Abstract
It is generally accepted that the inactive P420 form of cytochrome P450 (CYP) involves the protonation of the native cysteine thiolate to form a neutral thiol heme ligand. On the other hand, it has also been suggested that recruitment of a histidine to replace the native cysteine thiolate ligand might underlie the P450→P420 transition. Here we discuss resonance Raman investigations of the H93G myoglobin (Mb) mutant in the presence of tetrahydrothiophene (THT) or cyclopentathiol (CPSH), and on pressure-induced cytochrome P420cam (CYP101), that show a histidine becomes the heme ligand upon CO binding. The Raman mode near 220 cm−1, normally associated with the Fe-histidine vibration in heme proteins, is not observed in either reduced P420cam or the reduced H93G Mb samples, indicating that histidine is not the ligand in the reduced state. The absence of a mode near 220 cm−1 is also inconsistent with a generalization of the suggestion that the 221 cm−1 Raman mode, observed in the P420-CO photoproduct of inducible nitric oxide synthase (iNOS), arises from a thiol-bound ferrous heme. This leads us to assign the 218 cm−1 mode observed in the 10 ns P420cam-CO photoproduct Raman spectrum to a Fe-histidine vibration, in analogy to many other histidine bound heme systems. Additionally, the inverse correlation plots of the νFe-His and νCO frequencies for the CO adducts of P420cam and the H93G analogs provide supporting evidence that histidine is the heme ligand in the P420-CO bound state. We conclude that, when CO binds to the ferrous P420 state, a histidine ligand is recruited as the heme ligand. The common existence of a HXC-Fe motif in many CYP systems allows the C→H ligand switch to occur with only minor conformational changes. One suggested conformation of P420-CO involves the addition of another turn in the proximal L helix so that, when the protonated Cys ligand is dissociated from the heme, it can become part of the helix and the heme is ligated by the His residue from the adjoining loop region. In other systems, such as iNOS and CYP3A4 (where the HXC-Fe motif is not found) a somewhat larger conformational change would be necessary to recuit a nearby histidine.
Keywords: Cytochrome P450, cytochrome P420, H93G, myoglobin, resonance Raman, ligand switch
Introduction
The cytochrome P450 enzyme family (CYP) is composed of a broad range of heme-containing proteins that are involved in drug metabolism, toxicity, xenobiotic degradation, and biosynthesis1. One key structural feature of these proteins is the coordination of the thiolate anion of cysteine (Cys) to the heme iron as the fifth ligand in the active P450 form2-5. The biologically inactive conformation of a cytochrome P450 protein is typically denoted as the “P420” form and is characterized by a CO bound Soret peak near 420 nm, which is blue-shifted with respect to the peak at ~450 nm found in the active cytochrome P450. The inactive conformation can be formed from all known types of P450 using various methods6-10. It has recently been accepted that this spectral change is due to protonation of the cysteine thiolate, resulting in thiol ligation to the heme iron10-12. On the other hand, it has also been suggested that the P450 → P420 transition involves a ligand switch from a cysteine (Cys) to a histidine (His) ligated heme13. The similarity of the absorption spectra between the P420 form of cytochrome P450 and other proximal histidine ligated heme proteins14 reinforces such a correlation. Wells et al.13 provided key evidence of histidine ligation in the CO bound form of P420 by observing a strong νFe-His mode at 218 cm−1 in the 10 ns transient Raman spectra of the P420cam-CO photoproduct with an intensity equivalent to that of MbCO. Moreover, the equilibrium resonance Raman spectrum of the P420cam-CO adduct is virtually identical to that of MbCO, lending further support to the histidine ligation model, at least when CO is bound.
On the other hand, heme model compound studies4, 5 along with spectroscopic comparisons between chloroperoxidase and cytochrome P420cam have led to suggestions12 that a thiolate-thiol transition might accompany reduction of the ferric P420 (even though a residual low-spin thiolate population is also observed12). More recently, Perera et al.11 showed that the proximal ligand mutant (H93G) of deoxy myoglobin (Mb) can bind thiol and thioether compounds with high affinity, Kd ~ 10 μM. This conclusion is based on the observation of changes in the absorption spectra of the reduced H93G Mb mutant upon titration with tetrahydrothiophene (THT) and cyclopentathiol (CPSH)11. However, it must be pointed out that the changes of the absorption spectra upon ligand binding are quite small and this could be due to either direct heme ligation or to perturbations of the electrostatic environment surrounding the heme15, 16. The observed optical changes are not large enough to provide unambiguous evidence for heme ligation by either THT or CPSH.
Recently, Sabat et al.10 suggested that thiol could be the proximal heme axial ligand in the inactive (P420) form of inducible nitric oxide synthase (iNOS). This protein is analogous to the P450 class, based upon thiolate ligation to the heme in its active state. The conclusion to exclude histidine as the proximal ligand in the 5 ns transient Raman spectra of the CO adduct was based on the absence of the expected ~1 cm−1 H/D isotopic shift for the 221 cm−1 mode, which is usually assigned to the Fe-His vibration17-22. Based on the absence of a H/D isotopic shift, it was concluded that the 221 cm−1 mode observed in the inactive iNOS-CO photoproduct spectrum was neither a Fe-His nor a Fe-SH stretching mode (the latter ligation state was expected to generate an even larger isotopic shift). As a result, Sabat et al. assigned the 221 cm−1 mode to a new mode associated with the thiol-ligated ferrous heme chromophore. However, if this assignment is correct, the 221 cm−1 heme mode should be observed in the thiol-bound reduced state of the inactive P420 iNOS and other reduced P420 systems or analogs. Because the Raman spectrum of reduced P420 iNOS is not available10, we turned to the Raman spectrum of reduced P420cam to try and find the predicted thiol-bound heme mode at 221 cm−1. However, the Raman spectra of reduced P420cam from either this or a prior13 study does not reveal the presence of such a mode.
Therefore, in order to resolve the various characterizations of the heme proximal ligand in P420 systems, we have used resonance Raman spectroscopy to study the CO bound and ferrous forms of P420cam and H93G Mb; the latter, in the presence and absence of THT and CPSH. There is no 221 cm−1 mode observed in ferrous P420cam or in the reduced H93G Mb with or without the THT or CPSH ligands. Moreover, it is also noteworthy that the experiments show no difference, within the resolution of ±1cm−1, between the resonance Raman spectra of reduced H93G Mb with or without the THT and CPSH. These observations do not yield supportive evidence that either THT nor CPSH directly ligates the heme iron of the reduced H93G Mb. Additional experiments are presented that probe the inverse correlation of the νFe-CO and νCO Raman modes and are consistent with histidine as the proximal heme ligand in CO bound P420. As discussed below, we conclude that the ~220 cm−1 modes observed in P420-CO, iNOS-CO, and H93G-CO photoproduct Raman spectra are, in fact, signatures of Fe-His ligation in the CO bound complexes.
We also construct a kinetic scheme that describes the photostationary states of P420-CO and H93G-CO and accounts well for the various Raman observations consistent with previously determined rate constants. A fundamental hypothesis underlying the kinetic model is that CO binding to heme greatly increases the “acidity” of the ferrous iron atom so that it efficiently recruits the strong sigma donating histidine ligand. Earlier work has shown that the affinity of CO bound heme for imidazole is ~105 times larger than its affinity for a weak ligand such as water23. This concept has been used previously, both in the context of low pH MbCO ligation kinetics24 and in prior studies of ligand switching in H93G-CO25, 26; it is extended here to account for Raman observations in the P420 system.
Materials and Methods
All chemicals used in this study were purchased from Sigma Aldrich. Imidazole-free sperm whale H93G myoglobin was prepared as previously described11, 27. High purity P420cam was prepared in the absence of camphor by pressure treatment of P450cam as previously reported8, 9, 12. We have shown in previous studies that high hydrostatic pressure will dissociate bound substrate28 and that the pressures required to initiate P420 inactivation are less in the absence of camphor. Thus, in order to generate a clean sample of P420, the substrate free form of the protein was used. The reduced H93G Mb and P420cam samples were prepared in 0.05 M KPi pH 7.0 buffer and the protein concentrations were adjusted to 100 μM. A small amount of saturated sodium dithionite solution (1% by sample volume) was used to reduce the samples. In order to prepare the samples of H93G Mb with thiol or thioether, either 20 mM of tetrahydrothiophene (THT) or cyclopentanethiol (CPSH) in ethanol stock solution was added to the reduced H93G Mb solution11. The final concentration of THT and CPSH in the H93G Mb solution was 200 μM, which should lead to >95% binding with <1% bis-thiol formation, assuming the Kd=10 μm as taken from the work of Perera et al.11.
