Abstract
Imatinib, a breakpoint cluster region (BCR)-Abelson murine leukemia (ABL) tyrosine kinase inhibitor (TKI), has revolutionized the treatment of chronic myelogenous leukemia (CML). However, development of multidrug resistance (MDR) limits the use of imatinib. In the present study, we aimed to investigate the mechanisms of cellular resistance to imatinib in CML. Therefore, we established an imatinib-resistant human CML cell line (K562-imatinib) through a stepwise selection process. While characterizing the phenotype of these cells, we found that K562-imatinib cells were 124.6-fold more resistant to imatinib than parental K562 cells. In addition, these cells were cross-resistant to second- and third-generation BCR-ABL TKIs. Western blot analysis and reverse transcription-polymerase chain reaction(RT-PCR) demonstrated that P-glycoprotein (P-gp) and MDR1 mRNA levels were increased in K562-imatinib cells. In addition, accumulation of [14C]6-mercaptopurine (6-MP) was decreased, whereas the ATP-dependent efflux of [14C] 6-MP and [3H]methotrexate transport were increased in K562-imatinib cells. These data suggest that the overexpression of P-gp may play a crucial role in acquired resistance to imatinib in CML K562-imatinib cells.
Keywords: Human chronic myelogenous leukemia, multidrug resistance, imatinib, P-glycoprotein, drug transporters
Chronic myelogenous leukemia (CML) is thought to arise from pluripotent hematopoietic stem cells with a clonal disorder resulting from the t(9;22)(q34;q11) reciprocal chromosome translocation[1]–[3]. Imatinib, also known as imatinib mesylate, signal transduction inhibitor-571 (STI571), Gleevec, and Glivec[4],[5], is a potent selective small molecule tyrosine kinase inhibitor (TKI). As the first approved medication that targets the ATP-binding region of breakpoint cluster region (BCR)-Abelson murine leukemia (ABL) tyrosine kinase[6],[7], imatinib acts by reversing the effects of the BCR-ABL oncoprotein in CML through inhibition of BCR-ABL autophosphorylation, thereby preventing subsequent substrate phosphorylation, inhibiting cell proliferation, and inducing apoptosis[8]–[10]. Imatinib has revolutionized the treatment of CML. Indeed, 95% of CML patients in chronic phase achieve remission with imatinib[5],[11],[12]; however, many patients with accelerated phase or blast crisis develop resistance to imatinib[5],[13]. The proposed mechanisms underlying imatinib resistance in CML include impaired drug binding due to BCR-ABL kinase domain point mutations, amplification of the BCR-ABL gene, and overexpression of the multidrug resistance 1 (MDR1) gene in tumor cells[7],[14]–[17].
Overexpression of P-glycoprotein (P-gp), which is encoded by the MDR1 gene, has been frequently implicated in resistance to chemotherapeutic drugs[17],[18]. Distribution of imatinib to the brain has been identified to be limited by P-gp-mediated efflux[19]. In addition, both P-gp and multidrug resistance protein (MRP)-1 have been shown to directly interact with imatinib[20], though Mukai et al. [21] reported that imatinib interacts with P-gp but not MRP1. Imatinib has also been shown to be a substrate of P-gp[16],[22],[23]. Burger et al.[24], who provided the first evidence that imatinib is a substrate of breast cancer resistance protein (BCRP/ABCG2), found that chronic imatinib exposure leads to reduced intracellular accumulation of imatinib by induction of P-gp and BCRP[25] In contrast, Ferrao et al.[26] suggested that overexpression of P-gp is unlikely to be a major mechanism of imatinib resistance, and Hirayama et al.[27] reported that BCRP does not contribute to imatinib resistance in K562 cells.
To clarify these inconsistent and/or controversial study results, the roles of P-gp, MRP1, and BCRP in imatinib resistance need to be further explored. We have recently demonstrated that overexpressed MRP4 plays a major role in reducing the accumulation of [14C] 6-mercaptopurine (6-MP) and/or its metabolites by functioning as an efflux pump, thereby conferring resistance to 6-MP in acute lymphoblastic leukemia (ALL)[28]. In contrast, Zeng et al.[29] reported that overexpressed P-gp in the surface membrane of MDR1-transfected murine lymphocytic leukemic L1210/MDRC.06 cells is responsible for resistance to 6-MP. In a cell model obtained by in vitro passing of K562 cells in progressively increasing doses of doxorubicin, overexpression of MDR1 gene was shown to confer resistance to imatinib[16]. Using human BCR/ABL-positive CML K562 cells as the in vitro model, we investigated the mechanisms of cellular resistance to imatinib in CML and our results show that overexpression of P-gp induces acquired resistance to imatinib in CML.
Materials and Methods
Reagents
[14C]6-MP (1.887 Bq/mmol) was purchased from Moravek Biochemicals (Brea, CA). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Hyclone (Logan, UT). 9-(2-phos-phonylmethoxyethyl)adenine (PMEA) was purchased from Gilead (Forest City, CA). Imatinib, nilotinib, dasatinib, and bosutinib were purchased from Chemie Tek (Indianapolis, IN). Coomassie brilliant blue (CBB) stain solution was purchased from Bio-Rad (Hercules, CA). Monoclonal antibodies against P-gp were purchased from Signet Laboratories Inc. (Dedham, MA), and monoclonal antibodies against BCRP/ABCG2 were purchased from A.G. Scientific Inc. (San Diego, CA). The monoclonal antibodies against MRP1[30] and MRP4[31],[32] have been described previously. Cisplatin, 6-MP, 6-thioguanine (6-TG), 2-mercaptopurine (2-MP), creatine phosphokinase, cytarabine (AraC), 1- (4,5-dimethyl-thiazol-2-yl)-3,5-diphenylformazan (MTT), EDTA, etoposide, methotrexate (MTX), and vincristine were obtained from Sigma-Aldrich (St. Louis, MO).
