Abstract
The undifferentiated form of nasopharyngeal carcinoma (NPC) is the most common malignant head and neck cancer in South China, especially in Cantonese populations. However, few NPC cell lines have been established from the patients in this region. In this study, we established a new NPC cell line, termed SUNE2, from a Cantonese patient with undifferentiated NPC. This cell line had extremely low concentrations of Epstein-Barr virus (EBV) DNA in long-term culture and expressed low levels of latent membrane protein 1 (LMP1), latent membrane protein 2A (LMP2A), BamH1-A right frame 1 (BARF1), EBV-encoded RNA-1 (EBER1), and EBV-encoded RNA-2 (EBER2) in early passages. SUNE2 cells also showed much stronger transforming ability than 5-8F cells in colony formation assays and anchorage-independent growth assays in soft agar, and they only need 2 weeks to form tumors in nude mice. In summary, the SUNE2 cell line is a new in vitro model that can be used for further research on the mechanisms underlying the occurrence and development of NPC.
Keywords: Nasopharyngeal carcinoma, Epstein-Barr virus, SUNE2 cell line
Nasopharyngeal carcinoma (NPC) is one of the most common malignant cancers in China, especially in Guangdong Province[1]. Epstein-Barr virus (EBV) is a human gamma herpes virus that can exist in humans for a long time without producing any symptoms[2]. EBV infection is closely related to the high incidence of undifferentiated NPC, particularly in the central region of Guangdong Province in South China[1],[3]. Cantonese people account for a significant portion of the population in areas where EBV-associated NPC is prevalent. However, few NPC cell lines are available from Cantonese NPC patients. NPC has been classified into three major histological subtypes depending on the degree of differentiation. NPC cases in China and Southeast Asia are mostly type III, which is EBV-associated, undifferentiated non-keratinizing carcinoma[4]. Nearly all undifferentiated NPCs present with EBV DNA and gene products in tumor cells[5]. The expression of EBV latent genes EBV nuclear antigen 1 (EBNA1), latent membrane protein 1 (LMP1), latent membrane protein 2A (LMP2A), and BamH1-A right frame 1 (BARF1), as well as the EBV-encoded RNA (EBERs) and Bam A rightward transcripts (BARTs) displays a restricted pattern (latency II) in NPC[6],[7]. Southern blot hybridization results confirm that the resident viral genomes from NPC tissues are of monoclonal origin, indicating that EBV infection takes place before a single malignant cell proliferates to drive tumor development[8]. EBV is considered to play an important role in the development of NPC. However, the relationship between EBV and NPC has remained unclear. Although more than 20 NPC cell lines have been reported in recent years[9], C666 is the only cell line that does not lose the EBV genome upon cell culture passaging[10]. The limited representative EBV infection model has hampered research on the relationship between EBV and NPC. Therefore, the novel NPC cell line SUNE2, which is derived from a Cantonese patient, will be very useful for NPC study.
Materials and Methods
Cell culture and establishment of the novel NPC cell line SUNE2
One mouse embryonic fibroblast cell line (NIH3T3), three poorly differentiated NPC cell lines (CNE-2, 5-8F, and C666), and two EBV-positive B-cell lines (B95-8 and Raji) were maintained in our laboratory. NIH3T3 was maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Grand Island, NY). CNE2, 5-8F, C666, B95-8, and Raji were maintained in RPMI-1640 (Invitrogen, Grand Island, NY) with 10% FBS at 37°C in a humidified 5% CO2 incubator.
The SUNE2 cell line was derived from a NPC primary culture. The biopsy was performed on a 38-year-old female Cantonese patient diagnosed as having an undifferentiated squamous cell carcinoma at stage III (T3N1M0) according to the Union for International Cancer Control (UICC) staging system. Informed consent has been signed by this patient to allow using her tissue to establish the cell line. The titer of serum IgA antibody against EBV viral capsid antigen (VCA-IgA) was 1:80, but EBV early antigen (EA-IgA) was undetectable. The copy number of circulating cell-free EBV DNA was 3.16 × 103 copies/mL. After being removed from the patient, the biopsy specimen was immediately immersed in serum-free medium and then washed with PBS containing penicillin (100 units/mL) and streptomycin (100 units/mL) 3 times before culture. The specimen was then cut into pieces with scissors and plated in dishes with 1 mL RPMI-1640 containing 10% FBS and incubated at 37°C in an atmosphere of 5% CO2. When the pieces of tissue were attached, the residual medium was discarded completely and replaced with fresh keratinocyte/serum-free (KSF) medium (Invitrogen, Grand Island, NY). The culture was then propagated and passaged in KSF medium or RPMI-1640. The XUNE2 cell line was derived from the xenograft at passage 3 through primary culture, and was cultured in RPMI-1640 supplemented with 5% FBS.