Resonance Raman spectra were obtained using a standard setup with 90° light-collection geometry and a single grating monochromator model SP-2500i, Princeton Instruments, Acton, MA. An optical polarization scrambler was inserted in front of the monochromator to obtain the intensity of the scattered light without bias from the polarization-sensitive grating. The monochromator output was coupled to a thermoelectrically cooled charge-coupled detector (PIXIS 400B, Princeton Instruments). To improve detection in the low-frequency region of Raman shifts, an interferometric notch filter (Kaiser Optical Systems, Ann Arbor, MI) was used to extinguish the elastically and quasi-elastically scattered laser light. Samples were excited with a 413.1 nm laser line generated by a krypton laser (Innova 300, Coherent) using a power of 11 mW at the sample or with a 442 nm laser line from a HeCd laser (Melles Griot) at powers up to 32 mW. In order to study the photolabile CO adducts, a cylindrical quartz cell with 10 mm diameter was mounted to a home-built spinning system and used for the Raman measurement. The spinning speed was set at 6000 rpm for all experiments except static measurements. All Raman spectra were frequency calibrated using pure fenchone with ~ 1cm−1 spectral resolution.
Results
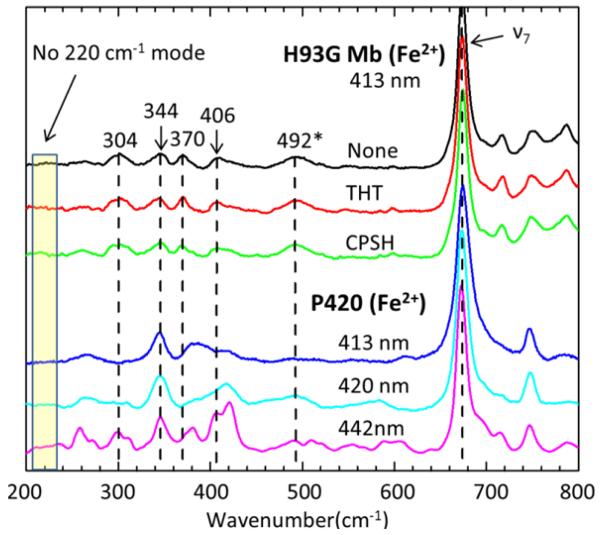
Resonance Raman spectra of deoxy H93G and its THT and CPSH adducts are compared with ferrous P420cam in Fig. 1. The corresponding high frequency region (1300 cm−1 - 1700 cm−1) region, including the ν4, ν3, ν2 and ν10 bands, is shown in Fig. S2 of the Supporting Information. All spectra in Fig. 1 are normalized to the ν7 band. No differences are observed in the Raman spectra of reduced H93G Mb, H93G(THT) Mb and H93G(CPSH) Mb, although the absorption spectra in the Soret region show subtle, but clear, differences11 (see also Fig. S1, Supporting Information). The concentrations of THT and CPSH used in these measurements should lead to ~ 95% binding according to Kd =10 μM as reported by Perera R. et al.11. Based on prior Raman studies of ligand binding to the H93G Mb mutant29, we expect to observe changes in the resonance Raman spectra when ligand binding to the heme takes place. Given the fact that there is no observable change in the resonance Raman spectra upon THT or CPSH binding, it seems possible that these thioether and thiol compounds might be binding to a site in the H93G protein that is close enough to affect the Soret band shape and position (~2 nm shift), but perhaps they are not replacing the heme water ligand that is normally present in reduced H93G Mb.
Figure 1.
Low frequency resonance Raman spectra of reduced H93G Mb, its THT and CPSH adducts, and ferrous P420cam. The excitation wavelength is 413 nm and the laser power at the sample is 11mw for the H93G samples, while for the P420 sample the excitation is 20mw at 413nm (blue), 25 mW at 420 nm (cyan) and 32mw at 442 nm (magenta). The sample cell is spinning at 6000 rpm. All spectra are normalized to the ν7 band. The 492 cm−1 feature also appears in Fig. 3 and is not assigned to a νFeCO mode.
The Raman spectra of reduced P420cam and the H93G derivatives also show no evidence of a mode near ~220 cm−1. Sabat et al. 10 observed a Raman mode at 221 cm−1 in the 5 ns photoproduct Raman spectrum of the inactive iNOS P420-CO adduct and, because a H/D isotopic shift was not detected, they assigned the 221 cm−1 mode to a heme vibration activated by thiol ligation. However, if thiol is ligated to the heme in the reduced state, one would expect this mode to be present in the equilibrium Raman spectrum of the reduced P420 sample (i.e., the equilibrium species should display essentially the same modes, although slightly shifted, when compared to the 5 ns transient photoproduct species). Note that the P420 sample was probed at several wavelengths (413 nm, 420 nm13, and 442 nm) and there is no evidence of a mode near 220 cm−1. The absence of a 221 cm−1 mode in the Raman spectrum of the equilibrium reduced P420cam and the H93G P420 analog samples does not support its assignment to a thiol-bound reduced heme mode10. On the other hand, if the usual assignment of this mode to the Fe-His vibration is made, its observation in the 10 ns transient Raman spectra provides strong evidence for a heme-histidine bond in the CO-bound forms of P42013 and, by analogy, iNOS10. Such an assignment is also consistent with previous transient Raman studies of the H93G-CO photoproduct26.
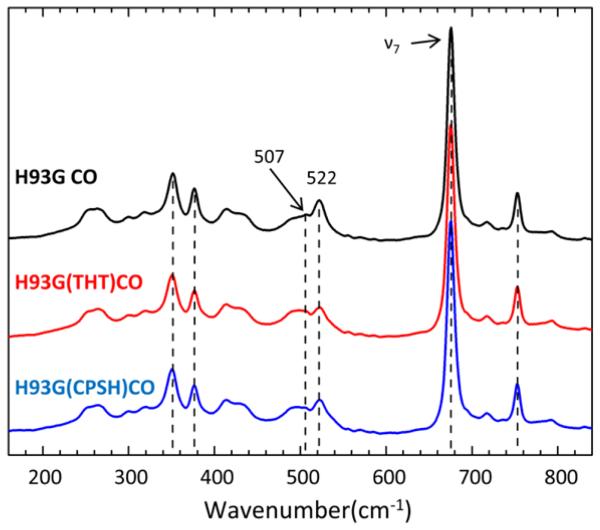
The low frequency resonance Raman spectra of CO bound H93G, H93G(CSPH), and H93G(THT) Mb, excited at 413 nm, are shown in Fig. 2. All spectra are normalized to the ν7 band. There is essentially no difference between the three samples, except for small changes in the relative amplitudes of the 507 cm−1 and 522 cm−1 modes, which are associated with the Fe-CO stretching frequency. The broad mode at 492 cm−1 seen in both Figs. 1 and 2 is not isotopically sensitive (Fig. 3) and therefore not assigned to a Fe-CO mode. The spectra of the corresponding high frequency region, including the ν4, ν3, ν2 and ν10 bands, are displayed in Fig. S3 of the Supporting Information and are identical for all three samples.
Figure 2.
Resonance Raman spectra of CO bound H93G, H93G (CSPH), and H93G (THT) excited at 413 nm. The incident laser power at the sample is 11mw. The sample is spinning at 6000 rpm. All spectra are normalized to the ν7 band.
Figure 3.
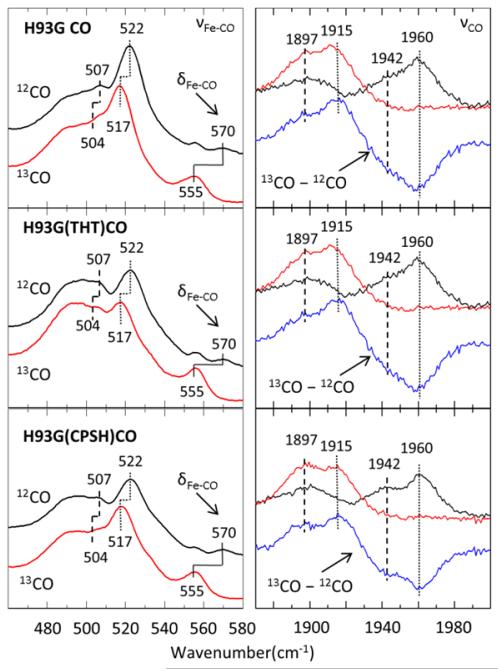
Resonance Raman spectra of the νFe-CO and νCO modes of 12CO and 13CO bound H93G, H93G (THT) and H93G (CPSH). The low frequency Fe-CO stretching and bending region is in the left panels. The high frequency CO stretching is in the right panels and the 13CO – 12CO difference is shown in blue. Excitation wavelength is 413 nm and laser power at the sample is 11mw. The sample was in a cell spinning at 6000 rpm. Dashed and dotted lines indicate νFeCO modes of H93G(His)CO and H93G(H2O)CO respectively.