Cell culture
BCR/ABL-positive CML cell line K562 (American Type Culture Collection, Manassas, VA), hereafter termed K562 cells, is a human cell line that was originally derived from a CML patient in blast crisis. An imatinib-resistant subclone (K562-imatinib) was selected from K562 cells by growth in the presence of increasing concentrations of imatinib, up to a final concentration of 30 mmol/L reached over a 3-month period. K562-imatinib cells were cultured in drug-free medium for at least 2 weeks before being used for the experiments. K562-imatinib cells exhibited a stable phenotype as shown by the MTT assay after being cultured in the absence of drugs for 3 months. HEK293/pcDNA and HEK293/ABCG2-R2 cells were kindly provided by Dr. Susan Bates and Dr. Robert Robey (NCI, NIH, Bethesda, MD) and have been described previously[33]. All cell lines were subcultured twice weekly at 37°C in a 5% CO2 humidified atmosphere in DMEM supplemented with heat-inactivated 10% FBS.
Analysis of drug sensitivity by MTT assay
Cell viability was determined by a modified MTT cytotoxicity assay as described previously[34]. In brief, cells were plated into 96-well tissue culture plates (1.2 × 104 cells/well) in 0.2 mL of medium. After the cells were incubated at 37°C in a 5% CO2 humidified atmosphere in DMEM supplemented with heat-inactivated 10% FBS for 70 h, 20 µL of MTT (2 mg/mL in PBS) was added to each well and the plates were incubated for another 2 h. Cells were collected in microcentrifuge tubes and media were removed by centrifugation at 1500 ×g for 2 min. Cell pellets were washed twice with ice-cold PBS and 100 µL of dimethylsulfoxide (DMSO) was added into each tube at room temperature to solubilize formazan crystals. The dissolved formazan was then transferred into fresh 96-well plates, and the absorbance was determined at 570 nm with an OPSYS Microplate Reader (DYNEX Technologies Inc., Chantilly, VA).
Analysis of accumulation and efflux of [14C]6-MP
Drug accumulation and efflux experiments were performed by slight modification of a method previously described[31]. In brief, for accumulation experiments, 2 × 106 cells/well of K562 or K562-imatinib cells were seeded in triplicate in 24-well plates and incubated at 37°C with 10 µmol/L [14C]6-MP in a complete medium for 60 min. Cells were collected in microcentrifuge tubes and media were removed by centrifugation at 1500 ×g for 2 min. Cell pellets were washed 3 times with ice-cold PBS, and then radioactivity was measured by liquid scintillation counting. For efflux experiments, 2 × 106 cells/well of K562 or K562-imatinib cells were seeded in triplicate in 24-well plates and were incubated at 37°C in an energy depletion medium (glucose-free, pyruvate-free DMEM containing 10% dialyzed FBS and 5 mmol/L sodium azide) containing 10 µmol/L [14C]6-MP for 60 min. The cells were then washed 3 times with ice-cold PBS and were incubated at 37°C for 30 and 60 min in complete medium without radiolabeled drug. Cell-associated radioactivity was determined at the end of 60-min incubation in an energy depletion medium and at various subsequent time points in the complete medium.
Preparation of membrane vesicles and Western blot analysis
Membrane vesicles were prepared by the nitrogen cavitation method as described previously[31]. Briefly, cells from culture were washed twice with ice-cold PBS and once with vesicle buffer [10 mmol/L Tris-HCl (pH 7.4), 0.25 mol/L sucrose, 0.2 mmol/L CaCl2], and then equilibrated at 4°C under nitrogen pressure at 400 psi (25 kg/cm2) for 15 min. The cell homogenate was added with ethylenediamine tetraacetic acid (EDTA) to a final concentration of 1 mmol/L, diluted with dilution buffer [10 mmol/L Tris-HCl (pH 7.4), 0.25 mol/L sucrose], and centrifuged at 1500 ×g for 10 min to remove nuclei and unlysed cells. The supernatant was layered onto a 35% sucrose cushion [10 mmol/L Tris-HCl (pH 7.4), 35% sucrose, 1 mmol/L EDTA] and centrifuged at 16 000 ×g for 30 min. The interface was collected and then centrifuged at 100 000 ×g for 45 min. The vesicle pellet was resuspended in dilution buffer by using a 26-gauge needle. The protein concentrations were determined using the Bradford method[35] and vesicles were stored at -80°C until use. For Western blot analysis, 20-30 mg protein was loaded per lane, resolved by 4%–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose filters. P-gp, BCRP/ABCG2, MRP1, and MRP4 were detected using monoclonal antibodies (at dilutions of 1:200, 1:500, 1: 1000, 1:2000, respectively) and horseradish peroxidase-conjugated secondary antibodies (at a dilution of 1: 1000). Enhanced chemiluminescence solution (Amersham Biosciences Corp., Piscataway, NJ) was used for visualization. Because actin, the normally used control, was undetectable in the samples prepared from the membrane vesicles, CBB staining was used to demonstrate approximately equal loading.