Tumorigenesis in nude mice
Cells at passage 6 were mixed with 25% extracellular matrix before being injected into 4–6 weeks old BALB/c athymic nude mice (nu/nu) obtained from the Animal Center of Southern Medical University, Guangzhou, China. All mice were housed and maintained under specific pathogen-free conditions and used in accordance with institutional guidelines and protocols approved by the Use Committee for Animal Care. After 2 weeks, tumors larger than 1 cm were removed from nude mice and immersed in different solutions for protein, RNA, or DNA preparation. In addition, paraffin-embedded tumors were prepared, and tumor sections were stained with hematoxylin & eosin (HE) according to standard protocols. Then, the sections were observed under microscopy.
In situ hybridization and immunohistochemical analysis of EBERs
Approximately 1 × 105 cells (SUNE2 and C666) were pelleted at 800 ×g for 5 min and washed with phosphate-buffered saline (PBS). The cell pellet was fixed in 4% polyoxymethylene in PBS for 10 min at room temperature, and dehydrated through a series of increasing ethanol concentrations, ending with 100% ethanol for 5 min. EBERs in cultured cells were analyzed using Epstein-Barr Virus Probe ISH Kit and detected with the BCIP/NBT Alkaline Phosphatase Substrate Detection System (NCL-EBV-K, Novocastra, Newcastle upon Tyne, UK) according to the manufacturer's protocols. EBERs in paraffin sections of primary tumor specimens and xenografts were analyzed using another Epstein-Barr Virus Probe ISH Kit which was better for tumor specimens and detected with the HRP/DAB Detection System (ISH-5021, PanPath, Amsterdam, Netherlands). Immunohistochemical (IHC) staining was performed using Zymed Histostain™-Plus Kits (Zymed, South San Francisco, CA, USA) according to the manufacturer's protocols.
Western blotting and immunostaining analysis
Raji, B95-8, C666, and SUNE2 cells were harvested and lysed in lysis buffer and were heated for 10 min at 98°C. Protein concentration was detected using the BCA Protein Assay Kit (Pierce Chemical Co., Rockland, IL). Equal amounts of proteins were separated on SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes. After blocking, membranes were incubated with anti-EBNA1 (a gift from Jaap Middeldorp) or anti-LMP1 (Dako, Carpinteria, CA) at a 1:1000 dilution overnight at 4°C. Anti-α-tubulin mouse monoclonal antibody (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used to confirm equal loading. After incubation with secondary antibody, resultant signals were detected using enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's protocols. For immunostaining, cells were fixed and stained using anti-keratin (Zhongshan Golden Bridge Biotechnology Co. Ltd., No. ZM-0069, Beijing, China), anti-LMP2A (Proteintech Group, Wuhan, China), or anti-BZLF1 (1:2000) (Dako, Glostrup, Denmark) at room temperature followed by incubation with Alexa Fluor546-conjugated secondary antibody (Invitrogen, Carlsbad, CA, USA) at a dilution of 1:500 for 1 h at room temperature. Nuclei were counterstained with DAPI and slides were examined using an Olympus confocal imaging system.
Colony formation assay
Cells were seeded in triplicate at 200 cells/well in 6-well plates, and then cultured in RPMI-1640 for 7 days. After most of the colonies had expanded to more than 100 cells, they were washed three times with PBS, fixed in methanol for 10 min, dyed with crystal violet for 15 min at room temperature, and then washed out the dye with pure water. The plates were photographed and the colonies were compared and statistically analyzed using the t-test. Three independent experiments were carried out to ensure reproducibility.
Anchorage-independent growth assay
Six-well plates were covered with a layer of 0.6% agar in RPMI-1640 medium supplemented with 20% FBS. Cells were prepared in 0.3% agar and seeded in triplicate at 5 × 103 cells/mL(1 mL/well). The plates were incubated at 37°C in a humidified atmosphere of 5% CO2 for 4 weeks. Resultant colonies were photographed at an original magnification of 200× under phase contrast microscopy. Each experiment was repeated at least three times.