Figure 3 shows the resonance Raman spectra for the νFe-CO and νCO modes of the 12CO (black) and 13CO (red) adducts of H93G, H93G(THT), and H93G(CPSH) Mb. The lower frequency Fe-CO stretching region is shown in the left panels, while the higher frequency CO stretching is displayed in the right panels. The isotopic shifts confirm that there are two peaks corresponding to the νCO stretching. For the 12CO sample there is a strong νFe-CO peak at 522 cm−1 that shifts to 517 cm−1 and a broader feature at 507 cm−1 where the shift is less obvious, but becomes apparent upon fitting the data as discussed in the Supporting Information (Fig. S4). The corresponding νCO stretching modes are located at 1960 cm−1 and 1942 cm−1, respectively. The νCO Raman frequencies agree very well with the infrared measurements reported previously25. The 507 cm−1 mode is associated with the histidine bound Fe-CO mode and its shift upon isotopic labeling is not so easily seen due to its breadth and weaker resonance enhancement. (Note that the Soret absorption band of CO bound heme is blue-shifted by approximately 5 nm when a weak ligand such as water replaces imidazole30. This means that, based on the Raman excitation profile of MbCO16, the resonance enhancement at 413 nm will favor the Fe-CO mode of the water bound heme relative to that of the histidine bound population). There is also interference from the broad feature near 492 cm−1, as seen in Fig. 1 and in the fit to the CO bound lineshape (Fig. S4). This peak has been previously reported for WT deoxyMb31, 32 and its H93G mutant25 and, since it shows no isotopic shift, it is not assigned to a Fe-CO mode. On the other hand, there is photolytic activity in the region near 507 cm−1 (vide infra), so we are confident that it represents the position of a Fe-CO oscillator. Upon 13CO substitution, the respective νCO modes, 1960 cm−1 and 1942 cm−1, show clear shifts down to 1915 cm−1 and 1897 cm−1, respectively.
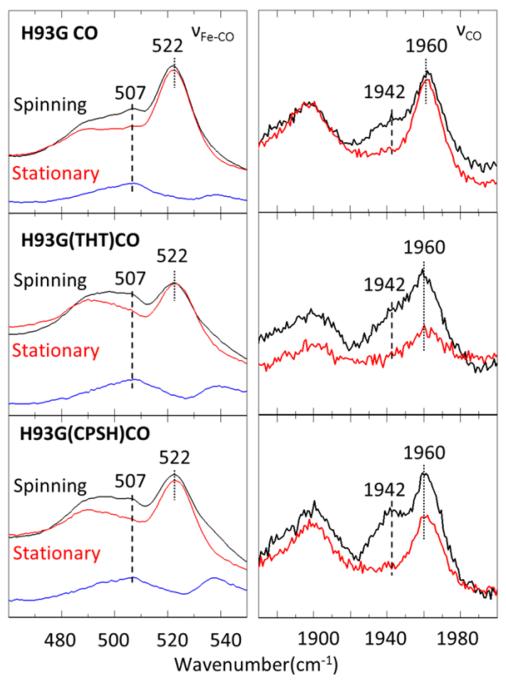
Figure 4 shows the resonance Raman spectra of the same samples as in Fig. 3, but it demonstrates the effect of photolysis when the spinning cell is stopped. The spectra in black have the sample in the quartz cell spinning at 6000 rpm, while the red spectra are accumulated in a static cell. The panels on the left side show the 460-550cm−1 νFe-CO stretching region, while the panels on the right side display the 1870~2000 cm−1 νCO stretching bands. As expected, the ν4 bands demonstrate that the ratio of the CO dissociated (5C) to the CO bound (6C) species is increased in the static cell (see Fig. S5 in Supporting Information). In the static-cell spectra, the intensities of the 507 cm−1 band and the 1942 cm−1 band decrease simultaneously relative to the 522 cm−1 and 1960 cm−1 bands. Thus, we associate the 507 cm−1 νFe-CO peak with the 1942 cm−1 νCO peak as belonging to the same FeCO species. A similar association holds for the 522 cm−1 and 1960 cm−1peaks. Moreover, the data indicate that the 507/1942 cm−1 FeCO species has a slower CO geminate rebinding rate (and thus a smaller relative population in the photostationary state of the static cell) than the species at 522 cm−1 and 1960 cm−1.
Figure 4.
Resonance Raman spectra of CO bound H93G with and without THT/CPSH. Spectra in black are taken with the sample spinning at 6000 rpm, while the red spectra are with a static cell. Panels on the left side show the 460-550cm−1 νFe-CO stretching band and the difference spectra in blue reveal the presence of the 507 cm−1 mode because of its increased photolysis compared to the 522 cm−1 species. Panels on the right side display the νCO stretching band and the analogous increased photolysis of the 1942 cm−1 species compared to the 1960 cm−1 species. The excitation wavelength is 413 nm and the laser power at the sample is 11mw.
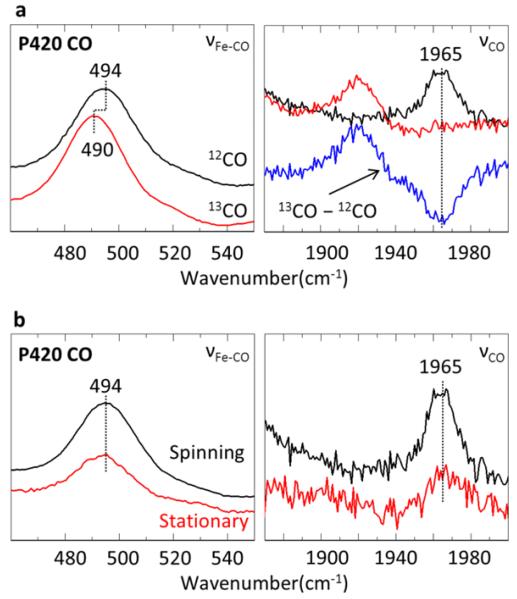
The resonance Raman spectra of the P420cam 12CO and 13CO adducts, showing the νFe-CO and νCO peaks, are displayed in Fig. 5a. To our knowledge, these are the first νFe-CO and νCO frequency correlations that have been made for a P420 system. The νCO difference spectrum between 13CO-12CO is shown in blue. For 12CO, the νFe-CO stretching peak is found at 494 cm−1, while the νCO mode is at 1965 cm−1. The νCO mode from the resonance Raman spectrum matches the IR measurments very well33, 34. In Fig. 5b we show the stationary vs. spinning cell comparisons where a loss of Fe-CO and CO band intensities is observed in the stationary cell. Again for P420, as observed in the H93G adducts, the ratio of the CO dissociated (5C) to the CO bound (6C) species is increased in the stationary cell. This is evidenced by the appearance of a ν4 band at 1361 cm−1 in Fig. S5 (Supporting Information) that confirms the presence of significant 5C photoproduct (rather than a 4C photoproduct)31. There is no sign of a mode near ~220 cm−1 in the photostationary state data (Fig. S5). Since this mode is clearly observed and assigned to νFe-His in the 10 ns transient Raman spectrum13, its absence in Fig. S5 is attributed to a ligand switch between His and another ligand with a rate that is faster than the cw photoexcited CO escape rate into solution. It should also be noted that the ν4 band is located at 1354 cm−1 in the 10 ns transient spectrum13 and this band is shifted to 1361 cm−1 in Fig. S5. This is also consistent with a ligand switch and probably involves water replacement as an intermediate heme ligand in the photocycle. A kinetic model that accounts for these observations, along with the previously observed kinetic rates for CO binding to P42035, is given in Scheme I below.
Figure 5.
(a) Resonance Raman spectra of the P420 12CO and 13CO adducts at λ=413nm. The power at the spinning sample cell is 11mw. The νFe-CO region is on the left and the νCO is on the right with the 13CO-12CO Raman difference spectra shown in blue. (b) Resonance Raman Spectra of P420 CO. Spectra in black are taken with the sample spinning at 6000 rpm, while the red spectra are with a static cell.
