Abstract
The TAS1R genes encode heterodimeric receptors that mediate umami (hTAS1R1 + hTAS1R3) and sweet (hTAS1R2 + hTAS1R3) sensations. The question of interest for this study is if TAS1R1 variation associates with differences in overall taste intensity. We leveraged an existing database of adults (n = 92, primarily European American) to test associations between 2 TAS1R1 single nucleotide polymorphisms (SNPs) (intronic rs17492553, C/T and exonic rs34160967, G/A) and intensity of 4 prototypical tastants (NaCl, sucrose, citric acid, and quinine), applied regionally to fungiform and circumvallate loci, and sampled with the whole mouth. Both SNPs were associated with modest shifts in perceived intensities across all taste qualities. Three genotype groups were represented for the intronic SNP—minor allele homozygotes (TT) averaged 40% lower intensities than did CC homozygotes for all regionally applied tastants, as well as whole-mouth NaCl and citric acid. Similar, but less pronounced, intensity differences were seen for the exonic SNP (GG homozygotes reported greater intensities than did the AA/AG group). Our predominantly European American cohort had a low frequency of AA homozygotes, which may have attenuated the SNP-related differences in perceived intensity. These preliminary findings, if replicated, could add TAS1R1 polymorphisms to the repertoire of genotypic and phenotypic markers of heightened taste sensation.
Key words: bitter, rs17492553, rs34160967, salty, sour, supertasting, sweet, taste genetics
Introduction
Behavioral evidence that individuals vary in the perceived intensity of tastes and oral somatosensations dates before the 20th century (Bailey and Nichols 1888). Historically, attention has been focused on characterizing this variation with the ability to taste the bitterness of phenylthiocarbamide (PTC) and chemically related propylthiouracil (PROP) (Blakeslee 1932; Guo and Reed 2001; Wooding 2006). Subsequent research has linked PTC/PROP bitter phenotypes and TAS2R38 polymorphisms to food choice, food intake, and chronic disease risk (reviewed by Duffy 2007; Tepper 2008). Other phenotypic markers of variation in oral sensation have emerged, including thermal taste (Green and George 2004), bitterness of vanilloid receptor agonists (Green and Hayes 2004), and density of fungiform papillae (FP) (Miller and Reedy 1990; Essick et al. 2003). Likewise, additional polymorphisms in the TAS2R family (Hayes et al. 2011) and in other genes (Perry et al. 2007; Mandel et al. 2010; Calò et al. 2011; Pirastu et al. 2012; Dias et al. 2013) have been linked to variation in oral sensation and food liking (Duffy et al. 2009; Duffy et al. 2010; Hayes et al. 2013).
The human TAS1R taste receptor gene family (TAS1R1, TAS1R2, and TAS1R3) on chromosome 1, like the TAS2R family of bitter-taste receptor genes, is highly polymorphic (Kim et al. 2006; Raliou et al. 2009a). A heterodimeric G protein-coupled receptor consisting of T1R2 and T1R3 (encoded by the TAS1R2 and TAS1R3 genes) mediates sweet taste perception in humans (Li et al. 2002), whereas the T1R1/T1R3 heterodimer responds to glutamate and other 5′ ribonucleotides (Li et al. 2002; Zhao et al. 2003; Chandrashekar et al. 2006; Kim et al. 2006). Two metabotropic glutamate receptors, mGluR4 and mGluR1, respond to glutamate in rodents (Chaudhari et al. 2000; Toyono et al. 2003; San Gabriel et al. 2005), and possibly in humans (Kurihara and Kashiwayanagi 2000; Raliou et al. 2009b), with corresponding genes GRM4 and GRM1 located on chromosome 6 in humans. However, the T1R1/T1R3 complex appears more broadly tuned in rodents, responding to a range of amino acids (Nelson et al. 2002; Chaudhari et al. 2009). Because amino acids elicit a variety of taste qualities (e.g., in humans, glycine is sweet, phenylalanine is bitter, and glutamate is meaty), these findings in rodents suggest that T1R1 or T1R3 polymorphisms may be important to overall versus quality-specific differences in taste perception. Indeed, a recent study with mice (Kusuhara et al. 2013) reported that chorda tympani nerve responses to sucrose and other sweeteners, but not salty, sour, and bitter compounds, were significantly smaller in the TAS1R1 knockout mice than in the heterozygous mice, suggesting that T1R1 is important to taste functioning beyond umami perception.
Taste findings from rodent, however, may not generalize to humans (Ishimaru et al. 2012). TAS1R1, in particular, is only expressed in the anterior tongue of rodents, whereas in primates (macaques), a close human relative with the same omnivorous diet, TAS1R1 is expressed in both fungiform and circumvallate papilla, where it perhaps has different roles or is involved in different transduction mechanisms (Hevezi et al. 2009; Ishimaru et al. 2012). Intriguingly, TAS1R1 and other TAS1Rs are pseudogenized in mammals with extremely narrow diets, such as bottlenose dolphins, sea lions, and common vampire bats (Jiang et al. 2012; Zhao et al. 2012). TAS1R1 pseudogenes may play a role in the reduced or no sensitivity to sweet- and bitter-taste stimuli in dolphins and sea lions (Friedl et al. 1990; Jiang et al. 2012) or poorly developed taste in blood-feeding vampires (Thompson et al. 1982; Ratcliffe et al. 2003).
