Abstract
The growing demand for new therapeutic strategies in the medical and pharmaceutic fields has resulted in a pressing need for novel druggable targets. Paradoxically, however, the targets of certain drugs that are already widely used in clinical practice have largely not been annotated. Because the pharmacologic effects of a drug can only be appreciated when its interactions with cellular components are clearly delineated, an integrated deconvolution of drug-target interactions for each drug is necessary. The emerging field of chemical proteomics represents a powerful mass spectrometry (MS)-based affinity chromatography approach for identifying proteome-wide small molecule-protein interactions and mapping these interactions to signaling and metabolic pathways. This technique could comprehensively characterize drug targets, profile the toxicity of known drugs, and identify possible off-target activities. With the use of this technique, candidate drug molecules could be optimized, and predictable side effects might consequently be avoided. Herein, we provide a holistic overview of the major chemical proteomic approaches and highlight recent advances in this area as well as its potential applications in drug discovery.
Keywords: Chemical proteomics, drug target profiling
Drug discovery is an inherently complex process with a history spanning thousands of years. However, the elusive mechanisms of action and limited specificity of compounds hamper their further application in clinical practice. A large number of drugs have been found to act upon multiple targets, inevitably resulting in side effects and drug resistance during treatment. The most notorious drug in history, thalidomide, which was used to alleviate morning sickness during pregnancy, was found to cause fetal malformations and multiple birth defects[1]. In addition, the difficulty in matching numerous complicated drugs with the desired physiologic effects is best characterized by the endless battle against drug resistance in antibiotics as well as in anti-cancer therapies.
It has been appreciated that the more we grasp the molecular mechanisms of potent drugs, the more we realize that these drugs are unexpectedly promiscuous to their relevant targets and the more we become aware of the obstacles lying ahead. However, as a highly efficient and high-throughput approach, the wide use of chemical proteomics in drug target identification has enhanced our confidence in improving our understanding of the molecular mechanisms of these drugs. With the aid of chemical proteomics, an unprecedented number of biological targets have been tested, and various technologies emerging today provide us with a superior platform to further investigate drug targets. Today, mass spectrometry (MS)-based global proteomic approaches, which analyze protein-protein interactions under distinct circumstances, are also widely used to identify novel drug targets in the pharmaceutical industry. More recently, two techniques have been developed to complement the more global approaches: (1) a compound-centric approach, which concentrates on characterizing the properties of drug-target interactions, and (2) activity-based protein profiling (ABPP), which focuses on the enzymatic activity of particular proteins. Here, we give an overview of the large-scale approaches of chemical proteomics as well as the above two techniques and their application in novel drug target profiling.
Global Proteomic Approaches
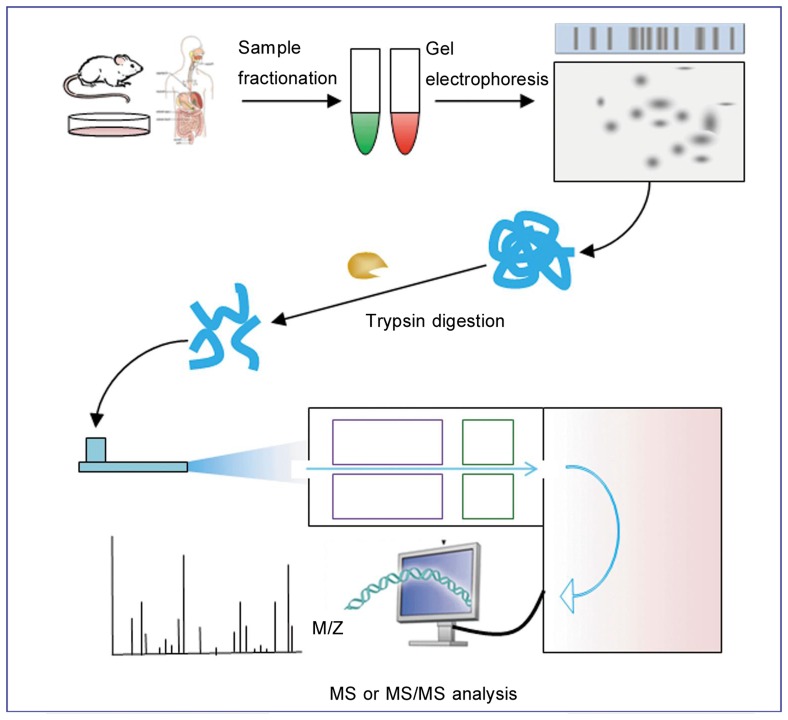
The 2002 Nobel Prize in Chemistry recognized the critical role of MS-based approaches in the analysis of complex proteins[2], especially in screening for novel drug targets. Hitherto, this technique had been successfully applied in protein analyses such as protein identification, post-translational modifications (PTMs) of proteins of interest, and protein-protein interactions. Generic MS-based proteomics covers five procedures (Figure 1). In addition, microarray technology renders possibly the synchronous analysis of multiple parameters through a single experiment[3]. Protein microarray is suitable for investigating enzyme-substrate as well as protein-protein interactions. Therefore, protein microarray can also be a potent tool in appraising drug targets.
Figure 1. Generic mass spectrometry (MS)-based proteomic experiment.
MS-based proteomic analysis usually begins with sample fractionation coupled with subsequent gel electrophoresis, which allows for the separation of different proteins. Spots of interest are further subjected to trypsin digestion, MS analysis, and protein database mining.
While the majority of existing approaches aim to discover new protein targets for a specific drug type, novel drug target discovery also has other uses in which the proteins identified might be involved in a certain disease or pathologic state. One notable advantage of global proteomic profiling methods over other methods lies in that proteomic approaches require no purification and are unbiased because the unmodified drug interacts with its endogenous targets. Based on this rationale, global proteomic methods have been widely used in the pharmaceutic industry. For example, multidimensional protein identification technology (MudPIT) is typically used to gain more knowledge regarding the mechanisms of action between natural products and targets involved in the maintenance of a particular phenotype. The MS analysis method mentioned above constitutes a powerful and universal method for such unbiased studies[4]. Currently, it is possible to explore the interactions between drugs and their targets using MS[5] and protein microarray[6] techniques. Other quantitative proteomic approaches could be used for the identification of novel drug targets, including methods involving differential labeling with stable isotopes, such as isotope-coded affinity tag (ICAT)[7] or isobaric tags for absolute and relative quantification (iTRAQ)[8]. These techniques are usually used to simultaneously quantify alterations in protein abundance. In addition, another approach named difference in gel electrophoresis (DIGE) can detect changes in protein expression levels, and these proteins can likewise be labeled with fluorescent probes. For example, the expression of urinary proteins was analyzed using DIGE and MALDI-MS[9]. This study group identified meprin-1α as a potential drug target for sepsis-induced acute renal failure (ARF)[10]. However, the DIGE method could hardly detect low-abundance proteins. In addition, membrane proteins cannot be easily analyzed by this approach. The histones isolated from individual biological replicates can be digested to their tryptic peptides, followed by LC-FT-MS/MS analysis during different MS workflow protocols[11]. In addition, techniques such as stable-isotope labeling by amino acids in cell culture (SILAC)[12] and two-dimensional gel electrophoresis (2-DE)[13] are also important tools in global proteomic approaches.