Real-time quantitative PCR analysis
Total RNA from different passages of SUNE2 cells was extracted using TRIzol reagent. After reverse transcription of the total RNA, the resultant cDNA was used as a template for detecting different EBV gene transcripts (Table 1). Real-time quantitative PCR and data collection were performed with an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA). The housekeeping gene GAPDH was used as an internal control to normalize the expression levels of different genes. The primers used for the amplification of the indicated genes are listed in Table 1.
Table 1. Primers used in real-time quantitative polymerase chain reaction (PCR).
| Gene | Primer sequences |
| GAPDH | Upstream: 5′-CTCCTCCTGTTCGACAGTCAGC-3′ |
| Downstream: 5′-CCCAATACGACCAAATCCGTT-3′ | |
| BamHI-W | Upstream: 5′-GGGTGCAGTAACAGGTAATC-3′ |
| Downstream: 5′-ATTCGCCTCTAAAGTTTTGA-3′ | |
| EBER1 | Upstream: 5′-GAGGTTTTGCTAGGGAGGAGAC-3′ |
| Downstream: 5′-GAAGACGGCAGAAAGCAGAGT-3′ | |
| EBER2 | Upstream: 5′-AACGCTCAGTGCGGTGCTA-3′ |
| Downstream: 5′-GCCGAATACCCTTCTCCCA-3′ | |
| EBNA1 | Upstream: 5′-GTAGGGGATGCCGATTATTTTG-3′ |
| Downstream: 5′-CTCCTTGACCACGATGCTTTC-3′ | |
| EBNA2 | Upstream: 5′-CATCTGCTATGCGAATGCTT-3′ |
| Downstream: 5′-ATGTGGCTGGACCAACCTG-3′ | |
| LMP1 | Upstream: 5′-GTATTGGCACAAGATGGAAAGC-3′ |
| Downstream: 5′-CAACTACCAGGCAGATGAGGC-3′ | |
| LMP2A | Upstream: 5′-ACGATGGCGGAAACAACTC-3′ |
| Downstream: 5′-GGGTCCTCATAAGGCGGTG-3′ | |
| BARF1 | Upstream: 5′-GCCTCTAACGCTGTCTGTCCA-3′ |
| Downstream: 5′-GCTCCCATCCTTTTCCTTCATA-3′ | |
| BZLF1 | Upstream: 5′-CCCAGTCTCCGACATAACCC-3′ |
| Downstream: 5′-CAGGCTGTGGAACACCAATG-3′ | |
| BRLF1 | Upstream: 5′-CGAGGACGGGATAGGTGAAC-3′ |
| Downstream: 5′-CGGCAAGCAGGTAGTGGAAC-3′ |
Results
Establishment of a new NPC cell line SUNE2
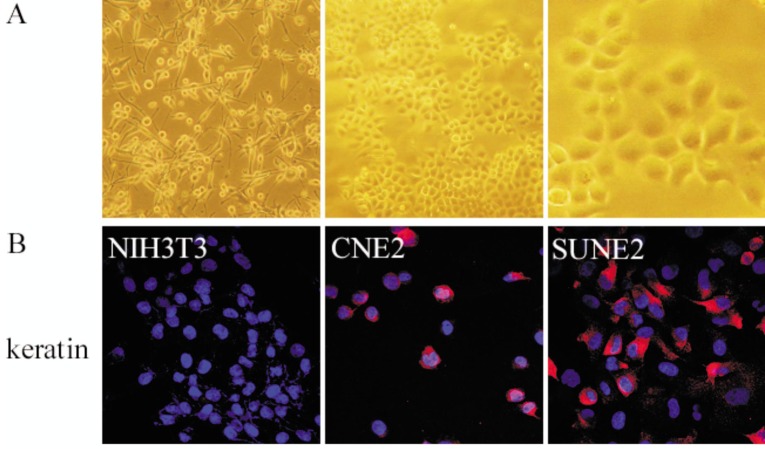
A new cell line named SUNE2 was established from tissue collected during a biopsy procedure from a Cantonese patient diagnosed with NPC. SUNE2 cells had been passaged more than 55 times in vitro. Surprisingly, nearly every cell showed certain feelers that likely underlie the propensity for these cells to form tumors, probably causing the strong transforming ability or tumor formation (Figure 1A, left panel). When KSF medium was changed to RPMI-1640 with 5% FBS, SUNE2 cells showed a phenotype similar to that of differentiated cells, became large and squamous, and nearly all cells firmly stick to the plate (Figure 1A, middle and right panels). Positive keratin immunoreactivity in SUNE2 cells suggested that SUNE2 was of epithelial origin (Figure 1B, right panel). NIH3T3 mouse embryo fibroblast cells served as negative controls (Figure 1B, left panel) and CNE2 cells served as positive controls for Keratin staining (Figure 1B, middle panel).