Variation in TAS1R1 in humans has been tied with variation in sensitivity to umami, a taste exemplified by glutamate, with a fraction of individuals as monosodium glutamate (MSG) hypotasters or nontasters (Lugaz et al. 2002; Chen et al. 2009; Raliou et al. 2009b; Pepino et al. 2010; Singh et al. 2010). In vitro studies show functional variation in ability to bind MSG with amino acid substitutions in T1R1 and T1R3 receptors (Shigemura et al. 2009; Raliou et al. 2011). Recently, interest has grown in the contributions of TAS1R1 (Entrez GeneID: 80835) and TAS1R3 (Entrez GeneID: 83756) polymorphisms to functional differences in umami and sweet perception in vivo. Although evidence to date relates TAS1R3 variation to sweet (Fushan et al. 2009) and umami taste perception (Chen et al. 2009; Raliou et al. 2009b; Shigemura et al. 2009), findings with TAS1R1 polymorphisms have been mostly limited to MSG recognition or detection thresholds.
Inconsistent findings across human studies suggest the need for further investigation of TAS1R1 in human taste perception. Two nonsynonomous single nucleotide polymorphisms (SNPs) in TAS1R1, A110V (rs41278020, C/T), and A372T (rs34160967, G/A) have been associated with differences in MSG taster or nontaster status (Raliou et al. 2009b). The published findings for the A372T SNP are conflicting, and homozygosity for its minor allele (genotype AA) is less frequently seen in European (3% in French [Raliou et al. 2009b]) versus Asian (18% in Japanese [Shigemura et al. 2009]) populations. In a very large sample (>3000) of adults from France (Raliou et al. 2009b), MSG nontasters were less likely to have the A allele, yet the significance in the chi-square analysis of allelic distribution was driven primarily by AG heterozygotes. In our reanalysis of the published distribution of MSG threshold categories by TAS1R1 A372T (rs34160967, G/A) genotype from a study of adults from Japan (Shigemura et al. 2009), AG heterozygotes were significantly more likely to fall into the MSG-insensitive category than either AA or GG homozygotes.
In an unpublished human psychophysical study (Rawal et al. 2009), we leveraged an existing database to test the ability of A372T SNP (rs34160967, G/A) and another intronic SNP (rs17492553, C/T) in the TAS1R1 gene to explain differences in liking and taste qualities from glutamate-rich foods that also are salty, sour, and bitter (soy sauce, grapefruit juice, and asparagus). The intronic SNP was investigated because of its high minor allelic frequency and its location; it lies in the third intron of the TAS1R1 gene, 14bp from the intron 3/exon 4 junction (IVS3-14) (NM_138697.3), and 1155bp downstream of the A372T SNP. Although the perception of umami sensations were not assessed, we found that genotype differences in preference of these complex foods were associated with perceived sourness and sweetness.
This paper extends our preliminary findings to test if one or both of these TAS1R1 SNPs explain differences in taste intensity, assessed in a protocol that was the foundation for the protocols in the National Institutes of Health Toolbox (Coldwell et al. 2013) and the National Health and Nutrition Examination Survey (Duffy et al. 2012). Our main finding was that homozygotes for the intronic SNP minor allele (TT) reported lower taste intensities than did CC homozygotes for all tastants, and heterozygotes for exonic SNP (AG) reported lower intensities than GG homozygotes for all regionally applied tastants except quinine. These preliminary findings, if replicated, could add TAS1R1 polymorphisms to the repertoire of genotypic and phenotypic markers of heightened taste sensation (Green and George 2004; Green and Hayes 2004; Calò et al. 2011).
Materials and methods
Subjects
A convenience sample of reportedly healthy, nonsmoking adults was recruited from the University of Connecticut community to participate in an observational study of variation in oral sensation, diet, and health. Exclusion criteria included pregnancy, severe food allergies, and thyroid disease. The study sample included 92 adults, primarily of European ancestry (84.8%), female (76%), and middle aged (mean 40.9±12.2 SD). Other ethnicities represented in the sample were Black (5.4%), Hispanic or Latino (5.4%), Asian (3.3%), and other (1.1%). All procedures were approved by the local institutional review board; adults provided informed and written consent and were paid for their participation. All of the data were collected across 2 experimental sessions in the Duffy laboratory, followed by a visit with phlebotomist for venipuncture for DNA extraction.
Intensity scaling
Adults used a general labeled magnitude scale (gLMS) (Bartoshuk et al. 2004) to rate the taste intensity of oral stimuli. As an intensity scale, the gLMS is a vertical scale ranging from “no sensation” (0) at the bottom to “strongest imaginable sensation of any kind” (100) at the top and with other adjectives (“barely detectable,” “weak,” “moderate,” “strong,” and “very strong”) placed in a quasi-logarithmic fashion. Prior to using the gLMS to report intensities, subjects received a verbal orientation to gLMS scaling and were trained on its proper use. As part of the orientation, the subjects practiced rating intensities of recalled nonoral experiences (remembered sound and light sensations). Subjects were first asked to determine which adjective/descriptor on the scale most closely described the intensity of the sensation and then to rate it in the larger context of all sensations. They were asked to rate the sensation by clicking either near or between the adjectives that most closely approximated the strength of their sensation. Upon clicking, the computer generated a numerical value based on the distance along the scale from a zero point (no sensation), which was recorded manually by the experimenter.