RT-PCR assay
The procedures and protocols from the Perfect RNA™ kit Handbook were followed. Total cellular RNA was isolated from K562 and K562-imatinib cells by using Perfect RNA™ kit from the Eppendorf Company (Westbury, NY). The total RNA concentration and purity were determined by measuring absorbance at 260 nm and 280 nm with the UV T60 spectrophotometer. Then, the integrity of total RNA was checked with agarose gel electrophoresis and ethidium bromide staining. For reverse transcription-polymerase chain reaction (RT-PCR), 1 mg total RNA was used for cDNA synthesis with the cMaster RTplusPCR System and cMaster RT kit. The primer sequence of MRP4 was sense 5′-TG ATGAGCCGTATGTTTTGC-3′ and antisense 5′-CTTCGG AACGGACTTGACAT-3′. The primer sequence of the internal control, glyceraldehyde 3-phosphate dehydro-genase (GAPDH), was sense 5′-GCCAAAAGGGTCATCATCTC-3′ and antisense 5′-GTAGAGGCAGGGATGATGTTC-3′. The primer sequence of MDR1 (P-gp) was sense 5′-ATATCAGCAGCCC-ACATCAT-3′ and antisense 5′-GAAGCACTGGGATGTCCGGT-3′. One step RT-PCR was carried out for 35 cycles as follows: reverse transcription at 50°C for 30 min, initial denaturation at 94°C for 2 min, template denaturation at 94°C for 15 s, primer annealing at 52°C for 20 s, and primer extension/elongation at 68°C for 30 s. The PCR products were separated by denaturing agarose gel electrophoresis. The gel was stained with 1 mg/mL ethidium bromide, and the bands were visualized using the UV light.
Vesicular transport experiments
The uptake of [3H]methotrexate (MTX) into membrane vesicles was studied using the rapid filtration method as previously described[33]. Membrane vesicles prepared from HEK293/pcDNA and HEK293/ABCG2-R2 cells were used as negative and positive controls, respectively. Transport experiments were carried out in medium containing membrane vesicles (10 µg), 0.25 mol/L sucrose, 10 mmol/L Tris-HCl, pH 7.4, 10 mmol/L MgCl2, 4 mmol/L ATP or 4 mmol/L AMP, 10 mmol/L phosphocreatine, 100 µg/mL creatine phosphokinase, and 0.5 µmol/L radiolabeled MTX, in a total volume of 50 µL. Reactions were carried out at 37°C for 10 min and were stopped by the addition of 3 mL of ice-cold stop solution (0.25 mol/L sucrose, 100 mmol/L NaCl, and 10 mmol/L Tris-HCl, pH 7.4). For the rapid filtration step, samples were passed through 0.22 µm GVWP filters (Millipore Corporation, Billerica, MA) presoaked in the stop solution under vacuum. The filters were washed 3 times with 3 mL of ice-cold stop solution and dried at room temperature for 30 min. Radioactivity was measured with a liquid scintillation counter. Rates of net ATP-dependent transport were determined by subtracting the values obtained in the presence of 4 mmol/L AMP from those obtained in the presence of 4 mmol/L ATP.
Statistical analysis
The data were analyzed by unpaired Student's t test using Microsoft Office Excel 2007 and P values <0.05 were considered significant.
Results
Imatinib-resistant K562 cells are cross-resistant to BCR-ABL TKIs and other nucleosides
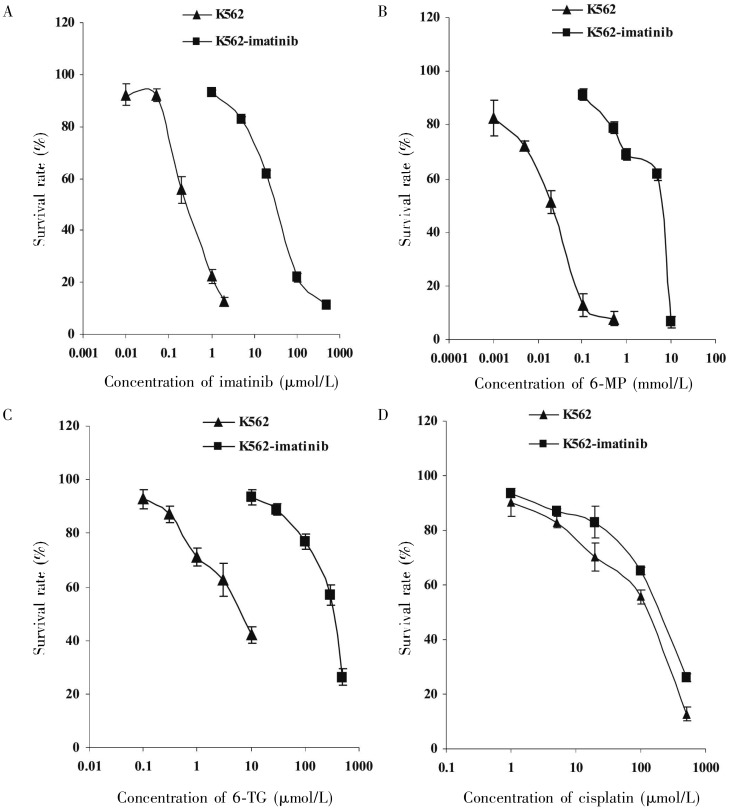
To investigate the mechanisms of cellular resistance to imatinib in CML, we first established an imatinib-resistant cell line using CML K562 cells. These cells were made resistant to imatinib by stepwise selection in imatinib. Analysis of drug sensitivity using the MTT assay indicated that K562-imatinib cells were 124.6-fold more resistant to imatinib than parental K562 cells (Table 1, Figure 1A, P < 0.001). K562-imatinib cells were also highly cross-resistant to other BCR-ABL TKIs including nilotinib (320.7-fold), dasatinib (4179.1-fold), and bosutinib (5543.5-fold) (Table 1, P < 0.001). In addition, K562-imatinib cells conferred resistance to nucleobase analogs 6-MP (267.7-fold), 6-TG (80.2-fold) (Table 1, Figures 1B and 1C, P < 0.001), and PMEA (2-fold, P < 0.05) but not 2-MP (Table 1, P > 0.05). For nucleoside analogs, K562-imatinib cells were slightly resistant to AraC (Table 1), but these effects were not statistically significant (P > 0.05). In addition, K562-imatinib cells were resistant to vincristine and cisplatin (Table 1, Figure 1D, P < 0.05).