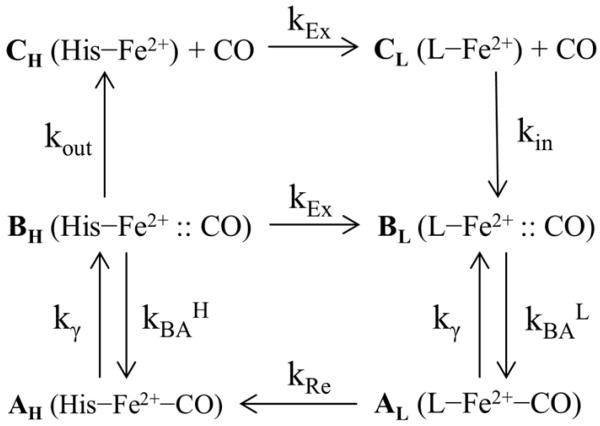
Scheme I.
Kinetic model for photostationary states in H93G-CO and P420-CO with H indicating a proximal histidine ligand and L indicating a water (or, on longer timescales a thiol) proximal ligand. The A-states have CO bound, the B-states have CO in the distal pocket, while CO has escaped into solution in the C-states. The H93G-CO sample reveals poplulations of AH, AL, and CL while only AH and CL populations are observed in P420-CO (unless the states AH and AL have identical FeCO frequencies). This kinetic scheme is reduced to an effective three state system and analyzed more completely in the Supporting Information.
Discussion
The Raman spectra of the CO adducts of the H93G Mb mutant with and without added THT and CPSH are presented in Figs. 3 and 4. From the variation in photoactivity, we conclude that the νFe-CO modes at 507 cm−1, and 522 cm−1 are associated with the νCO modes found at 1942 cm−1 and 1960 cm−1, respectively. These data points, along with others30, 36-38, are plotted on a νFe-CO vs νCO correlation diagram shown in Fig. 6. This allows us to compare the CO adducts of P420cam and the H93G compounds with the large data base of other CO bound heme species that have been studied using the correlation method16, 36.
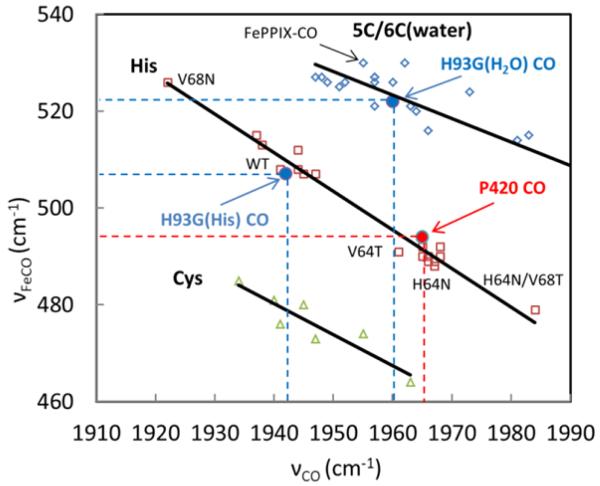
Figure 6.
Correlation plot of νFeCO and νCO with data from ref. 29-32. The red color open squares are for histidine (or imidazole) ligated heme systems. The open green triangles represent thiolate ligated heme systems. The open blue diamonds represent heme systems with a weak or absent proximal ligand. Three solid lines labeled 5C/6C(water), His, and Cys are the lines fitted to the data points. The solid blue and red dots are data points for H93G-CO and P420-CO from this work. The notation H93G(H2O)CO and H93G(His)CO correspond to the histidine and water ligated populations of H93G-CO observed in the H93G Raman spectra.
The νFe-CO and νCO modes typically follow an inverse π back-bonding relationship36 as shown in Fig. 6. Back-donation of iron dπ electrons to the CO π* orbitals strengthens the Fe-CO bond and weakens the C-O bond. Trans ligands with a stronger σ donation will weaken the Fe-CO bond more than expected, due to the change in π backbonding resulting from sigma donor competition for the iron dZ2 orbital37. Thus, CO adducts with a strong trans ligand lie lower on the plot. For the same reason, CO adducts with weaker trans ligand lie higher on the plot (e.g., the water-ligated FePPIX-CO data point30 is denoted by the arrow). Mb variants with a neutral trans His ligand and different distal pocket mutations are spread along the line labeled His. The position of the various CO adducts on the line reflects the polarity of their distal binding pocket. CO adducts with distal residues that produce strong H-bond interactions, like V68N, or the Mb “closed” distal pocket (A1) state16 lie higher on the line, whereas those with nonpolar distal residues, like H64V or the Mb “open” distal pocket (A0) state16, lie lower on the line. The 507/1942 cm−1 modes of the H93G MbCO complex are assigned to 6-coordinate low-spin histidine bound forms with a “closed” distal pocket, while the 522/1960 cm−1 modes are assigned to a water bound form25 (In principle, the 522/1960 cm−1 modes could also result from a five coordinate CO bound species; however, the photostationary state conditions do not produce a 4C photoproduct ν4 band. Rather, a ν4 band at 1359 cm−1 is observed, which is consistent with the assignment25 of water as the ligand trans to CO in these complexes).
The νFe-CO and νCO modes of cytochrome P420cam are found at 494 cm−1 and 1965 cm−1. These values agree with independent Raman and IR data13, 33. The 494/1965 cm−1 point is found on the inverse correlation diagram at a position that is consistent with a trans His ligand and an open distal pocket. This supports the assignment of histidine as the proximal ligand in P420cam-CO, but it is not fully conclusive because of recent work on model ferrous porphyrin complexes in thioether solvents that has suggested thiol complexes may have a similar back-bonding correlation37.
Interestingly, as seen in Fig. 5, the Fe-CO and CO frequencies of P420 do not change upon photoexcitation in a stationary cell even though a significant photoproduct population is created (as evidenced by the ν4 band at 1361 cm−1 observed in Fig. S5). Analogous to the H93G system, the P420-CO shows no indication of a Fe-His mode under the photostationary conditions (Fig. S5). This suggests that, as for H93G-CO25, 26, there is a rapid loss of the His ligand upon CO photodissociation (i.e., kEx >> kγ (kout/kBAH+kout+kEx) in Scheme I below). Moreover, there must be a relatively rapid reset rate, kRe, to the initial His bound ligation state (AH) following CO binding.
In contrast to H93G-CO, water does not appear as a photostationary state ligand (L) in P420-CO, although it probably participates as an intermediate in the ligand exchange process denoted by kEx in Scheme I. Assuming water functions as a transient ligand (L) in the photocycle, it is potentially detectable as a trans ligand in the CO bound state, AL, when the system is driven into a photostationary state equilibrium. In such a case, we might expect to see intermediates with frequencies (analogous to the 522/1960 cm−1 modes) that appear on the upper line in Fig. 6. However, the CO geminate recombination rate and geminate yield are very large for P42029, possibly exceeding those observed for CooA39. (The missing amplitude in the early kinetic work30 makes precise evaluation difficult, but the actual geminate amplitude for CO binding in P420 may approach 99%). This has the effect of reducing the effective rate of CL formation from AH, as well as the rate of AL formation from AH via the BH-BL channel in Scheme I. Under this condition very little AL population will exist in the photostationary state (see Supporting Information for a more detailed analysis).
The other possibility to account for the fixed positions of νFe-CO and νCO in the stationary cell (Fig. 5b) would be for the state AL in Scheme I to have Fe-CO and CO frequencies that are identical to the histidine-ligated CO bound state (AH). In principle, the latter possibility might be realized if thiol (not water) was the ligand (L). However, it would be highly coincidental if thiol and histidine ligation lead to exactly the same FeCO frequencies. Moreover, direct thiol ligation (without a water ligand intermediate) would also require very rapid protein rearrangements during the ligand switching process. Finally, the hypothesis of a single thiol ligand can also be ruled out because of the strong Fe-His mode observed in the 10 ns transient spectra of P420-CO. We can use the ν7 band as a reference to determine that the strength of the transient Fe-His mode in P420CO is equivalent to that of MbCO13. This observation is the “smoking gun”, demonstrating that thermal equilibrium must favor the AH state in the P420cam system.
In the myoglobin H93G mutant, the native proximal ligand is replaced by Gly and the only possible candidates for histidine ligation are His97 on the proximal side and His64 on the distal side. The binding of His64 was previously excluded by resonance Raman spectra of the CO bound H64V/H93G double mutant, which is very similar to that of H93G MbCO25. Based on transient Raman spectra that detects the 220 cm−1 Fe-His mode, His97 was assigned as the prime candidate to be the trans ligand when CO binds and acidifies the heme iron26. When CO is photolyzed, the time resolved step-scan infrared data indicate26 that the histidine ligand is replaced by water on a time-scale that is faster than ~106 s−1 and that the histidine ligand does not rebind until CO bimolecular rebinding takes place25. Thus, in the static cell, the population of the His bound form will be less than that in the spining cell because the continuous photolysis leads to a larger proportion of the fast rebinding water bound species. The water bound heme has a lower proximal barrier and rebinds CO much more rapidly than the His bound heme40. This is why only the νCO and νFeCO peaks, associated with the water bound heme population (AL), are observed in the photostationary state when the spinning cell is stopped.