Procedure
Intensity of taste stimuli on regional areas
Subjects reported the intensities for 1M NaCl, 1M sucrose, 32mM citric acid, and 1mM quinine hydrochloride painted with a cotton swab on the anterior and posterior tongue (innervated by the chorda tympani branch of cranial nerve VII and the glossopharyngeal branch of cranial nerve IX, respectively)—and sampled with the whole mouth to stimulate cranial nerves VII, IX, and X. Because applying tastants with a cotton swab may cause additional somatosensory stimulation and distort ratings, the cotton swabs were amply saturated with the taste solution prior to application on the tongue, and care was taken to “draw” the solution across the tongue tip. For the whole mouth, subjects were asked to take 10ml of the solution into his or her mouth, rinse for approximately 5 s, and then expectorate. Between each stimulus, the subjects rinsed their mouth with deionized (>15 MΩ) water to remove any residual stimulus.
Intensity of tones as a nonoral standard
Subjects reported the intensities of a series of 1000 Hz tones ranging in 12-dB steps from 50 to 98 dB throughout the testing sessions. The tones were used as a cross-modal standard and used to statistically account for variability in intensity scale usage in the analyses.
TAS1R1 genotyping
Subjects had 15ml of peripheral blood drawn by venipuncture, collected into ethylenediaminetetraacetic acid vacutainer tubes at the University of Connecticut. The DNA was extracted from whole blood in accordance with standard methods that followed manufacturer’s instructions (Qiagen Gentra PureGene) with occasional modification for hemolyzed samples. The genomic DNA was then shipped to the University of Florida and genotyped for TAS1R1 intronic SNP (rs17492553) and exonic SNP (rs34160967) by ABI TaqMan–automated genotyping as reported previously (Hayes et al. 2011). The assay numbers for TaqMan were C__25991161_10 (rs17492553) and C__25997001_10 (rs3416097). The polymerase chain reaction (PCR) step of the genotyping employed conditions of 58°C for 50 cycles, with genotypes subsequently identified using an ABI Prism 7900HT instrument in the University of Florida Pharmacogenomics Center. For quality control, 4 samples were genotyped in duplicate with the same result. The quality control indicators from TaqMan genotyping showed excellent separation of the 3 genotypes for the entire panel. For 3 other samples, independent PCR and sequencing verified the genotypes. The resulting allele frequencies were in Hardy–Weinberg equilibrium for all of the SNPs. Thus, the genotyping fidelity appeared excellent, as has been our experience in previous work (Hayes et al. 2011).
Additional markers of variation in oral sensation
Because variation in oral sensation has been associated with PROP-tasting phenotype, TAS2R genotype and number of FP, these markers were tested for potential interactions with TAS1R1 genotypes in explaining differences in taste intensity. Established PROP-tasting TAS2R38 genotypes and haplotypes (rs713598, rs1726866, and rs10246939) (Kim et al. 2003) were obtained by the same methods described above for TAS1R1. Color videomicroscopy of the tongue tip was used to assess FP density in a 6-mm-diameter circular template, using methods described previously (Duffy et al. 2004). Counts from the left and right sides of the tongue tip were averaged to obtain the mean number of FP count per standard area. Subjects also reported the intensity of 3.2mM PROP solution sampled with the whole mouth in standardized protocol reported previously (Duffy et al. 2004).
Data analysis
Data analyses were conducted with SPSS (version 17.0); significance criterion was set at P ≤ 0.05. Linear regression analysis, accounting for outliers, was used to test associations between the TAS1R1 SNPs and taste intensity. An analysis of covariance (ANCOVA), controlling for age, sex, and intensity of tones as a cross-modal standard, was used to compare rs17492553 and rs34160967 genotype group differences in the intensities of NaCl, sucrose, citric acid, and quinine hydrochloride on the FP and circumvallate papillae, and as perceived with the whole mouth. We combined AA and AG as 1 genotype group due to small numbers in the AA group, reflecting 1 or 2 copies of the minor allele. The 2 AA homozygotes were omitted in a subanalysis with no change in the findings reported. Post-hoc analyses were conducted with t-tests based on the error term from the overall analysis of variance (ANOVA) (Keppel 1991).
The impact of TAS2R38 diplotype, PROP bitterness phenotype, and FP number was tested as separate covariates in the ANCOVA, as an additional independent variable in 2-way ANCOVA (for example, PROP taster group by TAS1R1 genotype for each SNP), and by removing PROP nontasters (PROP bitterness < moderate on the gLMS) or supertasters (PROP bitterness > very strong on the gLMS) in the analysis. These measures of variability in oral sensation did not influence the statistical significance or pattern of results when used as covariates. The interaction term was not significant in the individual 2-way ANOVA with TAS1R1 genotype (for each SNP) and either taste phenotypes or TAS2R38 genotype groups. Finally, removing PROP nontasters or supertasters from the analyses changed neither the significance nor the patterns of findings. The data from these subanalyses are not presented in the article.