Table 1. Drug resistance profile of K562-imatinib cells.
| Reagent | IC50a (µmol/L) |
Relative resistanced | |
| K562 | K562-imatinib | ||
| Imatinib | 0.35 ± 0.09 | 43.6 ± 7.6b | 124.6 |
| Nilotinib | 0.0195 ± 0.0008 | 6.2543 ± 0.5865b | 320.7 |
| Dasatinib | 0.0019 ± 0.0004 | 7.9403 ± 0.3963b | 4179.1 |
| Bosutinib | 0.0011 ±0.0002 | 6.0978 ± 0.5246b | 5543.5 |
| 6-MP | 22.6 ± 5.2 | 6049 ± 169b | 267.7 |
| 6-TG | 4.3 ±1.3 | 345 ± 113b | 80.2 |
| PMEA | 346 ± 111 | 675 ± 132c | 2.0 |
| 2-MP | 1369 ± 158 | 1355 ± 168 | 1.0 |
| AraC | 6.3 ± 2.0 | 10.6 ± 1.6 | 1.7 |
| Vincristine | 0.13 ± 0.04 | 0.22 ± 0.06c | 1.7 |
| Cisplatin | 149 ± 33 | 349 ± 27c | 2.3 |
aValues are presented as mean ± SD of at least three separate experiments and data were analyzed by unpaired Student's t-test. cP < 0.001, and bP < 0.05, compared with K562 cells. dRelative fold resistance was obtained by dividing the IC50 value of K562-imatinib cells by the IC50 value of K562 cells for a particular treatment. 6-MP, 6-mercaptopurine; 6-TG, 6-thioguanine; PMEA, 9-(2-phosphonylmethoxyethyl)adenine; 2-MP, 2-mercaptopurine; AraC, cytarabine.
Figure 1. Sensitivity of K562 cells (▴) and K562-imatinib cells (▪) to imatinib (A), 6-mercaptopurine (6-MP) (B), 6-thioguanine (6-TG) (C), and cisplatin (D).
The data were analyzed using the MTT cytotoxicity assay as described in “Materials and Methods” section. Data points are presented as means ± standard deviation (SD) of at least three separate experiments.
Decreased accumulation and increased efflux of [14C]6-MP in K562-imatinib cells
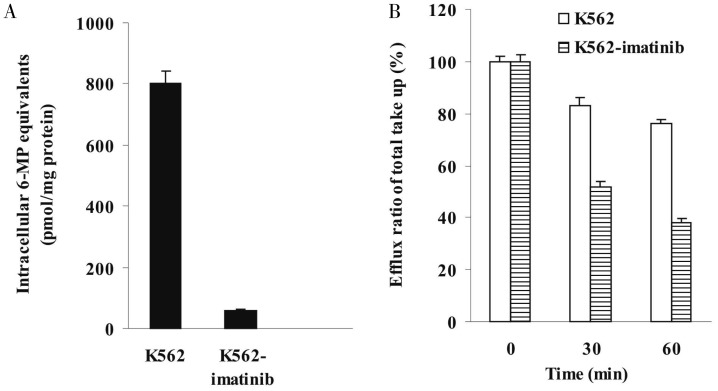
To determine whether decreased accumulation of the drug was involved in the resistance of K562-imatinib cells, accumulation of radioactivity derived from [14C] 6-MP was analyzed. Accumulation of [14C]6-MP and/or its metabolites was markedly reduced in K562-imatinib compared to control K562 cells (Figure 2A, P < 0.001), with the resistant cells accumulating only 7% of drugs compared to parental cells. To further dissect the basis of the decreased accumulation of [14C]6-MP equivalents in K562-imatinib cells, separate efflux experiments were performed. K562 and K562-imatinib cells were allowed to accumulate [14C]6-MP in energy depletion medium that prevented the operation of ATP-dependent efflux pumps. One hour after the treatment with [14C]6-MP, the intracellular accumulation was comparable in K562 and K562-imatinib cells. Cells were then switched to complete medium to allow efflux, and intracellular radioactivity were measured after 30 and 60 min. After 30 min efflux, 53% of the accumulated 6-MP was released from K562-imatinib cells, whereas only 16% of the accumulated 6-MP was released from K562 cells. After 60 min efflux, 62% of the accumulated 6-MP was released from K562-imatinib cells, whereas only 25% of the accumulated 6-MP was released from K562 cells (Figure 2B). These results indicate that increased efflux was involved in the reduced cellular accumulation in K562-imatinib cells.
Figure 2. Time-dependent accumulation and efflux of [14C]6-mercaptopurine (6-MP) and/or its metabolites in K562 and K562-imatinib cells.
A, accumulation of [14C]6-MP and/or its metabolites at 60 min in K562 and K562-imatinib cells in complete medium. Values are presented as mean ± SD of three separate experiments. Data were analyzed by unpaired Student's t-test. Accumulation of [14C]6-MP and/or its metabolites at 60 min in K562-imatinib cells decreased significantly compared with that in K562 cells (P < 0.001). B, time course of efflux of [14C]6-MP and/or its metabolites in K562 and K562-imatinib cells after various efflux times. Values are presented as mean + SD of three separate experiments. The percentages of retained 6-MP equivalents after 30 and 60 min efflux are indicated in K562 and K562-imatinib cells relative to that at 0 min. Data were analyzed by unpaired Student's t-test. After 30 and 60 min efflux, the retained 6-MP equivalents in K562-imatinib cells decreased significantly compared with that in K562 cells (P < 0.001).