The model for the CO photolysis and H2O-hisitidine ligand exchange in the H93G system in Scheme I is very similar to the ligand switch model presented in an earlier study25, but measurements of H93G-CO kinetics29 suggest that CO escape into solution must compete with the rate for histidine exchange with water in the pocket. The photostationary equilibrium spectra for H93G-CO display strong evidence that water ligates to the CO bound heme as suggested by earlier work25. Moreover, the rate of histidine recruitment by the water bound CO state (AL) to form AH is on the order of, or smaller than, the rate of AL and CL production from AH (see Supporting Information). The different photostationary state behavior of H93G compared to P420 can be traced to the geminate rebinding rate of AH in the respective systems and the fact that H93G retains a distal barrier that significantly slows its geminate rebinding compared to P420. We also note that the photostationary populations of AH and AL in Scheme I appear favor the AL state in H93G MbCO, even under spinning conditions. More details of the kinetic analysis in spinning and stationary conditions can be found in the Supporting Information.
The frequencies of the νFe-CO and νCO modes in H93G MbCO are unaffected by the addition of THT or CPSH. Comparison of the absorption spectra of H93G deoxyMb in 200 μM CPSH and THT solution to the pure H93G deoxyMb shows a clear difference, as can be seen in Fig. S1. These results are consistent with prior work11, but one can interpret the small absorption spectral change as THT and CSPH binding to the protein, close enough to affect the heme electrostatic environment15, 16, yet without direct ligation to the heme. While it is conceivable that both the thiol and thioether compounds bind to heme and precisely mimic the histidine and/or water-bound heme ligation states, it seems much more likely that these compounds are not actually ligating the heme iron. Rather, they could have a binding site nearby, close enough to account for the 2 nm shift in the Soret band, which provides the only evidence that these ligands are binding to the reduced H93G Mb system (recall from Figs 1, 2, S2, and S3 that there is no binding effect registered in the heme-specific resonance Raman spectra). A non-heme binding site for THT and CPSH is also consistent with the observation of two H93G MbCO species. Two H93G MbCO states (histidine and water) are observed in the Raman spectra as described above. If THT and CPSH were binding to the heme iron in the expected 1:1 stoichiometry, one would expect that a single set of νFe-CO and νCO modes would be observed.
In Fig. 2, the 522 cm−1 peak shows a somewhat lower relative intensity compared to the 507 cm−1 peak when THT and CPSH are added to H93G MbCO. This indicates that the His bound form, characterized by the 507 cm−1 peak intensity, has more relative population when the CPSH or THT are added to the solution. The system is undergoing a complex photon driven dynamics that involves competition between the photoexcitation rate, CO escape and bimolecular entry into the heme pocket, and the rate of recruitment of His97 as an iron ligand during the time CO is bound to the heme. Ligand switching models involving histidine and water have been proposed previously in the context of both H93G and low pH Mb24, 25, 31, 41. Upon CO binding to the water-ligated H93G Mb state, the iron seeks to bind a strong sigma donating ligand23 and recruits His97. Depending upon the photoexcitation rate, the CO escape and entry into the pocket, and the very different geminate rebinding rates for the water and His97 bound heme40, the two CO-bound populations will reach a photostationary equilibrium (e.g., see Scheme I). The presence of CPSH and THT in the heme pocket might be expected to modify the equilibrium between AL and AH, leading to the subtle changes in Fe-CO populations observed in Figs. 2 and 3.
In Fig. 1 there is no 220 cm−1 mode observed in the resonance Raman spectra of either reduced H93G Mb or reduced P420cam. This indicates that, in the absence of CO, histidine is not ligated to the reduced heme. The cysteine thiol ligand is the obvious candidate for ligation in the reduced state of P420. Although there is no direct spectroscopic evidence, the thiol ligation assignment in the ferrous form of P420 has been discussed in the context of similarities with CPO11, 12. The absence of water ligated CO bound signals in the photostationary state Raman spectra indicate that water does not form a particularly stable intermediate in P420-CO; however, it does not eliminate water as a possible transient ligand in states CL and AL of the photocycle.
Nanosecond transient Raman spectra of P420cam-CO13, iNOS P420-CO10 and H93G-CO25 adducts all show a strong 220 cm−1 mode, which is very similar to the MbCO 10 nanosecond transient Raman spectra13 and has been assigned in many heme protein systems as the Fe-His stretching vibration17-19, 22. If the 221 cm−1 mode, observed in the 5 ns photoproduct Raman spectrum of the P420 form of iNOS-CO, was a heme mode10 (rather than the Fe-His vibration), we should be able to observe it in the Raman spectrum of the reduced P420 samples/analogs in Fig. 1. The absence of a 221 cm−1 Raman mode in Fig. 1 is inconsistent with its assignment to a thiol ligated heme mode. This indicates that the transient photoproduct Raman spectra of P420 systems is actually revealing the presence of a proximal histidine ligand, which has been recruited in the CO bound state as shown in Scheme I. Such an analysis is consistent with the prior assignment for the 220 cm−1 mode observed in the H93G-CO system26. Thus, in thermal equilibrium (i.e., no photoexcitation or between the ns laser pulses), the histidine ligated P420-CO state is favored.
Since the νFe-His mode at ~220 cm−1 is not active in 6-coordinate CO adducts36 and the CO dissociated population rapidly replaces the histidine ligand with water, the photostationary experiments are not able to observe a νFe-His mode with either a spinning or a static cell. Thus, the 5 ns transient resonance Raman spectrum of P420-CO, which displays a 220 cm−1 mode similar in intensity to the MbCO photoproduct, provides the most direct evidence to assign histidine as the trans ligand in the CO bound P420 systems13. We suggest that the apparent absence of an isotopic shift for the 221 cm−1 mode in the inactive iNOS P420-CO photoproduct spectrum is due to the fact that the H/D shift is only expected10 to be 0.7 cm−1. The signal-to-noise of the transient Raman spectra10 of the iNOS P420-CO was significantly worse than that of the control experiment on MbCO where a ~ 1 cm−1 H/D shift was observed. The peak shift algorithm developed previously20 indicates that a 25 cm−1 (full width at half-height) Gaussian band that shifts by 0.7 cm−1 will generate a maximum-to-minimum in the Raman difference spectrum that is 8% of the measured peak height. Since the noise level of the transient difference spectrum between the protonated and deuterated iNOS P420-CO photoproduct exceeds this value by approximately a factor of two10, we do not believe that the 0.7 cm−1 isotopic shift of the Fe-His mode could be detected for iNOS P420 and therefore that the ~0.7 cm−1 H/D shift is present, but undetectable.
Finally, one must address the issue of whether there are His ligands available to undergo ligand switching with Cys in various P450 systems. Using the protein data base, we find that there is at least one His residue within 10 −15 Å of the heme iron in membrane bound P450 3A4, P450cam, and iNOS as shown in Figs. S7, S8, and S9 of the Supporting Information. These nearby histidine residues are candidates for binding to heme when P420 undergoes the tertiary structural changes associated with the binding of CO and the loss of the thiol ligand. These distances also have implications regarding the extent and type of the conformational fluctuations that are taking place in these systems as they become inactivated. In other heme proteins, such as chloroperoxidase42 and Rr-CooA43, axial thiolate ligands have been shown to undergo a ligand-switch to a nearby histidine following reduction of the heme iron. It has also been reported that the nitrophorin 1 H60C mutant loses its heme thiolate cysteine ligand upon heme reduction44. Generally, upon reduction of the heme iron, the neutralized heme core could trigger protonation of the thiolate ligand leading to its dissociation and the subsequent structural rearrangements that facilitate a functional and biologically relevant ligand switch43, 45. A variety of thiolate-heme proteins that have evolved to become heme-sensor proteins with functional ligand switching reactions have recently been reviewed46 and the P450/P420 reaction has many similarities.