Results
The genotype frequencies for both TAS1R1 SNPs were not significantly different from those reported for Americans of northern/western European descent listed at www.ncbi.nlm.nih.gov/SNP (dbSNP 2013) (Table 1). Furthermore, both SNPs were in Hardy–Weinberg equilibrium. For rs17492553, T was the minor allele in our sample, compared with C as reported in National Center for Biotechnology Information (NCBI) although this is likely just a sampling effect because both alleles are very close in frequency and not statistically different than the NCBI reference data. As expected based on published allele frequency differences between ethnic groups, our distribution for rs34160967 genotypes differed from the Japanese cohort utilized in the Shigemura et al. study (2009) (χ2 = 30.23, P < 0.0001), with relatively fewer AA and AG genotypes in our cohort (Table 1).
Table 1.
Genotype frequencies of TAS1R1 intronic (rs17492553) and exonic SNP (rs34160967) in the study sample versus published dbSNP databasea and other studiesb,c
| rs17492553 | Study sample | European American populationa | ||
|---|---|---|---|---|
| CC | 0.33 | 0.21 | ||
| CT | 0.43 | 0.50 | ||
| TT | 0.23 | 0.29 | ||
| rs34160967 | Study sample | European American populationa | Shigemura et al. (2009)b | Raliou et al. (2009b)c |
| AA | 0.02 | — | 0.18 | 0.02 |
| AG | 0.23 | 0.25 | 0.43 | 0.36 |
| GG | 0.75 | 0.75 | 0.39 | 0.62 |
aFrequencies from NCBI dbSNP database 2013. The study sample and published dbSNP database are of similar populations: rs17492553—χ2 (2) = 3.88, P = 0.14; rs34160967—Fisher’s exact (P = 0.44).
b,cFrom Fisher’s exact testing, the study sample distribution for rs34160967 was different than in adults recruited in Japan (Shigemura et al. 2009; P < 0.01) and not different from those recruited in France (Raliou et al. 2009b; P = 0.13).
Genotype variation in perceived intensities of regionally applied and whole-mouth taste sensations
The cohort showed variability in ratings for prototypical tastants applied to the FP and the circumvallate papillae and sampled with the whole mouth. Our cohort appeared to capture the usual variation in regionally applied tastants—the distribution of ratings was not statistically different from a large unpublished database (n = 450) comprising patients with chemosensory complaints and controls (Kolmogorov–Smirnov statistics, P > 0.05).
The mean intensity of tastants applied to the anterior tongue (FP) was near “moderate,” ranging from “no sensation” to above “very strong.” The variance across the distribution of ratings was lowest for sucrose; all other qualities had significantly higher variances (F-ratio > 2.0, P < 0.01). Mean posterior tongue (circumvallate) intensities were marginally larger than those for the FP; however, the magnitude varied by quality—paired t-tests showed significant differences by locus (P < 0.01), except for citric acid where a significant difference across regions was not observed (P = 0.34). The mean intensity ratings for the whole mouth were near “strong”; this was considerably higher than regional application on the anterior and posterior tongue (paired t-tests, P < 0.01). Again, sucrose showed the least variance out of the 4 prototypical tastants (F-ratio > 2.0, P < 0.01).
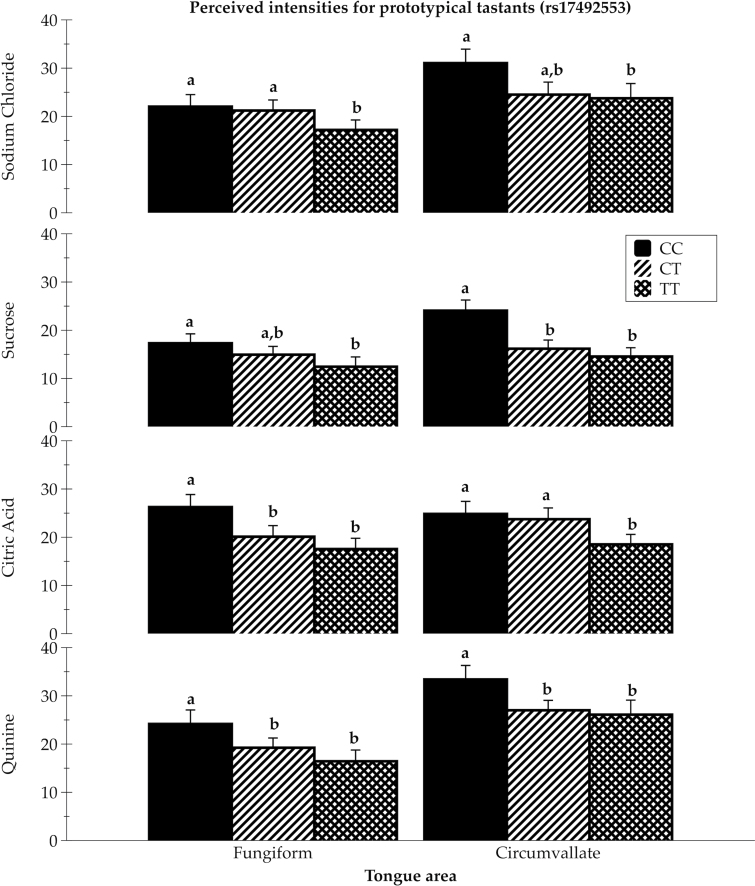
Mean differences across rs17492553 genotype groups for intensities of regionally applied tastants were significant in an ANCOVA model controlling for age, sex, and intensity of tones as a cross-modal standard. The TT homozygotes reported lower intensities than CC homozygotes for all 4 tastants, with heterozygotes falling between the two in most cases. This pattern was also present for whole-mouth ratings but was significant only for NaCl and citric acid. The intronic SNP (rs17492553, C/T) had more significant comparisons with the taste intensities than the exonic SNP (rs34160967, G/A), possibly because the study cohort showed representation across all 3 intronic SNP genotypes (CC, CT, and TT).