Activity-based Protein Profiling in Drug Target Identification
Although conventional global proteomic methods, which comparatively quantify the expression levels of transcripts and proteins, have yielded many useful insights, these platforms are, nevertheless, limited in their capacity to identify changes in protein activity that are caused by post-translational mechanisms. Moreover, cancer metabolism studies are further complicated by the potential for enzymes to perform distinct metabolic activities in tumor cells that might not be mirrored in normal physiology. Because of these challenges, novel proteomic techniques that will enable the accurate assessment of functions of proteins in complex biological systems like cancer cells are needed. ABPP has emerged as a key tool in the evolution of functional proteomics, and its implementation in the discovery and functional characterization of de-regulated enzymatic pathways in cancer has provided us with new insights into cancer therapy.
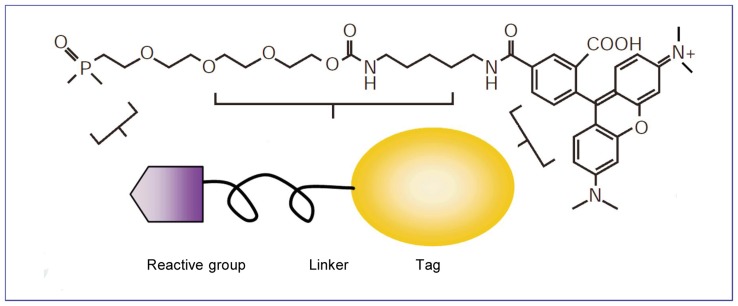
The core of the activity-based approach is the active site-directed covalent probe. The general structure of these probes in their most basic form can be categorized into three functionally distinct portions: a reactive component that covalently attaches to the active catalytic site of the target protein; a linker region that can modulate reactivity and specificity, conferring enough space for the binding of the reactive group and preventing steric hindrance; and a tag for further identifying and purifying modified enzymes (Figure 2).
Figure 2. The structure of an activity-based protein profiling (ABPP) probe, which comprises three general elements: an active site-directed reactive group for covalent attachment to the enzyme (shown here as an ethoxy fluorophosphonate group); a linker region to provide spacing and specificity (shown here as a polyethylene glycol group); and a tag for identification and/or purification (shown here as a rhodamine group).
Architecture of the reactive group
Obviously, the reactive group is the most important part of a probe, and designing a probe that covalently modifies the target protein is a great challenge. The difficulty lies in the duality of this functional group, as it must be both reactive towards a specific residue on a protein and inert towards other reactive species within the cell or cell extracts[14]. In general, different kinds of reactive groups have been developed for the study of different protein families. The reactive groups of the successfully designed chemical probes are based on the chemistries of covalent, mechanism-based inhibitors of various enzyme families. Usually, the reactive group is aimed to effectively target a protein family without binding to other proteins. These inhibitors rely on the mechanistic differences between individual enzyme families for selective targeting. For instance, serine and cysteine proteases use a catalytic amino acid nucleophile in their active site. Nevertheless, they possess distinct nucleophilic residues and different catalytic mechanisms. As protease inhibitors include a large source of reactive groups that have been designed based on subtle differences in reaction mechanisms for the major protease families, many chemical probes have been designed to target proteolytic enzymes[15]–[19].
In total, there are four general types of reactive groups that have been used in designing chemical probes: (1) mechanism-based probes (type I), in which the catalytic residue of the enzyme is normally involved in the attack of a substrate as its main nucleophile; (2) suicide substrates (type II) that contain a non-coated “masked electrophile”, which becomes activated upon cleavage by the enzyme, whereas the unmasked electrophile can react with nearby, non-catalytic, nucleophilic residues in the active site; (3) affinity alkylating probes (AFBPs) (type III) that contain affinity-based labeling groups, requires only a strong nucleophile or electrophile in the vicinity of the active-site pocket; (4) general alkylating probes (type IV), which are often used for probe design. This category of probes has recently been shown to be of great value for bulk proteomic analysis using MS-based methods[20].
However, under certain circumstances, the enzymes do not covalently bind to their targets and thus their reactive groups contain a chelator for noncovalent binding to the metal atom in the active site and a photoinducible chemical cross-linker for covalent binding to the enzyme active site upon ultraviolet irradiation[21].
Structure of the linker region
The linker region of a chemical probe connects the reactive group to the tag and serves several purposes. The primary function of the linker region is to provide enough space between the reactive groups. Besides, the linker region can prevent steric hindrance that would block the accessibility of the reactive group or of the tags used for purification. In addition, the linker region influences the specificity of the probe. For instance, we can now modify a recombinantly expressed ubiquitin with an electrophilic reactive group. In vitro, proteases can be covalently modified to use the resulting protein probe. This technique of making activity-based probes (ABPs) from recombinant proteins is useful in the design of probes for other enzymes or protein-binding domains that require substantial protein recognition elements for specificity.
Architecture of the tag
The tag on a chemical probe allows the quick and simple identification and purification of probe-modified proteins. In general, the tag acts as the key element that distinguishes an ABP from a stand-alone mechanism-based inhibitor, which primarily include biotin, fluorescent, and radioactive tags in the ABPP analysis. These tags facilitate the detection of probe-labeled enzyme targets, with similar mechanisms of affinity purification, gel-based screening assays, or imaging. In addition, they should be compatible with gel-based separation methods. The biotinylated tags are most frequently used for the enrichment, purification, and identification of target enzymes of ABPs[22]. In addition, radioactive and fluorescent tags can be visualized by the direct scanning of gels with a fluorescent scanner such as the Typhoon scanner. Therefore, fluorescent and radioactive tags might be more convenient and faster to analyze than biotin tags. In addition, during the application of fluorescent tags such as the AlexaFluors, fluorescent and radioactive tags show higher sensitivity and a wider dynamic range than the streptavidin-biotin detection method.