Dunford et al.47 recently reported that CYP121 from M. Tuberculosis can undergo reversible conversion from its P420 form back to the P450 form when the pH is raised from 6.5 to 10.5. The P420 form dominates at lower pH, while the P450 form dominates at higher pH. These observations were interpreted using a simple two-state model47 involving the reversible protonation of the proximal cysteine ligand. There are 4 histidine residues less than 15 Å from the heme iron in CYP121. However, the H343 residue is only 2 amino acids away from the proximal Cys ligand, forming a HXC-Fe sequence that is shared with both CYP101 and CYP51 (For most CYP51s, this sequence is conserved48, including the CYP51 from M. Tuberculosis studied by Dunford et al.). The proximity of His343 to the heme in CYP121 might facilitate its ligation to the iron atom in the event that protonation of Cys345 leads to its dissociation in the CO bound state. Upon deprotonation at higher pH, the Cys345 thiolate will again become a strong ligand that can potentially displace His343 so that the P450 form is reversibly recovered. The reversibility of this process will depend sensitively upon the strength of the His-heme-CO ligation and the new structural motif that is formed in the P420 state (vide infra). An important point is that a Cys(thiol)→His→Cys(thiolate) ligand switch mechanism provides an alternative interpretation of the pH dependent reversible P420/P450 conversion in CYP121 and would help to explain the irreversibility observed in CYP51 (assuming the P420 structure formed in CYP51 is particularly stable). Moreover, the relatively slow (0.72±0.02 s−1) and pH independent transition rate that is observed for P450→P420 conversion47 might actually be more consistent with a ligand switch process as the rate limiting step, rather than a simple thiolate protonation reaction. (In the context of Scheme I, this means that an additional column of states AT, BT, and CT, where T represents thiol, must be added in slow exchange with the photocycle states AL, BL, and CL, where L=H2O.) Additional investigations specific to the P420 forms of CYP121 and CYP51 are clearly needed to identify the proximal ligand in the CO bound forms at acid and alkaline pH.
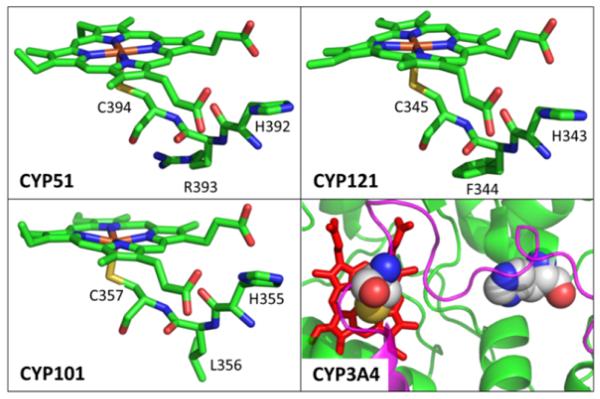
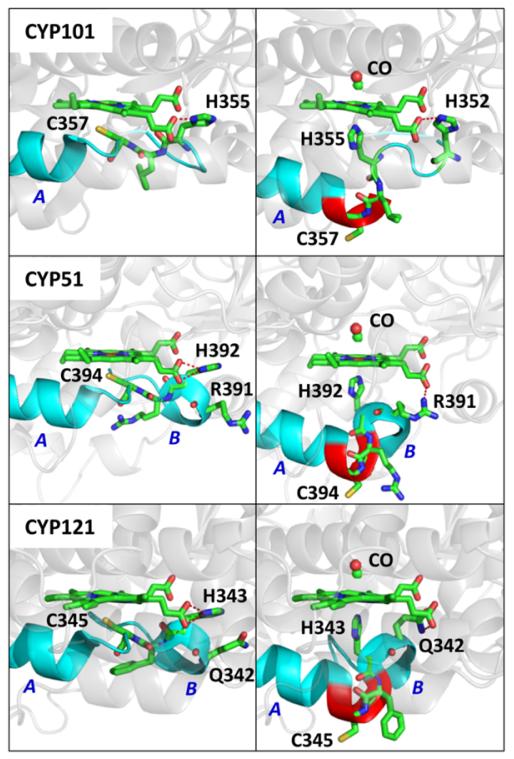
Along with CYP121 and CYP51, CYP101 shares the HXC-Fe motif with X representing Phe, Arg and Leu, respectively. The Cys loop HXC sequences in the heme proximal pocket of these proteins are shown in Fig. 7. Here we recognize that the His residue is actively engaged in H-bonding with the nearby propionate group of the heme. Some possible proximal pocket conformational changes that would allow binding of the nearby histidines to the heme iron are depicted in Fig. 8. Examination of the helix-loop transition region in CYP101, which lies between residues G359(helix) and L358(loop), suggests that upon dissociation of the protonated Cys357, the three adjacent loop residues L358, C357, and L356, could coil into the alpha helix, leaving His355 in prime position to bind to the iron atom as depicted in Fig. 8. Moreover, we find that another histidine (H352) lies downstream in the loop region and that it can be easily positioned to recover the necessary H-bond to the heme propionate. Thus, a relatively simple and energetically favorable conformational fluctuation, slightly extending the L-helix, could lead to the histidine-heme ligation following CO binding in CYP101. Larger scale renditions of this transition can be seen in section S10 of the Supporting Information.
Figure 7.
Crystal structure of CYP51 (PDB#1EA1), CYP121(PDB#2IJ7), CYP101(PDB#2CPP) and CYP3A4(PDB#3NXU). The HXC motif in CYP51, CYP121 and CYP101 are highlighted. The right bottom panel shows the loop (magenta) of the heme proximal pocket in CYP3A4. The C442 and H402 residues are shown with a space filling model. There is no HXC motif in CYP3A4.
Figure 8.
The three panels on the left side show the crystal structure of CYP101 (PDB#2CPP), CYP51(PDB#1EA1), and CYP121(PDB#2IJ7). The L-helix is extended by 5 residues past the Cys ligand in the CYP51 and CYP121 structures. The helix structures on the left and right side of the proximal cysteine ligand are labeled as A and B, respectively. The red dashed line indicates the hydrogen bond between “near” propionate group and H355, H392 and H343 in the three structures, respectively. The three panels on the right show the possible CO bound P420 structures for the three proteins. The proposed structures are formed by extending the A helix by three amino acids (highlighted in red). A remnant of the B helix structure is preserved in the proposed P420-CO structure for CYP51 and CYP121. In the proposed structures, H352, R391, and Q342 replace the histidine residues of the native structures by forming a hydrogen bonds (red dashed line) with the propionate group. Larger renditions and different angles showing the possible structures underlying the ligand switch can be found in the SI.
It is interesting to compare the CYP121 and CYP51 proteins, which also share the HXC motif. CYP121 displays a reversible pH dependent P450/P420 conversion while CYP51 does not47, 49. Evidently, CYP51 converts to its P420 form immediately upon reduction, even in the absence of CO 49, suggesting that its Cys heme ligand has a pK that is somewhat higher than for CYP121. A simple thiol/thiolate pH titration model for P420 conversion would predict that, upon raising the pH high enough (say above 10), the putative thiol Cys ligand in CYP51 should ultimately deprotonate and revert to a thiolate so that a reversible P420/P450 should also be observed in this system. On the other hand, the ligand switch model can easily explain the irreversible behavior. For example, if a particularly stable P420 structure is formed upon CO binding, the simple deprotonation of the Cys residue may not be enough to energetically reconfigure the protein structure and recover thiolate ligation to the heme iron.
In Fig. 8 we have shown a possible structural change for CYP51 conversion to P420. Here we find that, upon extension of the L-helix to include Cys394 and the orientation of His392 as the axial ligand, Arg391 is naturally prepositioned to form a strong H-bond with the heme propionate. Thus, the Cys(thiol)→His ligand switch in CYP51 may form a very stable alternative structure (e.g., by incorporating the Cys(thiol) into the adjacent alpha helix and forming a strong Arg391 H-bond with the heme propionate) leading to a situation that is energetically stable and not reversible by pH back-titration47. It should also be noted that the P420 form of CYP51 displays a partial reconversion to P450 upon loss of CO and re-oxidation49. Since the iron-histidine bond is weakened following CO dissociation and iron re-oxidation, it is evidently possible for the cysteine thiolate residue to successfully compete to once again become a heme ligand in the ferric state of CYP51. In contrast to CYP51, CYP121 undergoes a reversible pH titration between P420 and P45047, 49. The structures that we found for the P420-CO state in this system appeared to have adequate, but less satisfying, H-bonding to the heme propionate suggesting that it might have less energetic stabilization and therefore be more likely to undergo a reversible transition following pH back titration.