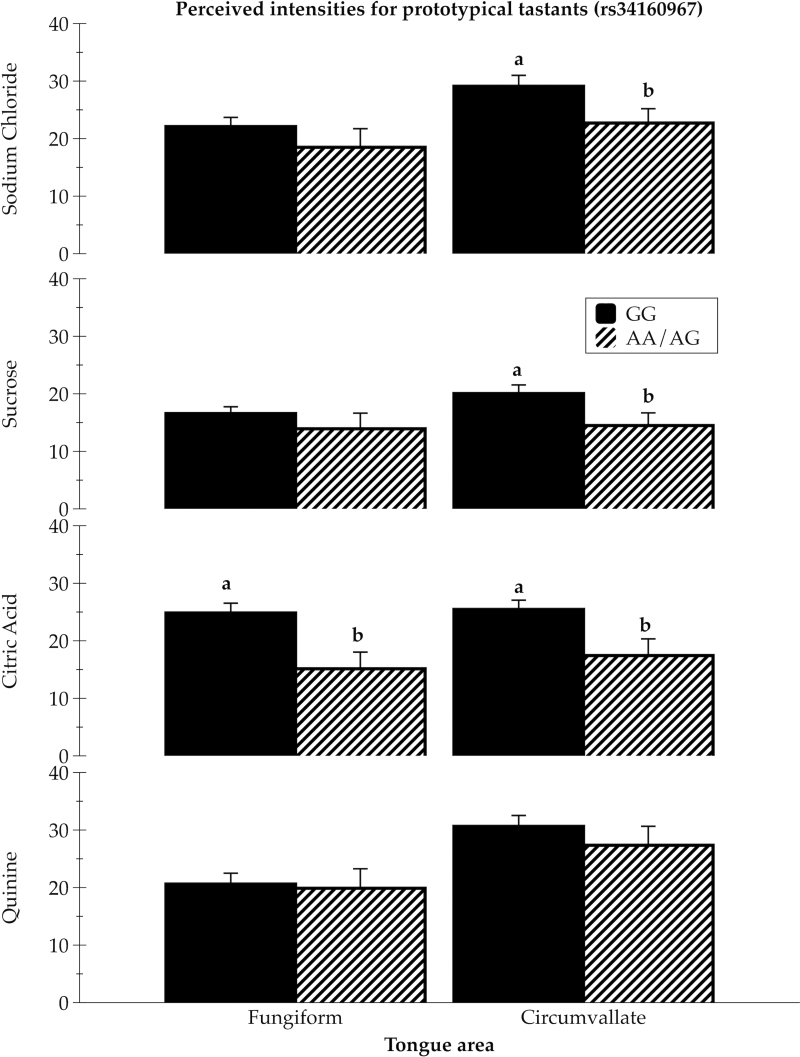
Although not significant, the exonic SNP rs34160967 also tended to associate with differences in the mean intensity of regionally applied tastants in ANCOVA controlling for age, sex, and intensity of tones as a cross-modal standard (F (4, 262) = 2.02; P < 0.10). The AA/AG genotype group reported lower intensities than GG homozygotes for all 4 tastants (both regionally applied and sampled with whole mouth) although not all of the stimuli applied regionally showed significant differences.
Salt
As shown in Figure 1, among rs17492553 genotype groups, TT homozygotes reported significantly lower intensities than did CC and CT genotypes for the regionally applied 1M NaCl on the FP. TT homozygotes also reported significantly lower intensities than did CC homozygotes for the regional application of 1M NaCl on the circumvallate papillae. Consistent with fungiform and circumvallate ratings, TT homozygotes reported significantly lower intensities than CC homozygotes for 1M NaCl sampled with the whole mouth (P < 0.02; data not shown).
Figure 1.
Intensity ratings for 1M sodium chloride (mean ± SEM), 1M sucrose (mean ± SEM), 32mM citric acid (mean ± SEM), and 1mM quinine hydrochloride (mean ± SEM), applied bilaterally to the tongue tip (fungiform papillae) and to the back of the tongue (circumvallate papillae) among 3 rs17492553 genotype groups (0.33 CC, 0.43 CT, and 0.23 TT), controlling for age, sex, and intensity of tones as a cross-modal standard; [F (6, 261) = 2.18, P < 0.04], with significant pairwise comparisons shown. Different superscript letters a, b indicate significant differences at P < 0.05.