Compound-Centric Chemical Proteomics
In contrast to ABPP, compound-centric chemical proteomics (CCCP) is largely about target discovery. In this context, the mechanism of action of a bioactive compound is inferred through the identification of interacting components, most often through affinity chromatography and advanced MS techniques. CCCP has successfully identified the cellular targets for β-lactones, anticancer agents, and a variety of natural products. This method includes classical drug affinity chromatography, which has been in use for decades[23]–[26], and statistics or bioinformatics for the subsequent identification of binding proteins. Modern scientists combine classical drug affinity chromatography with modern high-resolution MS analysis, leading to more precise and efficient profiling. Thus, this approach is partially dependent on the huge technical developments in the MS field, especially regarding the ever-increasing sensitivity and throughput seen in recent years. Nano-electrospray ionization (ESI)[27] and quantification methods (for example, stable isotope labeling[7],[28],[29]), high-resolution, high-sensitive detection such as quadrupole time-of-flight or linear ion trap (LTQ), Fourier transform ion cyclotron resonance (FT-ICR), and LTQ/orbitrap mass spectrometers have been developed successively. However, it is necessary for the technologies to be accessible to purpose-fitted laboratory information management systems, database management, and statistical evaluation.
Chemical proteomics also builds on the experience that the community has gathered through the characterization of protein-protein interactions using affinity purification and MS[28]. In general, protein analytical strategies are similar after the initial purification steps, and proteins detected early (idiotypic peptides, isoforms expressed, multiple reaction monitoring) can help identify proteins that interact with chemical compounds. However, there are still major differences between chemical and protein interaction proteomics. When complexes are purified after formed within cells, the characterization of protein interactions is most informative. Nevertheless, protein interaction with the chemical bait in chemical proteomics occurs in vitro. As a result of this difference, chemical proteomics can use tissue extracts acquired from diverse sources, including human subjects, which is otherwise only possible in those rare cases where one has access to high-specificity and high-affinity antibodies.
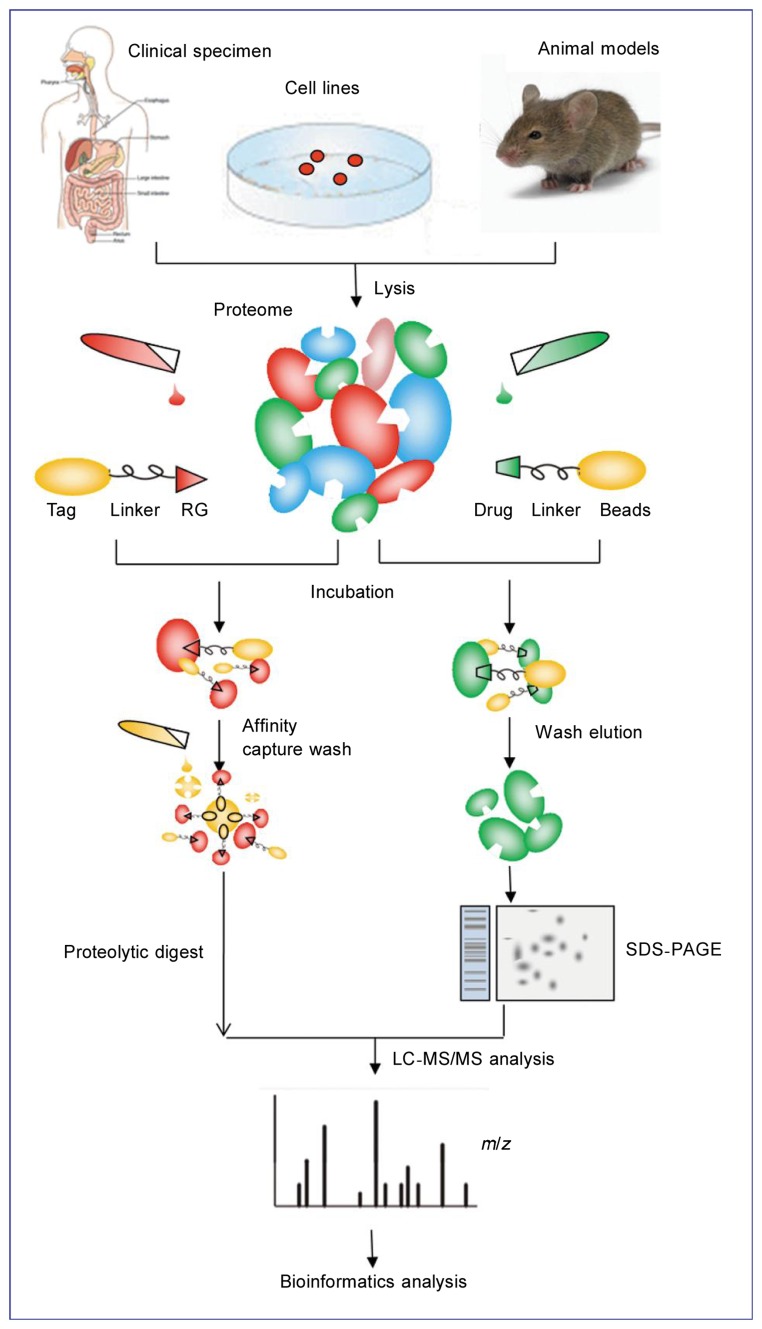
A classical chemical proteomic experiment starts with immobilizing a bioactive compound such as sepharose/agarose on a matrix (Figure 3). Sulfhydryl, amino, hydroxyl, and carboxyl groups (and so on) are all commonly used activated resins that allow for the attachment of specific chemical groups during this step. It is necessary to follow up the modification chemistry with a befitting biochemical or cell-based assay to ensure that activity is retained when a compound is modified for immobilization. In addition, cell extracts can be similarly prepared from cells or tissues. Secondly, these cell lysates are incubated with the affinity matrix and are sufficiently washed prior to elution. Nonspecific elution can be performed using detergents, salts, or denaturing agents. The specific elution can be achieved by competition with an excess of soluble compound or via the specific cleavage of an engineered linker[30]. A gel-based or a gel-free method such as “shotgun proteomics” processing, followed by protein digestion with a protease such as trypsin generates a complex peptide mix, which is subsequently analyzed by nano-HPLC coupled to nano-ESI-MS/MS. Afterwards, the results are searched against an appropriate protein database such as SwissProt with a search engine such as Mascot or Sequest.