Increased expression of P-gp in K562-imatinib cells
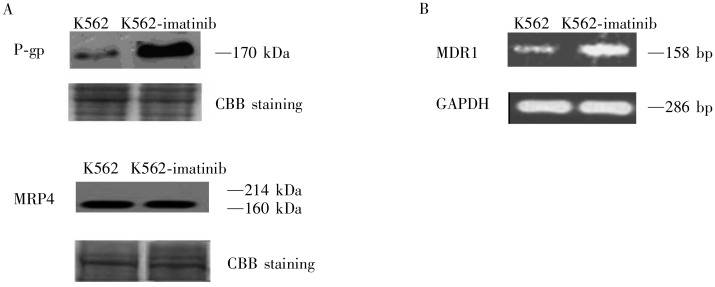
The increased efflux exhibited in K562-imatinib cells, in combination with cross-resistance to vincristine, suggested that P-gp might be involved in the resistance phenotype of the cell line. As shown in Figure 3A, P-gp was markedly overexpressed in K562-imatinib cells. However, expression of MRP4, a pump we previously demonstrated to be up-regulated in ALL cells that were made resistant to 6-MP[30], was similar in the two cell lines. Expression of MRP1 and BCRP/ABCG2 was undetectable in the two cell lines (data not shown).
Figure 3. Expression of P-glycoprotein (P-gp) and multidrug resistance protein 4 (MRP4) in K562 and K562-imatinib cells.
A, Western blot analysis of P-gp and MRP4 in membrane vesicle preparations from K562 and K562-imatinib cells. Membrane vesicles were prepared as described in Materials and Methods. Protein was resolved by SDS-PAGE on 4%-12% gel and electrotransferred to nitrocellulose membranes. Mobilities of the molecular mass markers are indicated in kilodaltons. The bottom panel is a section of an identical gel stained with coomassie brilliant blue (CBB) to demonstrate approximately equal loading. B, reverse transcription-polymerase chain reaction (RT-PCR) analysis of mRNA expression levels of MDR1 in K562 and K562-imatinib cells.
Increased expression of MDR1 mRNA levels in K562-imatinib cells
The RT-PCR assay was used to ascertain whether the mRNA level of MDR1 was up-regulated in the K562-imatinib cells compared with the K562 cells. As shown in Figure 3B, MDR1 mRNA levels were significantly increased in K562-imatinib cells compared with the K562 cells, which may contribute to the development of acquired resistance of K562-imatinib cells to imatinib.
Increased transport of [3H]MTX in K562-imatinib cell membrane vesicles
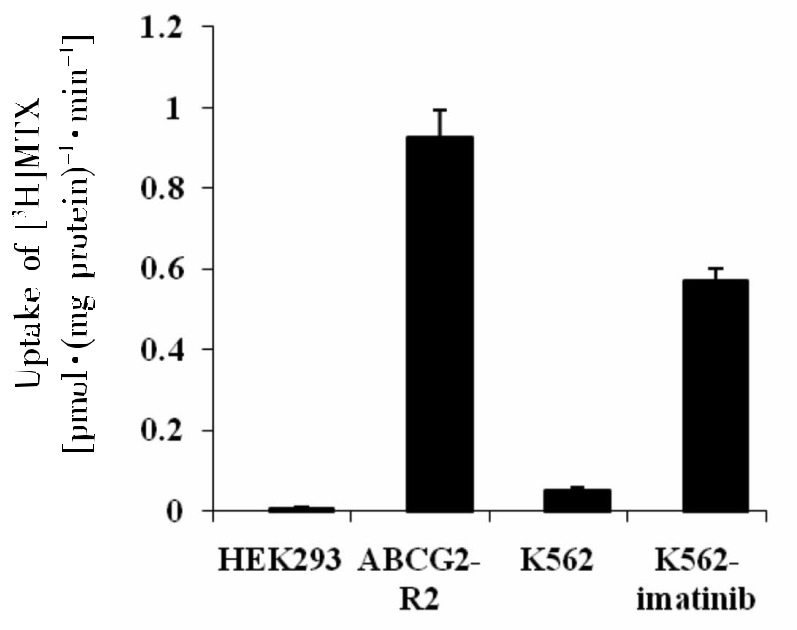
P-gp is an ATP-dependent membrane efflux pump capable of transporting anticancer drugs such as the established P-gp substrate MTX, leading to drug resistance. The ATP-dependent MTX transport with membrane vesicles prepared from K562-imatinib cells was significantly higher than that from K562 cells, with the transport rate of [3H]MTX being 0.57 and 0.05 pmol/mg protein per min by K562-imatinib and K562 membrane vesicles, respectively (Figure 4, P < 0.001). This result suggests that P-gp-mediated imatinib efflux could contribute to the decreased accumulation of imatinib in K562-imatinib cells, resulting in resistance to imatinib in K562-imatinib cells.
Figure 4. Uptake of [3H] methotrexate (MTX) into inside-out membrane vesicles derived from K562 and K562-imatinib cells.
Values are presented as mean ± SD of three separate experiments. Membrane vesicles prepared from HEK293/pcDNA and HEK293/ABCG2-R2 are used as negative and positive controls, respectively. Data were analyzed by unpaired Student's t-test. The ATP-dependent uptake of [3H]MTX in K562-imatinib cells increased significantly compared with K562 cells (P < 0.001).
