. [Google Scholar]
- 20.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 21.Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 23.Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64:8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinico-pathologic features in non-small cell lung cancers. Clin Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- 26.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 27.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the ideal/intact gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 28.Liang Z, Zhang J, Zeng X, et al. Relationship between EGFR expression, copy number and mutation in lung adenocarcinomas. BMC Cancer. 2010;10:376. doi: 10.1186/1471-2407-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Li Y, Chen G, et al. Detection and its clinical significance of EGFR gene mutation and gene amplification in 187 patients with non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2009;12:1219–1228. doi: 10.3779/j.issn.1009-3419.2009.12.01. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 30.Cappuzzo F, Ligorio C, Janne PA, et al. Prospective study of gefitinib in epidermal growth factor receptor fluorescence in situ hybridization-positive/phospho-akt-positive or never smoker patients with advanced non-small-cell lung cancer: the oncobell trial. J Clin Oncol. 2007;25:2248–2255. doi: 10.1200/JCO.2006.09.4300. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol. 2005;23:6838–6845. doi: 10.1200/JCO.2005.01.2823. [DOI] [PubMed] [Google Scholar]
- 32.Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yatabe Y, Takahashi T, Mitsudomi T. Epidermal growth factor receptor gene amplification is acquired in association with tumor progression of EGFR-mutated lung cancer. Cancer Res. 2008;68:2106–2111. doi: 10.1158/0008-5472.CAN-07-5211. [DOI] [PubMed] [Google Scholar]
- 34.Soh J, Toyooka S, Ichihara S, et al. Sequential molecular changes during multistage pathogenesis of small peripheral adenocarcinomas of the lung. J Thorac Oncol. 2008;3:340–347. doi: 10.1097/JTO.0b013e318168d20a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casorzo L, Corigliano M, Ferrero P, et al. Evaluation of 7q31 region improves the accuracy of EGFR fish assay in non small cell lung cancer. Diagn Pathol. 2009;4:36. doi: 10.1186/1746-1596-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]