Although many P450 systems share the HXC-Fe motif, some do not and their potential histidine ligands are not found as close to the proximal heme ligation site. Two important examples discussed above are iNOS and CYP3A4, where a larger tertiary structure change is necessary to bring a histidine close enough for heme ligation to occur. Figures 7, S7, S9, and S10 (Supporting Information) show nearby histidine residues that are ligation candidates for these systems. For iNOS, H661 (~10 Å away) and H407 (~ 16 Å away) are potential candidates for a ligand switch with Cys 415. For CYP3A4, the H402 (~14 Å away) is a possibility for ligand switching with Cys442 as seen in the lower right quadrant of Fig. 7. The fact that a strong 220 cm−1 mode is observed in the nanosecond transient Raman spectra of iNOS-CO indicates that, even for proteins without the HXC-Fe motif, a histidine ligand can possibly be recruited to form the final P420 state following CO binding. We also note that, in systems without the obvious HXC-Fe motif, it may be possible that other strong sigma donating ligands can act as a substitute for histidine binding to the CO ligated heme.
In summary, the conversion of the P450 thiolate to thiol appears to be an important step in the conversion of P450 to P420. However, upon CO binding the 10 ns transient Raman spectra demonstrate that a nearby histidine must be involved in a ligand switching equilibrium with the cysteine thiol. All indications are that this equilibrium favors a histidine bound P420CO ground state in CYP101. When there are His residues near the heme, as in the HXC-Fe loop, only relatively small tertiary structural changes are needed for the ligand switch (e.g., incorporation of C and X into the nearby helix structure can properly position the His for heme binding). The structures that are formed in these P420-CO states may have variable stability. In some cases, the process might be irreversible with pH titration (e.g., CYP51), depending upon the strength of the alpha helix interactions, the His-Fe-CO bond formation, and the heme propionate H-bond that is formed. On the other hand, if the P420-CO states are less stable, reversibility via pH titration would be more likely (e.g., CYP121). If the ligand switch requires a very large tertiary structural change, the process would be likely to proceed at a slower rate and may not be easily reversible.
Conclusion
We have used resonance Raman spectroscopy to study P420cam and the H93G Mb mutant, with and without the addition of THT and CPSH compounds. There is no evidence from the vibrational spectra to indicate that THT or CPSH ligate the heme iron. There is no 220 cm−1 Fe-His mode observed in the reduced samples, which indicates that histidine is not the heme ligand in the ferrous state. Thiol is the likely heme ligand in the thermally equilibrated ferrous state of the P420 systems, while water appears as the ligand in the reduced H93G system. The transient Raman spectra of the CO bound species indicate that a Fe-His bond is formed when CO binds to P420 systems and acidifies the heme iron. A histidine ligand for P420-CO is also indicated by the position of the υFe-CO and υCO frequencies on the inverse correlation plots although, strictly speaking, thiol cannot be ruled out by this correlation37. On the other hand, the invariance of the residual P420 FeCO frequencies when photostationary photoproduct states are formed indicates that the equilibrium between AH and AL in Scheme I favors the histidine ligated state, AH, unless a thiol ligated state, AL, yields exactly the same frequencies. The photostationary state Raman spectra of H93G MbCO indicate that, following CO binding to the water ligated heme, the rate for histidine recruitment is ~ 104 s−1 (see Supporting Information). Several P450 systems offer a common HXC-Fe motif that may facilitate Cys-His ligand switching, while other P450 systems must undergo slower and larger tertiary changes in order for the acidic iron of the CO-bound heme to replace the weakly bound thiol ligand with a nearby histidine or with some other strong sigma bonding donor.
The above considerations bring into focus some motivations for trying to better understand the P450-P420 reaction. For example, if a reversible helix-loop transition triggered by CO photolysis underlies the P450-P420 conversion, in even a sub-set of P450 systems, they would present interesting models for the study of protein conformational transitions using time-resolved spectroscopies. It is also conceivable that reversible transitions of this type might play a functional role in the regulation of the monoxygenase function. Finally, if it is possible to engineer the destabilization of the P420 form of the protein without similarly destabilizing the P450 form, one might envision P450 mutants that are more robust and less likely to convert to the P420 form. Mutations that remove propionate H-bonding in the P420 form (but not in the P450 form) provide one interesting target.
Supplementary Material
Acknowledgments
Funding Information: This work was supported by NSF MCB-0744738 (PMC) and NIH DK35090 (PMC), GM 26730 (JHD), GM31756 (SGS).
Abbreviations
- THT
tetrahydrothiophene
- CPSH
cyclopentathiol
- His
histidine
- Mb
myoglobin
Footnotes
A more detailed description of the kinetic model and additional supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 2.Champion PM, Stallard BR, Wagner GC, Gunsalus IC. Resonance Raman detection of an iron-sulfur bond in cytochrome P 450cam. J. Am. Chem. Soc. 1982;104:5469–5472. [Google Scholar]
- 3.Sono M, Andersson LA, Dawson JH. Sulfur donor ligand binding to ferric cytochrome P-450-CAM and myoglobin. Ultraviolet-visible absorption, magnetic circular dichroism, and electron paramagnetic resonance spectroscopic investigation of the complexes. J. Biol. Chem. 1982;257:8308–8320. [PubMed] [Google Scholar]
- 4.Collman JP, Sorrell TN. Model for the carbonyl adduct of ferrous cytochrome P 450. J. Am. Chem. Soc. 1975;97:4133–4134. doi: 10.1021/ja00847a046. [DOI] [PubMed] [Google Scholar]
- 5.Stern JO, Peisach J. A Model Compound Study of the CO-Adduct of Cytochrome P-450. J. Biol. Chem. 1974;249:7495–7498. [PubMed] [Google Scholar]
- 6.Gunsalus IC, Meeks JR, Lipscomb JD, Debrunner P, Munck E. In: Molecular mechanisms of oxygen activation. Hayishi O, editor. Academic Press; New York: 1974. p. 559. [Google Scholar]
- 7.Champion PM, Gunsalus IC, Wagner GC. Resonance Raman investigations of cytochrome P450CAM from Pseudomonas putida. J. Am. Chem. Soc. 1978;100:3743–3751. [Google Scholar]
- 8.Hui Bon Hoa G, Di Primo C, Dondaine I, Sligar SG, Gunsalus IC, Douzou P. Conformational changes of cytochromes P-450cam and P-450lin induced by high pressure. Biochemistry. 1989;28:651–656. doi: 10.1021/bi00428a035. [DOI] [PubMed] [Google Scholar]
- 9.Martinis SA. Ph.D. thesis. Department of Biochemistry, University of Illinois; Urbana-Champaign, IL: 1990. [Google Scholar]
- 10.Sabat J, Stuehr DJ, Yeh SR, Rousseau DL. Characterization of the Proximal Ligand in the P420 Form of Inducible Nitric Oxide Synthase. J. Am. Chem. Soc. 2009;131:12186–12192. doi: 10.1021/ja901016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perera R, Sono M, Sigman JA, Pfister TD, Lu Y, Dawson JH. Neutral thiol as a proximal ligand to ferrous heme iron: Implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. USA. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinis SA, Blanke SR, Hager LP, Sligar SG, Hui Bon Hoa G, Rux JJ, Dawson JH. Probing the heme iron coordination structure of pressure-induced Cytochrome P420cam. Biochemistry. 1996;35:14530–14536. doi: 10.1021/bi961511u. [DOI] [PubMed] [Google Scholar]
- 13.Wells AV, Li P, Champion PM, Martinis SA, Sligar SG. Resonance Raman investigations of Escherichia coli-expressed Pseudomonas putida cytochrome P450 and P420. Biochemistry. 1992;31:4384–4393. doi: 10.1021/bi00133a002. [DOI] [PubMed] [Google Scholar]
- 14.Antonini E, Brunori M, editors. Hemoglobin and myoglobin in their reactions with ligands. North Holland Publishing Company; Amsterdam: 1971. [Google Scholar]