Discussion
To investigate the cellular resistance mechanisms to imatinib in CML, we established an imatinib-resistant cell line (K562-imatinib) through stepwise drug selection of a CML cell line (K562 cells). Our results suggest that overexpression of P-gp induces acquired resistance to imatinib in CML. K562-imatinib cells, exhibiting MDR phenotype, were 124.6-fold more resistant to imatinib than the parental cells and cross-resistant to second (nilotinib and dasatinib) and third (bosutinib) generation TKIs, including 6-MP, 6-TG, PMEA, AraC, vincristine, and cisplatin (Table 1, Figure 1). Western blot results showed that P-gp was overexpressed in K562-imatinib cells compared with K562 cells, whereas MRP4 expression was similar in K562 and K562-imatinib cells (Figure 3A). The expression of MRP1 and BCRP/ABCG2 was undetectable in K562 and K562-imatinib cells (data not shown). MDR1 mRNA levels were overexpressed in K562-imatinib cells (Figure 3B). These results suggest that overexpression of P-gp can confer resistance to imatinib in K562-imatinib cells. These findings are consistent with a previous report which showed that resistance to imatinib in vincristine-resistant K562 cells is associated with overexpression of functional P-gp[36].
As shown in Figure 1B, K562-imatinib cells were cross-resistant to 6-MP, and the results from our recent data show that P-gp was up-regulated in 6-MP resistant CML cells[37]. In addition, Zeng et al.[29] found that high level of P-gp in the surface membrane of L1210/MDRC.06 cells is responsible for the resistance to 6-MP and other purine analogs.
Hart et al.[38] and Wuchter et al.[39] reported that P-gp might contribute to the poor prognosis of adult T-cell leukemia and adult AML. Kuwazuru et al.[40] examined the P-gp expression levels in fresh leukemia cells from CML patients in blast crisis and found that 6 of 11 patients (9 in the refractory state) were P-gp positive. In addition, they further revealed that P-gp expression levels correlate with patient response to chemotherapy[40]. However, none of these studies elucidated the mechanisms underlying the P-gp function.
We performed the 6-MP accumulation and efflux experiments to test the function of P-gp. Accumulation of [14C]6-MP and/or its metabolites was markedly reduced in K562-imatinib cells compared to that in parental K562 cells (Figure 2A). In efflux studies, only 47% and 38% of accumulated [14C]6-MP equivalents were retained in K562-imatinib cells at 30 and 60 min, respectively, whereas 84% and 75% of the accumulated [14C]6-MP equivalents were retained in K562 cells at the respective time points (Figure 2B). Our results indicate that cellular accumulation of [14C]6-MP equivalents in K562-imatinib cells was much lower than that in K562 cells, and efflux of [14C]6-MP equivalents from K562-imatinib cells was increased compared with that from K562 cells. Like imatinib[16],[20],[21], MTX is a substrate of P-gp[41],[42]. Therefore, using membrane vesicles, we performed an MTX transport assay to further test the function of P-gp. MTX transport into membrane vesicles from K562-imatinib cells was significantly higher than that into those from K562 cells (Figure 4). Although MTX is also a substrate of MRPs 1-5 and 8 as well as BCRP, only P-gp, not MRP1, MRP4, or BCRP/ABCG2, was up-regulated in K562-imatinib cells. So far, there are no reports showing that MRP2, 3, 5 or 8 are expressed in imatinib-resistant CML cells. Therefore, P-gp-mediated imatinib efflux could contribute to the decreased accumulation of imatinib in K562-imatinib cells, resulting in resistance to imatinib in K562-imatinib cells.
Other mechanisms may also be involved in imatinib resistance in CML including point mutations in the BCR-ABL kinase domain and amplification of the BCR-ABL gene[12]–[15]. At least 73 mutations leading to 50 amino acid substitutions have been described so far in CML resistance to imatinib therapy, and these mutations have been reported to increase the IC50 for imatinib[43]. However, several mutations in the kinase domain of the BCR-ABL Oncogene occur even before exposure to imatinib[44]. TKIs such as imatinib are reported not to directly induce mutations, but rather to select the mutations by providing a growth advantage to Philadelphia-positive subclones prior to the therapy or to the mutating cells originating in the highly unstable Philadelphia-positive stem cell compartment during TKI treatment[43]. Moreover, the highest sensitivity among the current methods for detecting kinase domain mutations in CML patients is only 15%–25%, and the high cost of these methods limits practical use[43]. Although Coutre et al.[45] showed that amplification of the BCR-ABL gene is associated with resistance of human leukemia cells to imatinib in vitro, it remains to be determined whether patients treated with imatinib will show increased amounts of phosphorylated BCR-ABL associated with disease relapse. Redaelli et al.[46] investigated the activity of bosutinib, dasatinib, imatinib, and nilotinib against a panel of 18 mutated forms of BCR-ABL associated with imatinib resistance in CML and Philadelphia-positive ALL patients. They found that a relative resistance against wild-type BCR-ABL greater than 2 was observed in 8 of 18 mutants in the case of bosutinib, 10 of 18 for dasatinib, and 13 of 18 for nilotinib and imatinib. For imatinib, bosutinib, and nilotinib, there was one highly resistant mutant in addition to the known T315I mutant (V299L for bosutinib and E255V for nilotinib), whereas T315I was the only mutant with a high relative resistance against dasatinib[46]. However, we tested the activity of bosutinib, dasatinib, imatinib, and nilotinib in the two cell lines and found that K562-imatinib cells showed high relative resistance to bosutinib, dasatinib, and imatinib. Therefore, it is unlikely that the cellular resistance to imatinib in K562-imatinib cells is due to point mutations. However, the possibility of involvement of T315I mutation cannot be ruled out. Further experiments are warranted to ascertain the role of this mutation in K562-imatinib cells. In addition, molecular experiments are needed to ascertain the involvement of signal transduction pathways such as alteration in Src-family kinases or up-regulation of p53/56 Lyn kinase, which has also been reported to be the cause of imatinib resistance[47].