For rs34160967 (G > A, exon 3), the AA/AG group reported significantly lower intensities than did GG homozygotes for the regionally applied 1M NaCl on the circumvallate papillae (Figure 2; P < 0.05).
Figure 2.
Intensity ratings for 1M sodium chloride (mean ± SEM), 1M sucrose (mean ± SEM), 32mM citric acid (mean ± SEM), and 1mM quinine hydrochloride (mean ± SEM), applied bilaterally to the tongue tip (fungiform papillae) and to the back of the tongue (circumvallate papillae) among 2 rs34160967 genotype groups (0.25 AA/AG and 0.75 GG), controlling for age, sex, and intensity of tones as a cross-modal standard; [F (4, 262) = 2.02, P < 0.10], with significant pairwise comparisons shown. Different superscript letters a, b indicate significant differences at least P < 0.05.
Sweet
As shown in Figure 1, among rs17492553 genotype groups, TT homozygotes reported significantly lower intensities than did CC homozygotes for the regional application of 1M sucrose on the FP. On the circumvallate papillae, both heterozygotes and TT homozygotes reported significantly lower intensities than did CC homozygotes for 1M sucrose. For the whole-mouth ratings, a similar, albeit nonsignificant, trend was seen for 1M sucrose, with TT homozygotes tending to report lower intensities than both CC and CT groups (data not shown).
In the rs34160967 (exon 3 SNP) analysis, the AA/AG group reported significantly lower intensities than did GG homozygotes for the regionally applied 1M sucrose on the circumvallate papillae (Figure 2; P < 0.05).
Sour
Similar to the trend seen with intensity ratings of 1M sucrose, among rs17492553 genotype groups, both TT and CT genotypes reported significantly lower intensities than CC homozygotes on the anterior tongue for 32mM citric acid (Figure 1). TT homozygotes also reported significantly lower perceived intensities than both CT and CC genotypes on the posterior tongue for citric acid. For the whole-mouth ratings, individuals with the TT genotype also reported significantly lower intensities than both CT (P < 0.01) and CC genotypes (P < 0.05) for 32mM citric acid (data not shown).
As shown in Figure 2, among rs34160967 genotypes, the AA/AG group reported significantly lower intensities than did GG homozygotes for 32mM citric acid on the anterior (P < 0.01) and posterior tongue (P < 0.01). The whole-mouth ratings for 32mM citric acid tended to show the same pattern, with AA/AG group reporting lower intensities than GG homozygotes (P < 0.07) (data not shown).
Bitter
Quinine intensity also differed across rs17492553 genotype groups with regional application (Figure 1). TT homozygotes and heterozygotes reported significantly lower intensities than CC homozygotes for 1mM quinine applied to the FP. Similar patterns were also observed for quinine applied to the circumvallate papillae, with TT homozygotes and heterozygotes reporting significantly lower intensities than CC homozygotes. Although not significant, a consistent trend was seen with whole-mouth ratings for 1mM quinine, with TT homozygotes reporting lower intensities than both CC and CT groups (data not shown).
Among rs34160967 genotypes, there was no evidence of differences in bitterness perceived from whole-mouth sampled and regionally applied 1mM quinine.
Discussion
Polymorphisms in the TAS2R38 gene and others in the TAS2R bitter receptors family are associated with variation in oral sensation, preference, and dietary behaviors. Although TAS1R1 polymorphisms have been tied to differences in threshold and intensity of umami stimuli, data from rodents (Nelson et al. 2002; Kusuhara et al. 2013) and mammals (Thompson et al. 1982; Friedl et al. 1990; Ratcliffe et al. 2003; Jiang et al. 2012), as well as from our laboratory (Rawal et al. 2009), suggest that these polymorphisms also associate with variation in taste perception beyond umami. In this study, we report from an existing database that an intronic (rs1749255) and exonic (rs34160967) SNP in TAS1R1 were associated with modest intensity differences of concentrated aqueous solutions across 4 prototypical tastants (sucrose, NaCl, citric acid, and quinine hydrochloride) applied regionally and/or with intensity differences of NaCl and citric acid perceived with the whole mouth. These differences were statistically independent of the effects of PROP taster status and density of FP on perceived taste intensity. If confirmed, this study’s findings provide impetus for future research to assess if TAS1R1 polymorphisms have an indirect or direct influence on overall taste functioning or are merely a genetic marker for differences in taste intensity.
Of the 2 TAS1R1 SNPs examined, all 3 genotype groups were represented for the intronic SNP (rs17492553, C/T) (Table 1), and CC homozygotes reported higher intensity than the TT homozygotes across all tastants applied to the tongue tip and posterior tongue and, in some cases, sampled with the whole mouth. The magnitude of difference averaged approximately 40%, ranging from about 28% to 61%. The exonic SNP (rs34160967, G/A, p.A372T) showed a similar association with taste intensity although the study cohort had only 2 AA homozygotes (Table 1). Individuals with one or more copies of the A minor allele reported lower intensities than GG homozygotes, significant only for some tastants (not quinine) and mostly at the circumvallate region. If the exonic SNP showed the same taste effects as the intronic SNP, the AA homozygotes would report the lowest taste intensity, which could increase the number of significant SNP–taste associations. However, in our reanalysis of the Shigemura et al. (2009) data, AG heterozygotes had higher MSG thresholds than either AA or GG homozygotes. Thus, we are uncertain about the impact of a greater frequency of AA homozygotes on the strength of association between the exonic SNP (rs34160967) and taste intensity. The taste intensity associations did not extend to other umami-related gene polymorphisms (unpublished analyses). Using the same database and analysis strategy, we failed to find significant associations between 3 GRM4 gene polymorphisms (rs2228623, rs963733, and rs2229901) and perceived intensities of the 4 prototypical tastants used here.