Figure 1. The morphology and keratin staining of the human nasopharyngeal carcinoma (NPC) cell line SUNE2.
A, SUNE2 cells grown in keratinocyte/serum-free (KSF) medium (×100, left panel) show feelers; SUNE2 cells grown in RPMI-1640 medium (×100, middle panel; ×400, right panel) firmly stick to the plate. B, SUNE2 cells are immunostained red in cytoplasm with anti-keratin, indicating their epithelial origin. NIH3T3 cells were used as a negative control, and CNE2 cells were used as a positive control.
SUNE2 cells are highly tumorigenic
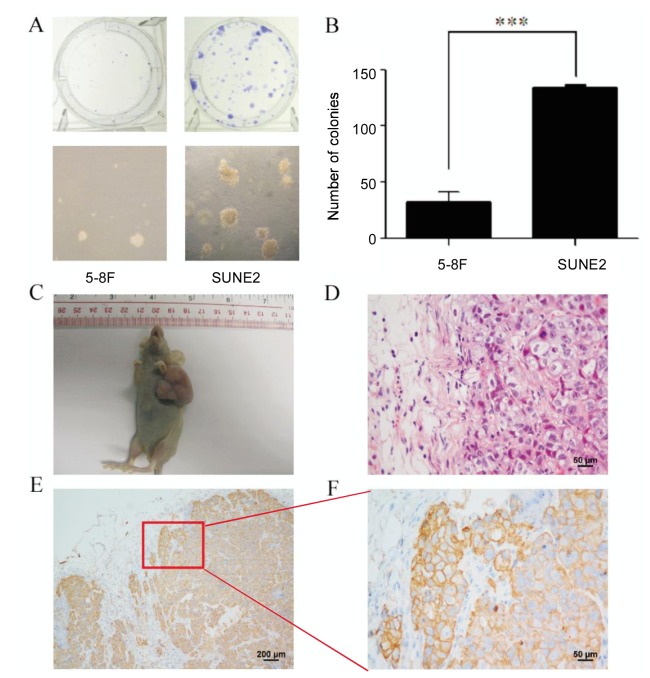
The proliferation of SUNE2 cells was significantly faster than other NPC cell lines, resulting in daily passaging of SUNE2 cells. To further determine the transforming ability of SUNE2 cells in vivo, we conducted a colony formation assay and an anchorage-independent growth assay. For these experiments, we used 5-8F cells as positive controls because of their high tumor formation ability. Colony formation assays showed that a single SUNE2 cell required one week to form a colony of more than 200 cells, whereas 5-8F cells needed a longer time to form a colony of similar size (Figure 2A, upper panel). Moreover, SUNE2 cells formed colonies that were much larger than 5-8F cells in anchorage-independent growth assays (Figure 2A, lower panel). The number of colonies of 5-8F cells was significantly lower than that of SUNE2 cells (Figure 2B). SUNE2 cells at passage 6 were transplanted into BALB/c mice, and xenografts at passage 3 were divided into three pieces. One xenograft piece was seeded into a culture dish to establish the primary culture cell line, named XUNE2; one was used to continue generating transplants; the final piece was fixed for further studies. To date, we have passaged the xenograft 13 times in BALB/c mice. Tumors in SUNE2 xenograft models grew signficantly larger and faster than controls in less than 2 weeks (Figure 2C). Paraffin-embedded sections of the xenograft were stained with HE (Figure 2D) and anti-keratin antibody (Figures 2E and 2F). We observed positive keratin staining, obvious tumor nests, oval nuclei, and indistinct cell borders, suggesting that SUNE2 cells were derived from undifferentiated epithelial cells and belonged to the non-keratinizing squamous cell carcinoma class of NPC (Figures 2D-F).
Figure 2. SUNE2 cells have a strong transforming ability.