In summary, the mechanisms of imatinib resistance in CML may be multi-factorial and certainly more complex than only point mutations or gene amplifications of BCR-ABL Our results clarify the contradictions in the previous findings on the expression of P-gp and its role in CML resistance to imatinib. Here, we report that imatinib-selected CML cells significantly overexpress P-gp at both the mRNA and protein levels. In addition, our finding suggests that P-gp confers acquired resistance to imatinib by functioning as an efflux transporter and reducing imatinib accumulation.
Acknowledgments
This study was supported by start-up funding from St. John's University (Z. S. Chen). We thank Dr. Susan E. Bates and Dr. Robert W. Robey (NIH, Bethesda, MD) for providing HEK293/pcDNA cells and HEK293/ABCG2-R2 transfected cells.
References
- 1.Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Goldman JM, Melo JV. Chronic myeloid leukemia-advances in biology and new approaches to treatment. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 3.Hehlmann R, Hochhus A, Baccarani M. Chronic myeloid leukemia. Lancet. 2007;370:342–350. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- 4.Roskoski R., Jr STI-571: an anticancer protein-tyrosine kinase inhibitor. Biochem Biophys Res Commun. 2003;309:709–717. doi: 10.1016/j.bbrc.2003.08.055. [DOI] [PubMed] [Google Scholar]
- 5.Walz C, Sattler M. Novel targeted therapies to overcome imatinib mesylate resistance in chronic myeloid leukemia (CML) Crit Rev Oncol Hematol. 2006;57:145–164. doi: 10.1016/j.critrevonc.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers CL. Disabling Abl-perspectives on Abl kinase regulation and cancer therapeutics. Cancer Cell. 2002;1:13–15. doi: 10.1016/s1535-6108(02)00022-3. [DOI] [PubMed] [Google Scholar]
- 7.Shah NP, Sawyers CL. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22:7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- 8.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 9.Deininger MW, Goldman JM, Lydon N, et al. The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL–positive cells. Blood. 1997;90:3691–3698. [PubMed] [Google Scholar]
- 10.Gambacorti-Passerini C, le Coutre P, Mologni L, et al. Inhibition of the ABL kinase activity blocks the proliferation of Bcr/Abl+ leukemic cells and induces apoptosis. Blood Cells Mol Dis. 1997;23:380–394. doi: 10.1006/bcmd.1997.0155. [DOI] [PubMed] [Google Scholar]
- 11.Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien SG, Guilhot F, Larson RA, et al. Investigators. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. New Engl J Med. 2003;348:994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 13.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 14.Weisberg E, Griffin JD. Mechanisms of resistance imatinib (STI571) in preclinical models and in leukemia patients. Drug Resist Updat. 2001;4:22–28. doi: 10.1054/drup.2001.0180. [DOI] [PubMed] [Google Scholar]
- 15.Gambacorti-Passerini CB, Gunby RH, Piazza R, et al. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome–positive leukemias. Lancet. 2003;4:75–85. doi: 10.1016/s1470-2045(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 16.Mahon FX, Belloc F, Lagarde V, et al. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood. 2003;101:2368–2373. doi: 10.1182/blood.V101.6.2368. [DOI] [PubMed] [Google Scholar]
- 17.Widmer N, Colombo S, Buclin T, et al. Functional consequence of MDR1 expression on imatinib intracellular concentrations. Blood. 2003;102:1142. doi: 10.1182/blood-2003-03-0993. [DOI] [PubMed] [Google Scholar]
- 18.Aller SG, Yu J, Ward A, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai H, Marbach P, Lemaire M, et al. Distribution of STI-571 to the brain is limited by P-glycoprotein-mediated efflux. J Pharmacol Exp Ther. 2003;304:1085–1092. doi: 10.1124/jpet.102.045260. [DOI] [PubMed] [Google Scholar]
- 20.Hegdus T, Orfi L, Septodi A, et al. Interaction of tyrosine kinase inhibitors with the human multidrug transporter proteins, MDR1 and MRP1. Biochim Biophys Acta. 2002;1587:318–325. doi: 10.1016/s0925-4439(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 21.Mukai M, Che XF, Furukawa T, et al. Reversal of the resistance to STI571 in human chronic myelogenous leukemia K562 cells. Cancer Sci. 2003;94:557–563. doi: 10.1111/j.1349-7006.2003.tb01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Illmer T, Schaich M, Platzbecker U, et al. P-glycoprotein– mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia. 2004;18:401–408. doi: 10.1038/sj.leu.2403257. [DOI] [PubMed] [Google Scholar]
- 23.Hamada A, Miyano H, Watanabe H, et al. Interaction of imatinib mesylate with human p-glycoprotein. J Pharmacol Exp Ther. 2003;307:824–828. doi: 10.1124/jpet.103.055574. [DOI] [PubMed] [Google Scholar]
- 24.Burger H, van Tol H, Boersma AW, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104:2940–2942. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 25.Burger H, van Tol H, Brok M, et al. Chronic imatinib mesylate exposure leads to reduced intracellular drug accumulation by induction of the ABCG2 (BCRP) and ABCBI (MDR1) drug transport pumps. Cancer Biol Ther. 2005;4:747–752. doi: 10.4161/cbt.4.7.1826. [DOI] [PubMed] [Google Scholar]