Figure 3. Comparison of the global proteomic approaches: activity-based probe profiling (ABPP) and compound-centric chemical proteomics (CCCP).
ABPP requires the formation of a covalent bond between the probe molecule and the protein, which is achieved either through the attachment of an irreversible inhibitor to a moiety in the active site of the enzyme or through the initial binding of a reversible inhibitor conjugated to a photo-reactive element and the subsequent formation of a covalent bond upon irradiation. To facilitate affinity chromatography, the reactive warhead is connected via a linker to a reporter tag such as biotin. After the cell lysate of interest is incubated with this ABP, targeted proteins are captured on an affinity matrix and digested with trypsin before MS, protein database mining, and further bioinformatic analysis. For the CCCP approach, the compound of interest (typically, with a known bioactivity) is conjugated chemically to an inert and biocompatible matrix in a way that does not interfere with its activity. The compound matrix is then incubated with the biological extract of choice. The captured proteins are eluted and processed by either SDS-PAGE or a gel-free method. Subsequent tryptic digestion produces a peptide mixture that is analyzed as described for ABPP.
One can characterize the target profile of a bioactive compound by following the CCCP approach. However, this method does not offer immediate information on the activation state of the identified proteins, in contrast to ABPP. Moreover, CCCP, as a more objective method, enables the identification of binders of biochemical classes that were previously unexpected[31]–[33], including those without enzymatic function. Hence, CCCP allows for high throughput and global profiling of novel drug targets. However, CCCP is only applicable to small molecules that can be chemically modified for immobilization and does not distinguish between specific versus nonspecific interactions.
Advantages and Disadvantages of Chemical Proteomics
The chemical proteomic approach has great strengths and is widely used in scientific research. It provides a powerful weapon to profile previously uncharacterized proteins via identifying drug-target interactions. Besides, this approach is not limited to panels of recombinant proteins; it can be used to probe the integral proteome or affirmatory sub-proteomes. It is particularly useful when the proteins are encountered by small molecules in their natural, un-engineered state. Moreover, chemical proteomics can be used in any cell type, tissue, or species of interest, ranging from humans to microorganisms. For this reason, this method renders the exploration of relevant disease drug mechanisms possible in physiologically and clinically relevant sources such as tumor tissues, and it can also be used for virulence profiling in clinical samples.
Like any other technologies, the chemical proteomic approach also suffers from certain limitations. One limitation is that a large amount of protein is required, depending on the specific cell types and protocols used. However, a recent research has demonstrated remarkable downscaling to 8 mg protein from HEK293 cells[34], 2 mg protein from HeLa S3 cells[35], and even 0.5 mg protein from K562 cells using this approach, thus greatly improving the applicability of this method. Another limitation of this method lies in that all proteins cannot be recovered after lysis, and determining the specific proteins that are lost from an individual lysate preparation is difficult. Membrane proteins, particularly those with several transmembrane domains, are a great challenge for analysis using this technology because it requires that proteins remain in their native conformation. However, a recent study has successfully identified a receptor tyrosine kinase that is a type I transmembrane protein[36]. Additionally, this method results in a high level of background because certain proteins are overabundant or too prone to interacting with either hydrophobic or charged surface. Other strengths and limitations are listed in Table 1.
Table 1. Advantages and disadvantages of chemical proteomics compared to conventional target identification and selectivity profiling techniques.
| Advantages | Disadvantages |
| Entire proteome or defined subproteomes | High background |
| Natural proteins | Active metabolites |
| Un-engineered states | Not associated with IC50 |
| Natural expression levels | Protein solubilization |
| Competitive cellular environment | Immobilization of chemical modification |
| Performed with any cell type or tissue | No difference between direct and indirect binders |
| Disease-relevant cells (such as tumor tissue) |
Application of Chemical Proteomics in Drug Target Discovery
While global proteomic studies have long played an important role in assessing protein structure, function, and cellular interactions, chemical strategies can lead to the efficient detection of specific classes of proteins and represent an effective tool for subsequent protein isolation from the proteome. The use of chemical proteomics has led to the discovery of a variety of compound targets (Table 2), in particular, the targets of kinase inhibitors and natural products.
Table 2. Drug targets identified through modern chemical proteomics.
| Drug | Involved targets | Chemical proteomics | Disease |
| Pyrido[2,3-d] pyrimidine | Src, PDGFR, FGFR, RICK, p38α | Affinity chromatography, nano-HPLC MS/MS, LC-MS/MS | Cancer |
| SB 203580 | RICK, CK1, GAK, PKNβ, JAK1 | Affinity chromatography, MS | Inflammatory diseases |
| Imatinib | BCR-ABL, ABL, c-KIT, PDGFR, NQO2, c-fms | LC-ESI-MS/MS, HPLC-MS, ELISA, western blotting | Chronic myeloid leukemia |
| Nilotinib | c-ABL, BCR-ABL, c-KIT, PDGFR, ARG NQO2, DDR1 | LC-ESI-MS/MS, HPLC-MS, immunoblotting | Chronic myeloid leukemia |
| Dasatinib | c-ABL, BCR-ABL, BCR-ABL, DDR1, BTK, TEC | LC-ESI-MS/MS, immunoblotting, SDS/PAGE, LC-MS/MS | Chronic myeloid leukemia |
| Bosutinib | ABL and SRC family kinases, STE and TEC family kinases, CAMK2G | Affinity chromatography, MS, kinobeads/iTRAQ | Chronic myeloid leukemia |
| (R)-Roscovitine | CDKs, PDXK | Affinity chromatography, electrophoresis, and western blotting | Cancers, neurodegenerative diseases, viral infections, and glomerulonephritis |
| GF109203X | PKC, Ste20-related kinase, adenosine kinase, quinine reductase type 2, voltage-dependent sodium channels, and the 5-HT 3 receptor | SDS-PAGE separation, MS, immunoprecipitation | Cancers, heart failure |
| Bisindolylmaleimide-III | PKC-α, GSK3-β, CaMKII, adenosine kinase, CDK2, quinine reductase type 2, PKAC-R, prohibitin, VDAC, and heme binding proteins | Mass spectrometry, affinity chromatography | Cancers, heart failure |
| SU6668 | β-PDGFR, VEGFR2, FGFR, Yes and Lyn, RSK3, AMPKα1, and ULK3 | 16-BAC/SDS-PAGE, MS, immunofluorescence | Cancer |
| Thalidomide | bFGF, CRBN | Immunofluorescence, immunoblotting, and ELISA | Myeloma, erythema nodosumleprosum, and leprosy |
| Arsenic trioxide | CDK6, cdc2, cyclin A, PML-RARa and PML | Western blotting, immunoprecipitation | Acute promyelocytic leukemia |
PDGFR, platelet-derived growth factor receptor; FGFR, fibroblast growth factor receptor; RICK, Rip-like interacting caspase-like apoptosis-regulatory protein kinase; CK1, Casein kinase 1; GAK, cyclin G-associated kinase; PKNβ, protein kinase N beta; JAK1, Janus kinase 1; NQO2, quinone oxidoreductase 2; DDR1, discoidin domain receptor-1; BTK, Bruton's tyrosine kinase; CAMK2G, calcium/calmodulin-dependent protein kinase type II gamma chain; CDKs, cyclin-dependent kinases; PDXK, pyridoxal kinase; PKC, protein kinase C; VDAC, voltage-dependent anion channel; AMPKα1, the AMP-activated protein kinase α1; ULK3, Unc-51-like kinase 3; bFGF, basic fibroblast growth factor; CRBN, cereblon.