A, colony formation assay (upper panel) and anchorage-independent growth of 5-8F and SUNE2 cells in soft agar (lower panel). SUNE2 cells form bigger and more colonies compared with 5-8F cells. B, quantitative analysis of colony numbers of 5-8F cells and SUNE2 cells with three independent assays. ***, P < 0.001. C, significant tumor formation in a nude mouse after injection with SUNE2 cells at passage 6. D, a paraffin section of a xenograft formed by SUNE2 cells was stained with HE (×400), showing obvious tumor nests, oval nuclei, and indistinct cell borders. E, a paraffin section was stained with monoclonal anti-keratin antibody (Zymed), and only the epithelial cells were stained brown in cytoplasm (×100). F, a representative field is shown by keratin staining of the xenograft (×400).
Existence of EBV in SUNE2 cells
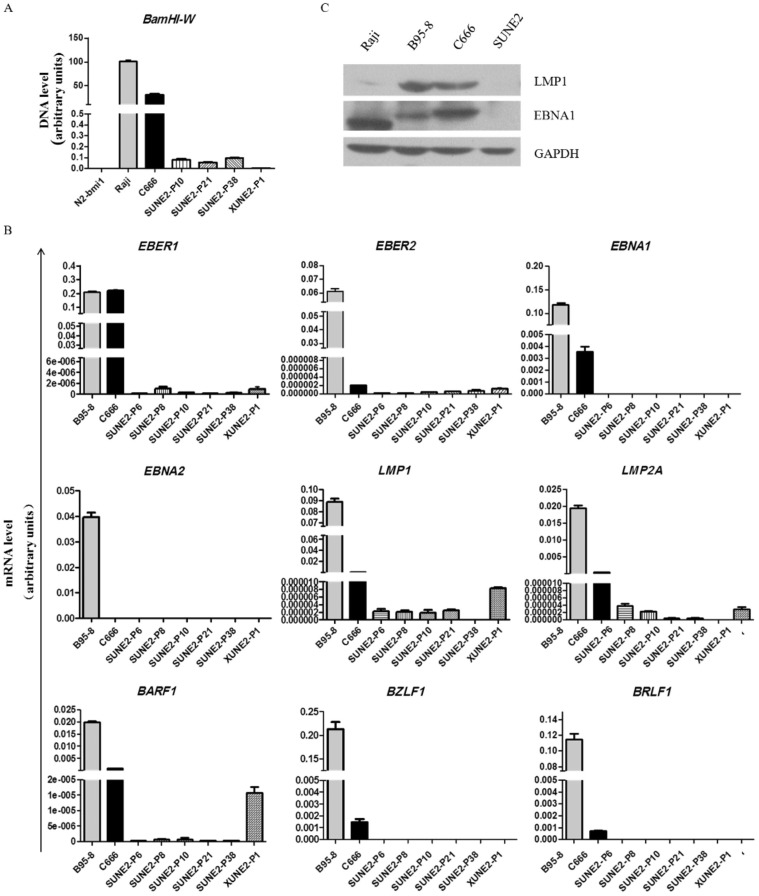
We detected the levels of the EBV DNA BamHI-W fragment and the EBERs in SUNE2 cells at different passages and in XUNE2-P1 (xenograft at passage 1) using real-time quantitative PCR. The results showed that a small amount of BamHI-W consistently existed in SUNE2 cells (Figure 3A) and revealed extremely low expression of EBER1 and EBER2 in continuous culture (Figure 3B, upper panel). Using EBV-positive B cell line B95-8 and EBV-positive NPC cell line C666 as positive controls, we further examined the expression of EBV latent genes and lytic genes in SUNE2 cells at passages 8, 10, 21, and 38 and in xenograft at passage 1 with real-time quantitative PCR. Expression of neither EBNA1 nor EBNA2 was detected (Figure 3B, upper and middle panels). Type ll-related genes like LMP1, LMP2A, and BARF1 were detectable but their expression levels were far lower in SUNE2 cells than in C666 or B95-8 cells (Figure 3B, middle and lower panels). The expression of LMP2A decreased gradually with the increase in passage number and became almost undetectable at passage 10. LMP1 expression did not significantly change until passage 21 and then disappeared at passage 38. Extremely trace amounts of BARF1 were detected at passage 38. High levels of LMP1, LMP2A, and BARF1 expression were observed in xenograft at passage 1 (Figure 3B, middle and lower panels). But we found no expression of LMP1 and EBNA1 in SUNE2 cells (Figure 3C). The expression of BZLF1 and BRLF1, two typical lytic genes, was negative in our samples (Figure 3B, lower panel).