- 26.Ferrao PT, Frost MJ, Siah SP, et al. Overexpression of P-glycoprotein in K562 cells does not confer resistance to the growth inhibitory effects of imatinib (STI571) in vitro. Blood. 2003;102:4499–4503. doi: 10.1182/blood-2003-01-0083. [DOI] [PubMed] [Google Scholar]
- 27.Hirayama C, Watanabe H, Nakashima R, et al. Constitutive overexpression of P-glycoprotein, rather than breast cancer resistance protein or organic cation transporter 1, contributes to acquisition of imatinib-resistance in K562 cells. Pharm Res. 2008;25:827–835. doi: 10.1007/s11095-007-9376-3. [DOI] [PubMed] [Google Scholar]
- 28.Peng XX, Shi Z, Damaraju VL, et al. Up-regulation of MRP4 and down-regulation of influx transporters in human leukemic cells with acquired resistance to 6-mercaptopurine. Leuk Res. 2008;32:799–809. doi: 10.1016/j.leukres.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Zeng H, Lin ZP, Sartorelli AC. Resistance to purine and pyrimidine nucleoside and nucleobase analogs by the human MDR1 transfected murine leukemia cell line L1210/VMDRC.06. Biochem Pharmacol. 2004;68:911–921. doi: 10.1016/j.bcp.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Breuninger LM, Paul S, Gaughan K, et al. Expression of multidrug associated protein in NIH/3T3 cells confers multidrug resistance associated with increased drug efflux and altered intracellular drug distribution. Cancer Res. 1995;55:5342–5347. [PubMed] [Google Scholar]
- 31.Chen ZS, Lee K, Kruh GD. Transport of cyclic nucleotides and estradiol 17-beta-D-glucuronide by multidrug resistance protein 4: resistance to 6-mercaptopurine and 6-thioguanine. J Biol Chem. 2001;276:33747–33754. doi: 10.1074/jbc.M104833200. [DOI] [PubMed] [Google Scholar]
- 32.Lee K, Klein-Szanto AJ, Kruh GD. Analysis of the MRP4 drug resistance profile in transfected NIH3T3 cells. J Natl Cancer Inst. 2000;92:1934–1940. doi: 10.1093/jnci/92.23.1934. [DOI] [PubMed] [Google Scholar]
- 33.Chen ZS, Robey RW, Belinsky MG, et al. Transport of methotrexate, methotrexate polyglutamates, and 17beta- estradiol 17-(beta-D-glucuronide) by ABCG2: effects of acquired mutations at R482 on methotrexate transport. Cancer Res. 2003;63:4043–4054. [PubMed] [Google Scholar]
- 34.Peng XX, Li YB. Induction of cellular glutathione-linked enzymes and catalase by the unique chemoprotective agent, 3H-1,2-dithiole-3-thione in rat cardiomyocytes affords protection against oxidative cell injury. Pharmacol Res. 2002;45:491–497. doi: 10.1006/phrs.2002.0991. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Assef Y, Rubio F, Colo G, et al. Imatinib resistance in multidrug-resistant K562 human leukemic cells. Leuk Res. 2009;33:710–716. doi: 10.1016/j.leukres.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 37.Peng XX, Shi Z, Tiwari, et al. Up-regulation of P-glycoprotein confers acquired resistance to 6-mercaptopurine in human chronic myeloid leukemia cells. Oncol Lett. 2011;2(3):549–556. doi: 10.3892/ol.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart SM, Ganeshaguru K, Hoffbrand AV, et al. Expression of the Multidrug resistance-associated protein (MRP) in acute leukemia. Leukemia. 1994;8:2163–2168. [PubMed] [Google Scholar]
- 39.Wuchter C, Leonid K, Ruppert V, et al. Clinical significance of P-glycoprotein expression and function for response to induction chemotherapy, relapse rate and overall survival in acute leukemia. Haematologica. 2000;85:711–721. [PubMed] [Google Scholar]
- 40.Kuwazuru Y, Yoshimura A, Hanada S, et al. Expression of multidrug transporter-glycoprotein, in chronic myelogenous leukemia cells in blast crisis. Br J Haematol. 1990;74:24–29. doi: 10.1111/j.1365-2141.1990.tb02533.x. [DOI] [PubMed] [Google Scholar]
- 41.Loscher W, Potschka H. Blood-brain barrier active efflux transporters: ATP-binding cassette gene family. NeuroRx. 2005;2:86–98. doi: 10.1602/neurorx.2.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Graaf D, Sharma RC, Mechetner EB, et al. P-glycoprotein confers methotrexate resistance in 3T6 cells with deficient carrier-mediated methotrexate uptake. Proc Natl Acad Sci USA. 1996;93:1238–1242. doi: 10.1073/pnas.93.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baccarani M, Pane F, Saglio G. Monitoring treatment of chronic myeloid leukemia. Haematologica. 2008;93:161–169. doi: 10.3324/haematol.12588. [DOI] [PubMed] [Google Scholar]
- 44.Jiang X, Saw KM, Eaves A, et al. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst. 2007;99:680–693. doi: 10.1093/jnci/djk150. [DOI] [PubMed] [Google Scholar]
- 45.le Coutre P, Tassi E, Varella-Garcia M, et al. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–1766. [PubMed] [Google Scholar]
- 46.Redaelli S, Piazza R, Rostagno R, et al. Activity of bosutinib, dasatinib, and nilotinib against 18 imatinib-resistant BCR/ABL mutants. J Clin Oncol. 2009;27:469–471. doi: 10.1200/JCO.2008.19.8853. [DOI] [PubMed] [Google Scholar]
- 47.Pene-Dumitrescu T, Smithgall TE. Expression of a Src family kinase in chronic myelogenous leukemia cells induces resistance to imatinib in a kinase-dependent manner. J Biol Chem. 2010;285:21446–21457. doi: 10.1074/jbc.M109.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]