Aberrant phosphorylation of protein kinases cause a wide range of diseases, from cancers to autoimmune disorders[37],[38]. Protein kinases are currently a prevalent target of drug development. With a growing demand for potent drugs to fight such widespread and debilitating diseases, dozens of kinase inhibitors have entered clinical trials in recent years, and many newly developed oral drugs have been used in clinics. As most of these inhibitors target the conserved ATP-binding pocket, specificity has become a severe bottleneck compromising the clinical application of these drugs. Chemical proteomics is now widely applied to identify possible targets of kinase inhibitors through affinity purification from cellular extracts, resulting in the identification of the whole spectrum of potential drug targets and providing a strong foundation for developing novel potent drugs that exert no side effects.
Although conventional high-throughput approaches are informative, they still have several limitations. Because of these limitations, chemical proteomics is receiving increasing attention recently and has been primarily used in the kinase research community. Moreover, because chemical proteomics can be applied directly to detect diseased cell extracts, the study of specific kinase inhibition mechanisms will be more straightforward and reliable. For instance, pyrido [2,3-d] pyrimidine is intensively used to examine related kinase targets by chemical proteomics. Wissing et al.[39] devised an advanced experimental procedure using chemical proteomics to explore the target profile of pyrido [2,3-d] pyrimidine. In detail, they immobilized pyrido[2,3-d] pyrimidine as a detector and conclusively identified more than 30 human protein kinase targets. In addition to the already known targets, including Src, platelet-derived growth factor receptor (PDGFR) and fibroblast growth factor receptor (FGFR)[40],[41], they found that serine/threonine kinases Rip-like interacting caspase-like apoptosis-regulatory protein kinase (RICK) and p38α could also potently interact with pyrido[2,3-d]pyrimidine. They also investigated SB 203580, another kinase inhibitor of p38[42]. Likewise, they immobilized SB 203580 in combination with several common approaches, such as MS, and identified new targets, including RICK, CK1, GAK, protein kinase Nβ (PKNβ), and Janus kinase 1 (JAK1). Their analysis suggested that RICK interacts more strongly with SB 203580 than GAK and CK1 do.
In addition, significant efforts have been exerted on identifying the well-known BCR-ABL tyrosine kinase inhibitor, imatinib, the second-generation drugs, nilotinib and dasatinib, and the dual SRC/ABL inhibitor, bosutinib, which are all used in clinics to treat chronic myeloid leukemia. Rix et al.[33] used affinity matrices in combination with liquid chromatography electrospray ionization tandem MS (LC-ESI-MS/MS) to identify the target profiles of imatinib, nilotinib, and dasatinib in K562 and CML primary cells. The data showed that, in addition to the known major targets of imatinib such as ABL, KIT, and PDGFR, an unexpected compound NAD(P)H:quinone oxidoreductase NQO2 also demonstrated a prominent interaction with imatinib in both K562 cells and CML patient samples. Moreover, a recent study using ELISA and western blotting showed that macrophage colony-stimulating factor receptor c-fms, which is a crucial regulator in the growth and differentiation of the monocyte-macrophage lineage, acted as a novel target of imatinib[43]. Unexpectedly, western blot analysis suggested that imatinib failed to influence c-fms protein expression but did inhibit c-fms phosphorylation, leading to inactivation of c-fms. Their results showed that c-ABL and BCR-ABL acted as interactors of nilotinib, and they further validated ARG (ABL2), NQO2, and the receptor tyrosine kinase DDR1 (discoidin domain receptor 1)[44] as significant targets of nilotinib. Researchers have previously used chemical proteomics to detect partial targets of nilotinib, such as BCR-ABL, c-ABL, c-KIT, and PDGFR[45],[46]. However, a current study performed by generation of comprehensive drug-protein interaction profiles with chemical proteomic assays showed that dasatinib targeted a wide range of kinases in vitro. Rix et al.[33] identified 24 kinase targets of dasatinib and found that c-ABL, BCR-ABL, and DDR1 were significantly interacted with dasatinib, as expected. In addition, other potent proteins were found to bind with dasatinib. These proteins were confirmed to include the TEC family kinases, BTK and TEC, by LC-MS/MS using total K562 cell lysates[47]. Overall, dasatinib was suggested to be much more promiscuous than the slightly selective kinase inhibitors imatinib and nilotinib. It has been shown that bosutinib, an anti-leukemia drug similar to dasatinib, also has a broad profile of kinase targets. Upon immobilizing c-bosutinib on N-hydroxyl succinimide (NHS)-activated Sepharose 4 Fast Flow combined with MS, Rix et al.[48] found that c-bosutinib not only effectively targets the ABL and SRC family kinases but also inhibits the STE and TEC family kinases. Interestingly, these researchers also found that CAMK2G, a Ca2+/calmodulin-dependent protein kinase involved in the proliferation of myeloid leukemia cells[49], was not a kinase target inhibited by dasatinib, but rather displays a specific affinity toward the kinase inhibitor, bosutinib. Therefore, dasatinib[47] and bosutinib[48],[50], unlike nilotinib or imatinib, might cause unpredicted side effects, resulting in the limited application of both drugs in clinic.