Figure 3. Expression of EBV-related genes and proteins in SUNE2 cells.
A-B, real-time quantitative PCR of BamHI-W (A) and EBER1, EBER2, EBNA1, EBNA2, LMP1, LMP2A, BARF1, BZLF1, and BRLF1 (B) in SUNE2 at different passages (P8, P10, P21, P38) and SUNE2 xenograft P1 (XUNE2-P1). B95-8 and C666 cells were used as positive controls. BamHI-W was detected with DNA template and the other genes were detected with cDNA templates by real-time quantitative PCR, and the housekeeping gene GAPDH was used as an internal control. C, Western blots showing EBNA1 and LMP1 expression in Raji, B95-8, C666, and SUNE2 cells. GAPDH was used as a loading control. Lysates from Raji, B95-8, and C666 cells were used as positive controls.
SUNE2 cells express EBERs but have no EBV-related protein expression
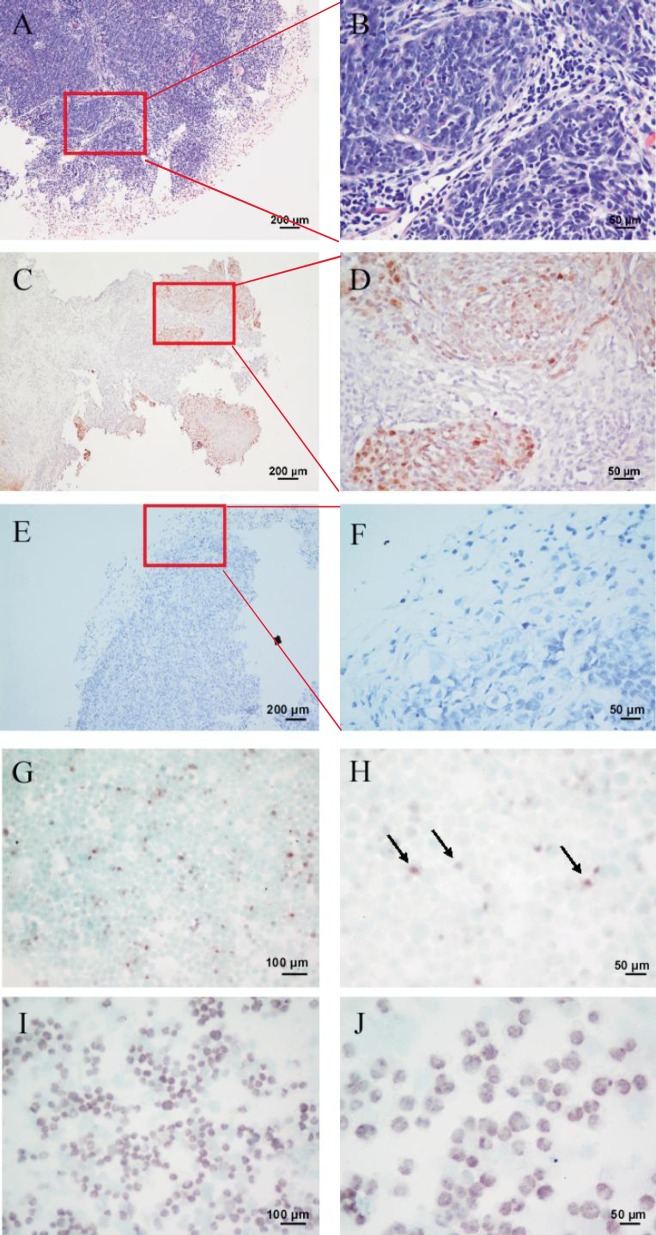
To further investigate the existence of EBV in xenografts and SUNE2 cells, we detected EBERs using in situ hybridization in paraffin-embedded sections and in SUNE2 cells. The C666 cell line was used as a positive control. HE staining (Figures 4A and 4B) and in situ hybridization (Figures 4C and 4D) of the original NPC sample showed that EBER signals were strong in tumor nests, negative in paraffin-embedded sections of the xenografts (Figures 4E and 4F), and weak in SUNE2 cells (Figures 4G and 4H, arrows) in contrast to strong EBER signals in C666 cells (Figures 4I and 4J). This result was consistent with real-time quantitative PCR data. Although very low expression of LMP1 was detected at the transcript level, we failed to detect LMP1 protein expression in SUNE2 cell lysates. Additionally, LMP2A and BZLF1 were analyzed by immunostaining, but the expression of both was negative (data not shown).