There is some agreement between the present findings in adult humans and reported literature on taste associations with TAS1R1. Our data parallel those in mammals who have very limited diets, some reduced taste functioning, and TAS1R1 psuedogenes (Thompson et al. 1982; Friedl et al. 1990; Ratcliffe et al. 2003; Jiang et al. 2012). Although rodents may not be a good model for human TAS1R1 (Ishimaru et al. 2012), our data were consistent with that of Kusuhara and colleagues (2013) who found that the TAS1R1 knockout mice, compared with the heterozygous mice, had smaller nerve responses to sucrose and other sweeteners on their anterior tongue. In contrast, they did not see any significant difference between the 2 mice for nerve responses to salty, sour, and bitter compounds, which may relate to differential expression pattern and functioning of TAS1R1 in rodents versus humans (Hevezi et al. 2009; Ishimaru et al. 2012).
There may be multiple explanations for why TAS1R1 SNPs were associated with taste intensity. One or both SNPs may have a direct mechanistic impact (apart or together) on taste intensity or an indirect impact through strong linkage disequilibrium with other, unmeasured polymorphisms in TAS1R1 or other genes. We are unaware of prior data regarding the functional significance of either SNP tested here. Because the exon 3 amino acid substitution (alanine to threonine) in the exonic rs34160967 SNP is a nonconservative amino acid change that also is not conserved across species, we are uncertain whether the amino acid substitution would affect T1R1 function. Interestingly, the rs34160967 SNP lies in exon 3 that is alternatively spliced, suggesting that part of the peptide encoded by this exon is less critical for T1R1 receptor functioning and instead is involved in other ways to influence taste functioning. Although unpublished, the alternative isoform due to the skip of exon 3 was reported to NCBI in 2008 by a Japanese group and sequenced from a human spleen full-length cDNA library (NCBI accession number AK2922014). The exonic SNP also could affect splicing (it appears to be immediately adjacent to an exonic enhancer identified by the bioinformatics program ESEfinder [Cartegni et al. 2003]) or the frequency of alternative splicing of exon 3.
Although difficult to interpret possible functions of most intronic SNPs, we can speculate a possible effect of rs17492553 SNP on alternative splicing of exon 3. The rs17492553 SNP is 1155bp downstream of SNP rs34160967, in the polypyrimidine tract of intron 3, which is important for recognition of branch point and lariat formation in normal splicing (Ast 2004). The tract is already somewhat weak, with no stretches of thymines longer than 3, and the first base of exon 4 is “weak” (C, found only 10–15% of the time; G is most frequent in the consensus). The first base of exon 3 also is weak (A), further enabling its alternative splicing. Together, our phenotype–genotype observations are consistent with the speculation that one or both SNPs impact the alternative splicing of exon 3, and that the relative expression of this exon may influence taste. Our findings suggest the need for functional analyses of each SNP and haplotypes, as well as deep sequencing of the haploblock region.
We also should consider that TAS1R1 may have minimal impact on taste intensity—the observed effects may have occurred via interaction with other gene(s) that impact taste function and intensity. For example, the GNAT3 gene is highly coexpressed with TAS1R1 (Ishimaru et al. 2012) and encodes G protein alpha subunit gustducin, a taste signaling molecule involved in transduction of sweet, bitter, and umami tastes (Glendinning et al. 2005). Genetic variations occurring at GNAT3 gene have been associated with sucrose sensitivity (Fushan et al. 2010), and alpha-gustducin knockout mice show diminished behavorial and gustatory nerve responses to sweet, bitter, umami, and even highly concentrated salty but not sour substances (Glendinning et al. 2005).
Our study had a number of limitations. The database included a relatively homogenous sample of adults of European ancestry and did not capture the variation in TAS1R1 genes seen in Asians (Shigemura et al. 2009), which restricts the generalizability of present findings. A prototypical umami stimulus like MSG also was not tested, which could have tested parallels between TAS1R1 variation and the intensity of salty, sweet, sour, bitter, and umami tastes. Future research should be extended to pure and simple glutamate stimuli like MSG or MSG + IMP/GMP solutions. We were also unable to perform haplotype analysis of the intronic and exonic SNP together because grouping of the 4 potential haplotypes resulted in some group sizes that were too small for meaningful analyses (data not shown). Preliminarily, the 2 SNPs exhibit a high degree of linkage disequilibrium (D′= 0.91; G and C tending to be on the same DNA strand). Haplotype analysis of a larger data set is a logical next approach because it is feasible that some haplotypes may affect transcription (e.g., exon 3 splicing) more than others.