(R)-roscovitine (CYC202), a CDK inhibitor, was tested with regard to its kinase target profile by combining chemical proteomics with a wide panel of purified kinases. As a result of these efforts, its target database now includes 151 kinases. Further characterization using electrophoresis and western blotting demonstrated that instead of selectively inhibiting CDKs, (R)-roscovitine can target a non-protein kinase, pyridoxal kinase (PDXK), which catalyzes the phosphorylation and activation of vitamin B6. The wide range of potential targets for CYC202 renders it impossible to enter into clinical treatment because of its potential side effects, and chemical proteomics should be widely used to study the novel targets of CYC202.
Another frequently targeted kinase family, the serine/threonine type protein kinase C (PKC) family[51], is a critical regulator involved in the activation of cellular functions and proliferation[52]. Bisindolylmaleimides, especially GF109203X, a commonly used inhibitor of PKC, have made a prominent contribution to the study of PKC biological activities and its related diseases. However, the large number of GF109203X targets is an obstacle in PKC research. Recently, Brehmer et al.[31] performed a study in which they immobilized bisindolylmaleimides III, VIII, and X to identify related kinase targets in COS-7 and HeLa cells. Subsequent analysis confirmed that, along with the protein kinases CDK and Ste20-related kinase, several non-protein kinases, including adenosine kinase and quinone reductase type-2, act as novel targets of bisindolylmaleimide inhibitors. As voltage-dependent sodium channels and the 5-hydroxytryptamine 3 receptor have been identified as potent targets of GF109203X[53],[54], the study of PKC enters a new era.
Several receptor tyrosine kinases (RTKs), such as vascular endothelial growth factor receptor (VEGFR), PDGFR, and FGFR also play a critical role in endothelial cell growth and angiogenesis[55],[56]. An anti-angiogenic small molecule drug, SU6668, has been used to study the mechanism of vascularization, but its wide range of targets makes these investigations difficult. SU6668 shows a high affinity towards βPDGFR and VEGFR2 but a low level of inhibition of FGFR[57]. More recently, Godl et al.[58] immobilized SU6668 in combination with chemical proteomic methods and discovered several significant targets. Interestingly, in addition to tyrosine kinases such as the Src-family members Yes and Lyn, these researchers unexpectedly identified various Ser/Thr kinase targets, including TBK1, two Aurora kinases (RSK3, AMPKa1), and ULK3. These results indicate that the Ser/Thr kinase is potentially important, and additional targets should be identified so that the side effects and molecular actions of the drugs can be clearly delineated.
In addition to kinase inhibitors, natural products are another focus of chemical proteomic research, given that the majority of newly discovered natural products are subjected to limited biological activity testing, mostly focusing on antibacterial or anticancer activity. Thus, the potential of most natural products as drugs or unique and specific tool compounds for the perturbation of biological pathways still remains largely untapped. For these reasons, using natural products in chemical proteomic experiments is a highly worthwhile and pioneering effort. A series of studies have been conducted to profile the targets of a variety of natural products such as ovalicin, myriocin, FR182877, and the macrolide pladienolide[30].
Recent Advances in Chemical Proteomics
Using specific chemical probes to survey the activities of certain classes of proteins has entered a rapidly evolutionary area of proteomics. These delicately designed probes can be used for multi-aspect proteomics, mainly in protein expression analysis and identification as well as cellular localization and regulation. During the last few years, the application of probes has become a key focus of pharmaceutic companies for novel drug development. However, designing probes of high efficiency and specificity is a primary challenge pharmaceutic companies are confronted with. Hence, it is not surprising that each new drug candidate that enters the clinic takes 250 full-time employee years and $70 million for drug manufacturers to develop.
Trifunctional probes were used to identify several protein targets involved in some pivotal cell processes. For example, cysteine proteases have been reported to play an essential role in apoptosis, cataract formation, and malarial infection, and selective probes have been designed to target this group of proteins. Other probes have been used to profile proteins involved in many physiologic and pathologic processes, such as tissue remodeling, peptide hormone signaling, and cancer. Metalloproteases (MPs), which also belong to the protease family and mainly include matrix metalloproteinases (MMPs) and angiotensin-converting enzymes (ACE)[59], are activated in various physiologic processes, such as blood pressure regulation and tissue modeling. As a result, numerous researchers are focusing on studying and designing specific probes against MPs in diseases including arthritis[60], Alzheimer's disease[61], cancer[62], and heart disease[63]. In contrast to common probes that possess electrophilic reactive groups that can label the conserved active site nucleophiles of specific classes of these enzymes, such as the serine and cysteine proteases and subunits of the proteasome, MPs use a zinc-activated water molecule for catalysis. Many selective inhibitors designed for MPs are substrate-based analogs that contain zinc-binding groups (ZBGs) such as sulfhydryls, formylhydrazines, aminocarboxylates, and the preferable hydroxamic acids[64]. Chan et al.[65] have already validated the high affinity of these probes to MPs through an experiment using peptide-based hydroxamate inhibitors in combination with chemical proteomic methods. They ultimately found that the designed probes targeted a broad profile of MPs. To describe the activity and inhibitor sensitivity of MPs in cell and tissue proteomes, Saghatelian et al.[21] further applied specific probes by coupling a zinc-chelating hydroxamate to a benzophenone photo-crosslinker. They confirmed the identification of highly up-regulated MPs in invasive cancer cells and the detection of MP inhibitor targets that are currently in clinical trials[21]. Trifunctional chemical probes were also applied to the detection and identification of enzyme activities from complex proteomes[66]. Two types of activity-based probes are widely used in chemical proteomics: fluorescent probes for the rapid identification of certain targets and biotinylated probes for target purification and detection. Recently, Gregory et al.[67] synthesized and applied both potent probes. They used biotinylated probes to purify sulfonate-reactive proteins and then identified them by MS methods. As a result, they detected several labeled protein activities for which molecular identities were sought, indicating that the combination of two probes helped identify targets in activity-based proteomic experiments.