- 15.Kushkuley B, Stavrov SS. Theoretical study of the distal-side steric and electrostatic effects on the vibrational characteristics of the FeCO unit of the carbonylheme proteins and their models. Biophys. J. 1996;70:1214–1229. doi: 10.1016/S0006-3495(96)79680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morikis D, Champion PM, Springer BA, Sligar SG. Resonance raman investigations of site-directed mutants of myoglobin: effects of distal histidine replacement. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 17.Nagai K, Kitagawa T, Morimoto H. Quaternary structures and low frequency molecular vibrations of haems of deoxy and oxyhaemoglobin studied by resonance Raman scattering. J. Mol. Biol. 1980;136:271–289. doi: 10.1016/0022-2836(80)90374-5. [DOI] [PubMed] [Google Scholar]
- 18.Friedman JM, Rousseau DL, Ondrias MR. Time-Resolved Resonance Raman Studies of Hemoglobin. Annu. Rev. Phys. Chem. 1982;33:471–491. [Google Scholar]
- 19.Friedman JM, Scott TW, Stepnoski RA, Ikeda-Saito M, Yonetani T. The iron-proximal histidine linkage and protein control of oxygen binding in hemoglobin. A transient Raman study. J. Biol. Chem. 1983;258:10564–10572. [PubMed] [Google Scholar]
- 20.Rousseau DL. Raman difference spectroscopy as a probe of biological molecules. J. Raman Spectrosc. 1981;10:94–99. [Google Scholar]
- 21.Argade PV, Sassardi M, Rousseau DL, Inubushi T, Ikeda-Saito M, Lapidot A. Confirmation of the assignment of the iron-histidine stretching mode in myoglobin. J. Am. Chem. Soc. 1984;106:6593–6596. [Google Scholar]
- 22.Friedman JM, Rousseau DL, Ondrias MR, Stepnoski RA. Transient Raman study of hemoglobin: structural dependence of the iron-histidine linkage. Science. 1982;218:1244–1246. doi: 10.1126/science.7146910. [DOI] [PubMed] [Google Scholar]
- 23.Rougee M, Brault D. Influence of Trans Weak or Strong Field Ligands Upon Affinity of Deuteroheme for Carbon-Monoxide - Monoimidazoleheme as a Reference for Unconstrained 5-Coordinate Hemoproteins. Biochemistry. 1975;14:4100–4106. [Google Scholar]
- 24.Sage JT, Li PS, Champion PM. Spectroscopic Studies of Myoglobin at Low pH - Heme Ligation Kinetics. Biochemistry. 1991;30:1237–1247. doi: 10.1021/bi00219a011. [DOI] [PubMed] [Google Scholar]
- 25.Franzen S, Bailey J, Dyer RB, Woodruff WH, Hu RB, Thomas MR, Boxer SG. A photolysis-triggered heme ligand switch in H93G myoglobin. Biochemistry. 2001;40:5299–5305. doi: 10.1021/bi0023403. [DOI] [PubMed] [Google Scholar]
- 26.Franzen S, Peterson ES, Brown D, Friedman JM, Thomas MR, Boxer SG. Proximal ligand motions in H93G myoglobin. Eur. J. Biochem. 2002;269:4879–4886. doi: 10.1046/j.1432-1033.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- 27.Barrick D. Replacement of the Proximal Ligand of Sperm Whale Myoglobin with Free Imidazole in the Mutant His-93.fwdarw.Gly. Biochemistry. 1994;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 28.Fisher MT, Scarlata SF, Sligar SG. High-pressure investigations of cytochrome P-450 spin and substrate binding equilibria. Arch. Biochem. Biophys. 1985;240:456–463. doi: 10.1016/0003-9861(85)90050-5. [DOI] [PubMed] [Google Scholar]
- 29.Cao W, Ye X, Sjodin T, Christian JF, Demidov AA, Berezhna S, Wang W, Barrick D, Sage JT, Champion PM. Investigations of Photolysis and Rebinding Kinetics in Myoglobin Using Proximal Ligand Replacements. Biochemistry. 2004;43:11109–11117. doi: 10.1021/bi049077g. [DOI] [PubMed] [Google Scholar]
- 30.Ye X, Yu A, Georgiev GY, Gruia F, Ionascu D, Cao W, Sage JT, Champion PM. CO Rebinding to Protoheme: Investigations of the Proximal and Distal Contributions to the Geminate Rebinding Barrier. J. Am. Chem. Soc. 2005;127:5854–5861. doi: 10.1021/ja042365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sage JT, Morikis D, Champion PM. Spectroscopic Studies of Myoglobin at Low Ph - Heme Structure and Ligation. Biochemistry. 1991;30:1227–1237. doi: 10.1021/bi00219a010. [DOI] [PubMed] [Google Scholar]
- 32.Hu S, Smith KM, Spiro TG. Assignment of Protoheme Resonance Raman Spectrum by Heme Labeling in Myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 33.O’Keefe DH, Ebel RE, Peterson JA, Maxwell JC, Caughey WS. An infrared spectroscopic study of carbon monoxide bonding to ferrous cytochrome P-450. Biochemistry. 1978;17:5845–5852. doi: 10.1021/bi00619a036. [DOI] [PubMed] [Google Scholar]
- 34.Mouro C, Jung C, Bondon A, Simonneaux G. Comparative Fourier transform infrared studies of the secondary structure and the CO heme ligand environment in cytochrome P-450cam and cytochrome P-420cam. Biochemistry. 1997;36:8125–8134. doi: 10.1021/bi9700173. [DOI] [PubMed] [Google Scholar]
- 35.Tian WD, Wells AV, Champion PM, Di Primo C, Gerber N, Sligar SG. Measurements of CO geminate recombination in cytochromes P450 and P420. J. Biol. Chem. 1995;270:8673–8679. doi: 10.1074/jbc.270.15.8673. [DOI] [PubMed] [Google Scholar]
- 36.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J. Inorg. Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Role of the axial ligand in heme-CO backbonding; DFT analysis of vibrational data. Inorg. Chim. Acta. 2000;297:11–17. [Google Scholar]
- 38.Ray GB, Li XY, Ibers JA, Sessler JL, Spiro TG. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J. Am. Chem. Soc. 1994;116:162–176. [Google Scholar]
- 39.Benabbas A, Karunakaran V, Youn H, Poulos TL, Champion PM. Effect of DNA binding on geminate CO recombination kinetics in CO-sensing transcription factor CooA. J. Biol. Chem. 2012;287:21729–21740. doi: 10.1074/jbc.M112.345090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye X, Ionascu D, Gruia F, Yu A, Benabbas A, Champion PM. Temperature-dependent heme kinetics with nonexponential binding and barrier relaxation in the absence of protein conformational substates. Proc. Natl. Acad. Sci. USA. 2007;104:14682–14687. doi: 10.1073/pnas.0702622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sage JT, Morikis D, Li PS, Champion PM. Low Ph Myoglobin Photoproducts. Biophys. J. 1992;61:1041–1044. doi: 10.1016/S0006-3495(92)81912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruia F, Ionascu D, Kubo M, Ye X, Dawson J, Osborne RL, Sligar SG, Denisov I, Das A, Poulos TL, Terner J, Champion PM. Low-frequency dynamics of Caldariomyces fumago chloroperoxidase probed by femtosecond coherence spectroscopy. Biochemistry. 2008;47:5156–5167. doi: 10.1021/bi7025485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inagaki S, Masuda C, Akaishi T, Nakajima H, Yoshioka S, Ohta T, Pal B, Kitagawa T, Aono S. Spectroscopic and redox properties of a CooA homologue from Carboxydothermus hydrogenoformans. J. Biol. Chem. 2005;280:3269–3274. doi: 10.1074/jbc.M409884200. [DOI] [PubMed] [Google Scholar]
- 44.Vetter SW, Terentis AC, Osborne RL, Dawson JH, Goodin DB. Replacement of the axial histidine heme ligand with cysteine in nitrophorin 1: spectroscopic and crystallographic characterization. J. Biol. Inorg. Chem. 2009;14:179–191. doi: 10.1007/s00775-008-0436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aono S, Ohkubo K, Matsuo T, Nakajima H. Redox-controlled ligand exchange of the heme in the CO-sensing transcriptional activator CooA. J. Biol. Chem. 1998;273:25757–25764. doi: 10.1074/jbc.273.40.25757. [DOI] [PubMed] [Google Scholar]
- 46.Igarashi J, Kitanishi K, Martinkova M, Murase M, Iizuka A, Shimizu T. The roles of thiolate-heme proteins, other than the P450 cytochromes, in the regulation of heme-sensor proteins. Acta Chim. Slov. 2008;55:67–74. [Google Scholar]
- 47.Dunford AJ, McLean KJ, Sabri M, Seward HE, Heyes DJ, Scrutton NS, Munro AW. Rapid p450 heme iron reduction by laser photoexcitation of mycobacterium tuberculosis CYP121 and CYP51B1: analysis of CO complexation reactions and reversibility of the p450/p420 equilibrium. J. Biol. Chem. 2007;282:24816–24824. doi: 10.1074/jbc.M702958200. [DOI] [PubMed] [Google Scholar]
- 48.Lepesheva GI, Waterman MR. Structural basis for conservation in the CYP51 family. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean KJ, Warman AJ, Seward HE, Marshall KR, Girvan HM, Cheesman MR, Waterman MR, Munro AW. Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions. Biochemistry. 2006;45:8427–8443. doi: 10.1021/bi0601609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.