Figure 4. Detection of EBERs in the primary tumor specimen, xenograft model, and SUNE2 cells.
A, a paraffin-embedded section of the biopsy specimen shows tumor nests (HE ×100). B, a representative field shows obvious tumor nests, oval nuclei, and indistinct cell borders (HE ×400). C, a paraffin-embedded section of the biopsy specimen is EBER-positive (ISH ×100). D, a representative field of Panel C shows high EBER expression (ISH ×400). E, EBER in situ hybridization in a paraffin-embedded section of the xenograft was analyzed using Epstein-Barr Virus Probe ISH Kit (ISH ×100). F, a representative field of panel E shows negative EBERs expression (ISH ×400). EBER signal detection in both the biopsy specimen and xenograft was performed with the HRP/ DAB coloration kit (dark brown) (C–F). G, EBER in situ hybridization in SUNE2 cells (ISH ×200). H, a representative field shows high EBERs expression (ISH ×400). I, EBER in situ hybridization in C666 cells, which were used as a positive control (×200). J, a representative field shows EBERs expression (ISH ×400). EBER signal in both the SUNE2 and C666 cell lines was detected with the BCIP/NBT Alkaline Phosphatase Substrate Kit (dark violet-blue) (G–I).
Discussion
EBV is known to play an important role in the pathogenesis of undifferentiated NPC. NPC cases in China are primarily undifferentiated, and this type of NPC is common in a specific geographic district[11]. We successfully established a new NPC cell line SUNE2 from a primary culture of undifferentiated epithelial cells collected from a Cantonese patient with the non-keratinizing squamous cell carcinoma type of NPC. Positive keratin immunostaining of the paraffin-embedded sections and cell climbing slides provided further evidence that the SUNE2 cell line was epithelial in origin. SUNE2 cells consistently maintain a small amount of the EBV DNA Bam Hl-W fragment and extremely low expression levels of EBER1 and EBER2 in long-term culture (at passage 38), indicating that SUNE2 cells retain EBV and EBV-related genes. The Hone-1[12],[13], CG-1[14], and C666[15] cell lines have been considered authentic, EBV-positive, human NPC cell lines. However, Hone-1 and CG-1 cells were initially EBV-positive but lost their EBV genome after long-term culture[16]. Similarly, CNE2, CNE3, HNE1, and SUNE1 cells were also derived from EBV-positive and poorly differentiated NPC primary cultures, but all of these cells also lost EBV after long-term in vitro culture[9],[17]. Only the C666 cell line has consistently harbored EBV.
The results of colony formation assays and anchorage-independent growth assays, as well as the obvious tumor growth in a xenograft model suggested that the proliferation rate and tumor formation ability of SUNE2 cells was significantly high. In situ hybridization for EBER detection did not show any signal in xenografts but showed weakly positive signals in SUNE2 cells. It is possible that SUNE2 cells may not require particularly high levels of EBV to retain their malignant phenotype when cultured in vitro. The results of real-time quantitative PCR showed that SUNE2 cells retain an extremely low level of EBV genome. These results may be caused by the low specificity of in situ hybridization or the lack of expression in the xenograft model. Latent and lytic gene transcript expression was detected with real-time quantitative PCR at different passages. The expression of EBV genes in SUNE2 cells seems to follow a latency type II pattern, with detectable expression of LMP1, LMP2A, and BARF1. However, no expression of EBNA1 and EBNA2 was detected, even at early passages. The expression of EBER1 and latent genes in XUNE2-P1 was higher than that in SUNE2 cells, likely because the in vivo microenvironment provides more suitable conditions for promoting retention of the EBV genome. Some reports have shown that virus particles were not detectable using electron microscopy in fresh NPC biopsies[18]–[20]. Nevertheless, infectious virus can be released from NPC cells in the supernatant after passage in a nude mouse[21], suggesting that the increased expression of EBV genes in XUNE2-P1 is probably resulted from the enrichment of EBV in vivo. However, the levels of the latent protein expression were too low to detect by either Western blotting or immunostaining in our cell and xenograft models.