As the most momentous and attractive regulators involved in various biological activity, the identification of protein kinases and protein phosphatases by chemical probes has received increasing attention. The protein kinase family, which consists of more than 500 members, includes crucial regulators of diverse cellular functions. However, due to the specificities and complexities of kinase structure, the detection of kinases by selective probes has not been effective. On the contrary, the protein phosphatase family is especially appropriate for testing due to its high affinity to type I reactive groups on chemical probes, which is due to the presence of nucleophilic groups on phosphatase cysteine residues[14]. However, the development of potent probes for phosphatases is still at the primary stages.
Chemical probes may be useful for the rapid development of novel drug targets and may result in the accelerated development of corresponding new drugs. However, recent studies have reported that only 122 of 483 known drug targets have been targeted by orally available small-molecule inhibitors that are marketed to treat human diseases. Today, it is still imperative to design efficient probes for drug target profiling to enable the rapid development of new drugs.
Conclusion and Future Perspectives
Chemical proteomics is an indispensable technology that is useful in clinical testing and drug development. In particular, the identification of kinase targets by chemical proteomics has been intensively investigated in various diseases, ranging from cancer to autoimmune disorders[37],[38], further highlighting the potential clinical significance of this platform. Recently, chemical proteomics has been applied both in vitro and in vivo to profile potential novel drug targets for a more precise understanding of drug side effects and drug resistance under certain disease states. Traditional global proteomic approaches are usually applied to assess the interactions of an unmodified drug with its endogenous targets without bias. However, this approach is rarely used to directly identify the drug-protein interactions. This deficiency in the field has been overcome by two novel chemical proteomic approaches developed by modern scientists. The CCCP method focuses on identifying the interacting proteins; thus, there is no requirement for the reactive groups to covalently attach to proteins. Therefore, the trifunctional probes used in CCCP, which can directly assess enzyme activity, provide us with the most immediate and refined information of the biological targets. On the other hand, compared with CCCP, ABPP can potently characterize the specific target classes and is therefore a recognized weapon in drug discovery. However, the design of trifunctional probes requires a significant amount of time and money. Although each of these three technologies is suitable for a limited range of conditions, they could be used in a wider context when combined with other technologies. It is anticipated that more effective approaches can be established so that, in combination with chemical proteomics, the profiling of novel drug targets will become more feasible and reliable, thus rendering future drugs more efficient and less toxic.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (No. 2011CB910703), the National 863 High Tech Foundation (No. 2007AA021205), and Chinese NSFC (No. 81072022).
References
- 1.Ito T, Ando H, Suzuki T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Templin MF, Stoll D, Schrenk M, et al. Protein microarray technology. Trends Biotechnol. 2002;20:160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- 4.Veenstra TD. Proteomic approaches in drug discovery. Drug Discov Today. 2007;3:433–440. [Google Scholar]
- 5.Chaurand P, Stoeckli M, Caprioli RM. Direct profiling of proteins in biological tissue sections by MALDI mass spectrometry. Anal Chem. 1999;71:5263–5270. doi: 10.1021/ac990781q. [DOI] [PubMed] [Google Scholar]
- 6.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ryan TE, Patterson SD. Proteomics: drug target discovery on an industrial scale. Trends Biotechnol. 2002;20:S45–S51. doi: 10.1016/s1471-1931(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 10.Holly MK, Dear JW, Hu X, et al. Biomarker and drug-target discovery using proteomics in a new rat model of sepsis-induced acute renal failure. Kidney Int. 2006;70:496–506. doi: 10.1038/sj.ki.5001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AY, Paweletz CP, Pollock RM, et al. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J Proteome Res. 2008;7:5177–5186. doi: 10.1021/pr800510p. [DOI] [PubMed] [Google Scholar]
- 12.Liang X, Hajivandi M, Veach D, et al. Quantification of change in phosphorylation of BCR-ABL kinase and its substrates in response to imatinib treatment in human chronic myelogenous leukemia cells. Proteomics. 2006;6:4554–4564. doi: 10.1002/pmic.200600109. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Li Z, Bai S, et al. Mechanism of cancer cell adaptation to metabolic stress: proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic siganling pathway. Mol Cell Proteomics. 2009;8:70–85. doi: 10.1074/mcp.M800195-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Jeffery DA, Bogyo M. Chemical proteomics and its application to drug discovery. Curr Opin Biotechnol. 2003;14:87–95. doi: 10.1016/s0958-1669(02)00010-1. [DOI] [PubMed] [Google Scholar]
- 15.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, et al. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chem Biol. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- 16.Bogyo M, McMaster JS, Gaczynska M, et al. Covalent modification of the active site threonine of proteasomal β subunits and the Escherichia coli homolog HsIV by a new class of inhibitors. Proc Natl Acad Sci USA. 1997;94:6629–6634. doi: 10.1073/pnas.94.13.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YS, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornberry NA, Peterson EP, Zhao JJ, et al. Inactivation of interleukin-1.beta. converting enzyme by peptide (acyloxy) methyl ketones. Biochemistry. 1994;33:3934–3940. doi: 10.1021/bi00179a020. [DOI] [PubMed] [Google Scholar]
- 19.Faleiro L, Kobayashi R, Fearnhead H, et al. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goshe MB, Smith RD. Stable isotope-coded proteomic mass spectrometry. Curr Opin Biotechnol. 2003;14:101–109. doi: 10.1016/s0958-1669(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Saghatelian A, Jessani N, Joseph A, et al. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci USA. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulick MG, Bogyo M. Application of activity-based probes to the study of enzymes involved in cancer progression. Curr Opin Genet Dev. 2008;18:97–106. doi: 10.1016/j.gde.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson NE, Foley KM, Stalder ES, et al. Identification, production, and use of polyol-responsive monoclonal antibodies for immunoaffinity chromatography. Methods Enzymol. 2009;463:475–94. doi: 10.1016/S0076-6879(09)63028-7. [DOI] [PubMed] [Google Scholar]
- 24.Harding MW, Galat A, Uehling DE, et al. A receptor for the immuno-suppressant FK 506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 25.Crews CM, Collins JL, Lane WS, et al. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J Biol Chem. 1994;269:15411–15414. [PubMed] [Google Scholar]