In summary, we have established a new NPC cell line SUNE2 from a Cantonese patient with undifferentiated NPC. These cells still retain an extremely low level of EBV DNA at passage 38, and the expression of EBV genes followed the latency type II pattern, with low levels of LMP1, LMP2A, BARF1, EBER1, and EBER2 in early passages. Taken together, these data suggest that the SUNE2 cell line will be a new in vitro model with which to further study the mechanisms of NPC occurrence and development.
Acknowledgments
We thank Jaap Middeldorp (Amsterdam Free University) for a kind gift of anti-EBNA1 (OTX-1). These studies were supported by grants from National Natural Science Foundation of China (No. 81025014, 91019015).
References
- 1.Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30(2):114–119. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deyrup AT. Epstein-Barr virus-associated epithelial and mesenchymal neoplasms. Hum Pathol. 2008;39(4):473–483. doi: 10.1016/j.humpath.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JI. Epstein-Barr virus infection. N Engl J Med. 2000;343(7):481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 4.Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12(6):431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 5.Stevens SJ, Verkuijlen SA, Hariwiyanto B, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int J Cancer. 2006;119(3):608–614. doi: 10.1002/ijc.21914. [DOI] [PubMed] [Google Scholar]
- 6.Bell AI, Groves K, Kelly GL, et al. Analysis of Epstein-Barr virus latent gene expression in endemic burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J Gen Virol. 2006;87(Pt 10):2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
- 7.Brooks L, Yao QY, Rickinson AB, et al. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: Coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66(5):2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 9.Gullo C, Low WK, Teoh G. Association of Epstein-Barr virus with nasopharyngeal carcinoma and current status of development of cancer-derived cell lines. Ann Acad Med Singapore. 2008;37(9):769–777. [PubMed] [Google Scholar]
- 10.Cheung ST, Huang DP, Hui AB, et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer. 1999;83(1):121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 11.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5(5):423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 12.Yao KT, Zhang HY, Zhu HC, et al. Establishment and characterization of two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus and derived from nasopharyngeal carcinomas. Int J Cancer. 1990;45(1):83–89. doi: 10.1002/ijc.2910450116. [DOI] [PubMed] [Google Scholar]
- 13.Glaser R, Zhang HY, Yao KT, et al. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc Natl Acad Sci U S A. 1989;86(23):9524–9528. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang YS, Lin SY, Lee PF, et al. Establishment and characterization of a tumor cell line from human nasopharyngeal carcinoma tissue. Cancer Res. 1989;49(23):6752–6757. [PubMed] [Google Scholar]
- 15.Hui AB, Cheung ST, Fong Y, et al. Characterization of a new EBV-associated nasopharyngeal carcinoma cell line. Cancer Genet Cytogenet. 1998;101(2):83–88. doi: 10.1016/s0165-4608(97)00231-8. [DOI] [PubMed] [Google Scholar]
- 16.Teng ZP, Ooka T, Huang DP, et al. Detection of Epstein-Barr virus DNA in well and poorly differentiated nasopharyngeal carcinoma cell lines. Virus Genes. 1996;13(1):53–60. doi: 10.1007/BF00576978. [DOI] [PubMed] [Google Scholar]
- 17.Weber AL, al-Arayedh S, Rashid A. Nasopharynx: clinical, pathologic, and radiologic assessment. Neuroimaging Clin N Am. 2003;13(3):465–483. doi: 10.1016/s1052-5149(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 18.Kieff E. Epstein-Barr virus—increasing evidence of a link to carcinoma. N Engl J Med. 1995;333(11):724–726. doi: 10.1056/NEJM199509143331110. [DOI] [PubMed] [Google Scholar]
- 19.Busson P, Ganem G, Flores P, et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer. 1988;42(4):599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 20.Gazzolo L, De-The G, Vuillaume M, et al. Nasopharyngeal carcinoma. Ii. Ultrastructure of normal mucosa, tumor biopsies, and subsequent epithelial growth in vitro. J Natl Cancer Inst. 1972;48(1):73–86. [PubMed] [Google Scholar]
- 21.Chow KC, Ma J, Lin LS, et al. Serum responses to the combination of Epstein-Barr virus antigens from both latent and acute phases in nasopharyngeal carcinoma: complementary test of EBNA-1 with EA-D. Cancer Epidemiol Biomarkers Prev. 1997;6(5):363–368. [PubMed] [Google Scholar]