- 26.Knockaert M, Gray N, Damiens E, et al. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chem Biol. 2000;7:411–422. doi: 10.1016/s1074-5521(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 27.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 28.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 29.Oda Y, Huang K, Cross FR, et al. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 31.Brehmer D, Godl K, Zech B, et al. Proteome-wide identification of cellular targets affected by bisindolylmaleimide-type protein kinase C inhibitors. Mol Cell Proteomics. 2004;3:490–500. doi: 10.1074/mcp.M300139-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Bach S, Knockaert M, Reinhardt J, et al. Roscovitine targets, protein kinases and pyridoxal kinase. J Biol Chem. 2005;280:31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 33.Rix U, Hantschel O, Dürnberger G, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib, reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 34.Aye TT, Mohammed S, van den Toorn HW, et al. Selectivity in enrichment of cAMP-dependent protein kinase regulatory subunits type I and type II and their interactors using modified cAMP affinity resins. Mol Cell Proteomics. 2009;8:1016–1028. doi: 10.1074/mcp.M800226-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ong SE, Schenone M, Margolin AA, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YX, Knyazev PG, Cheburkin YV, et al. AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 2008;68:1905–1915. doi: 10.1158/0008-5472.CAN-07-2661. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghoreschi K, Laurence A, O'Shea JJ. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nat Immunol. 2009;10:356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wissing J, Godl K, Brehmer D, et al. Chemical proteomic analysis reveals alternative modes of action for pyrido [2,3-d] pyrimidine kinase inhibitors. Mol Cell Proteomics. 2004;3:1181–1193. doi: 10.1074/mcp.M400124-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Panek RL, Lu GH, Klutchko SR, et al. In vitro pharmacological characterization of PD 166285, a new nanomolar potent and broadly active protein tyrosine kinase inhibitor. J Pharmacol Exp Ther. 1997;283:1433–1444. [PubMed] [Google Scholar]
- 41.Klutchko SR, Hamby JM, Boschelli DH, et al. 2-substituted aminopyrido [2,3-d]pyrimidin-7(8H) ones. Structure-activity relationships against selected tyrosine kinases and in vitro and in vivo anticancer activity. J Med Chem. 1998;41:3276–3292. doi: 10.1021/jm9802259. [DOI] [PubMed] [Google Scholar]
- 42.Godl K, Wissing J, Kurtenbach A, et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dewar AL, Cambareri AC, Zannettino AC, et al. Macrophage colony-stimulating factor receptor c-fms is a novel target of imatinib. Blood. 2005;105:3127–3132. doi: 10.1182/blood-2004-10-3967. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JD, Edman JC, Rutter WJ. A receptor tyrosine kinase found in breast carcinoma cells has an extracellular discoidin I-like domain. Proc Natl Acad Sci USA. 1993;90:5677–5681. doi: 10.1073/pnas.90.12.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter TA, Wodicka LM, Shah NP, et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabian MA, Biggs WH, Treiber DK, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 47.Hantschel O, Rix U, Schmidt U, et al. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci USA. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Remsing Rix LL, Rix U, Colinge J, et al. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–485. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 49.Si JT, Collins SJ. Activated Ca2+/calmodulin-dependent protein kinase II gamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 2008;68:3733–3742. doi: 10.1158/0008-5472.CAN-07-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bantscheff M, Eberhard D, Abraham Y, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 51.Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- 52.Cohen S, Braiman A, Shubinsky G, et al. Protein kinase C-theta in platelet activation. FEBS Lett. 2011;585:3208–3215. doi: 10.1016/j.febslet.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 53.Coultrap SJ, Sun H, Tenner TE, et al. Competitive antagonism of the mouse 5-hydroxytryptamine3 receptor by bisindolylmaleimide I, a “selective” protein kinase C inhibitor. J Pharmacol Exp Ther. 1999;290:76–82. [PubMed] [Google Scholar]
- 54.Lingameneni R, Vysotskaya TN, Duch DS, et al. Inhibition of voltage-dependent sodium channels by Ro 31-8220, a “specific” protein kinase C inhibitor. FEBS J. 2000;473:265–268. doi: 10.1016/s0014-5793(00)01532-5. [DOI] [PubMed] [Google Scholar]
- 55.Bergers G, Song S, Meyer-Morse N, et al. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111:1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciardiello F, Caputo R, Bianco R, et al. Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor. Clin Cancer Res. 2001;7:1459–1465. [PubMed] [Google Scholar]
- 57.Laird AD, Vajkoczy P, Shawver LK, et al. SU6668 is a potent antiangiogenic and antitumor agent that induces regression of established tumors. Cancer Res. 2000;60:4152–4160. [PubMed] [Google Scholar]
- 58.Godl K, Gruss OJ, Eickhoff J, et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 2005;65:6919–6926. doi: 10.1158/0008-5472.CAN-05-0574. [DOI] [PubMed] [Google Scholar]
- 59.Bode W, Maskos K. Structural basis of the matrix metalloproteinases and their physiological inhibitors, the tissue inhibitors of metalloproteinases. Biol Chem. 2003;384:863–872. doi: 10.1515/BC.2003.097. [DOI] [PubMed] [Google Scholar]
- 60.Martel-Pelletier J, Welsch DJ, Pelletier JP. Metalloproteases and inhibitors in arthritic diseases. Best Pract Res Clin Rheumatol. 2001;15:805–829. doi: 10.1053/berh.2001.0195. [DOI] [PubMed] [Google Scholar]
- 61.Hooper NM, Turner AJ. The search for alpha-secretase and its potential as a therapeutic approach to Alzheimer's disease. Curr Med Chem. 2002;9:1107–1119. doi: 10.2174/0929867023370121. [DOI] [PubMed] [Google Scholar]
- 62.Vihinen P, Kahari VM. Matrix metalloproteinases in cancer: prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 63.Sierevogel MJ, Pasterkamp G, de Kleijn DP, et al. Matrix metalloproteinases: a therapeutic target in cardiovascular disease. Curr Pharm Des. 2003;9:1033–1040. doi: 10.2174/1381612033455099. [DOI] [PubMed] [Google Scholar]
- 64.Skiles JW, Gonnella NC, Jeng AY. The design, structure, and therapeutic application of matrix metalloproteinase inhibitors. Curr Med Chem. 2001;8:425–474. doi: 10.2174/0929867013373417. [DOI] [PubMed] [Google Scholar]
- 65.Chan EW, Chattopadhaya S, Panicker RC, et al. Developing photoactive affinity probes for proteomic profiling: hydroxamate-based probes for metalloproteases. J Am Chem Soc. 2004;126:14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- 66.Adam GC, Sorensen EJ, Cravatt BF. Trifunctional chemical probes for the consolidated detection and identification of enzyme activities from complex proteomes. Mol Cell Proteomics. 2002;1:828–835. doi: 10.1074/mcp.t200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 67.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]