In conclusion, we provide human psychophysical evidence that TAS1R1 polymorphisms associate with modest differences in overall taste intensity, which were independent from other well-studied markers of variation in oral sensation. To date, the phenomenon of heightened taste sensations or “supertasting” has been consistently defined by the bitterness of PROP and density of FP although additional phenotypes and markers continue to emerge (Hayes and Keast 2011), including bitterness from capsaicin and other irritants (Green and Hayes 2004), perception of taste from thermal stimulation (Green and George 2004), and polymorphism in the gustin gene (Calò et al. 2011). The intronic SNP TAS1R1 C > T (rs17492553), alone or in combination with the exonic SNP, could add to the growing list of genetic markers for “supertasting.” Finally, the National Health and Nutrition Examination Survey for the first time includes a taste exam involving regional application of concentrated NaCl and quinine to the tongue tip (Duffy et al. 2012). Findings from this study suggest that observed variability in tongue tip taste intensity may result from taste gene polymorphisms and exposure to insults that may alter chorda tympani nerve taste (Bartoshuk et al. 2012).
Funding
This work was supported by US Department of Agriculture Hatch Project [CONS00827 and PEN04332] and the National Institutes of Health [DC008613 and DC010904].
Acknowledgements
We thank Michelle Burch, Will Eaton, and Dr Hua Li for technical assistance with the genetics experiments (University of Florida).
References
- Ast G. 2004. How did alternative splicing evolve? Nat Rev Genet. 5(10):773–782 [DOI] [PubMed] [Google Scholar]
- Bailey EH, Nichols EL. 1888. On the Sense of Taste. Science. 11(268):145–146 [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Catalanotto F, Hoffman H, Logan H, Snyder DJ. 2012. Taste damage (otitis media, tonsillectomy and head and neck cancer), oral sensations and BMI. Physiol Behav. 107(4):516–526 [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Duffy VB, Green BG, Hoffman HJ, Ko CW, Lucchina LA, Marks LE, Snyder DJ, Weiffenbach JM. 2004. Valid across-group comparisons with labeled scales: the gLMS versus magnitude matching. Physiol Behav. 82(1):109–114 [DOI] [PubMed] [Google Scholar]
- Blakeslee AF. 1932. Genetics of Sensory Thresholds: Taste for Phenyl Thio Carbamide. Proc Natl Acad Sci U S A. 18(1):120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calò C, Padiglia A, Zonza A, Corrias L, Contu P, Tepper BJ, Barbarossa IT. 2011. Polymorphisms in TAS2R38 and the taste bud trophic factor, gustin gene co-operate in modulating PROP taste phenotype. Physiol Behav. 104(5):1065–1071 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31(13):3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. 2006. The receptors and cells for mammalian taste. Nature. 444(7117):288–294 [DOI] [PubMed] [Google Scholar]
- Chaudhari N, Landin AM, Roper SD. 2000. A metabotropic glutamate receptor variant functions as a taste receptor. Nat Neurosci. 3(2):113–119 [DOI] [PubMed] [Google Scholar]
- Chaudhari N, Pereira E, Roper SD. 2009. Taste receptors for umami: the case for multiple receptors. Am J Clin Nutr. 90(3):738S–742S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Alarcon S, Tharp A, Ahmed OM, Estrella NL, Greene TA, Rucker J, Breslin PA. 2009. Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes. Am J Clin Nutr. 90(3):770S–779S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell SE, Mennella JA, Duffy VB, Pelchat ML, Griffith JW, Smutzer G, Cowart BJ, Breslin PA, Bartoshuk LM, Hastings L, et al. 2013. Gustation assessment using the NIH Toolbox. Neurology. 80(Suppl 3):S20–S24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dbSNP (Database of Single Nucleotide Polymorphisms) 2013. National Center for Biotechnology Information, National Library of Medicine [Internet]. Available from: URL http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=rs17492553; http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=rs34160967 Last accessed: June 2013.
- Dias AG, Rousseau D, Duizer L, Cockburn M, Chiu W, Nielsen D, El-Sohemy A. 2013. Genetic variation in putative salt taste receptors and salt taste perception in humans. Chem Senses. 38(2):137–145 [DOI] [PubMed] [Google Scholar]
- Duffy VB. 2007. Variation in oral sensation: implications for diet and health. Curr Opin Gastroenterol. 23(2):171–177 [DOI] [PubMed] [Google Scholar]
- Duffy VB, Doty RL, Hayes JE, Rawal S, Hoffman HJ. 2012. A chemosensory component in the 2012 National Health and Nutrition Examination Survey (NHANES): adults ages 40+ years [abstract]. Thirty-fourth Annual Meeting of the Association for Chemoreception Sciences Huntington Beach (CA) 29p; 2013 Apr 25–28; Chem Senses.; 10.1093/chemse/bjs091. 19 p. [Google Scholar]
- Duffy VB, Hayes JE, Davidson AC, Kidd JR, Kidd KK, Bartoshuk LM. 2010. Vegetable Intake in College-Aged Adults Is Explained by Oral Sensory Phenotypes and TAS2R38 Genotype. Chemosens Percept. 3(3–4):137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Sullivan BS, Faghri P. 2009. Surveying food and beverage liking: a tool for epidemiological studies to connect chemosensation with health outcomes. Ann N Y Acad Sci. 1170:558–568 [DOI] [PubMed] [Google Scholar]
- Duffy VB, Peterson JM, Bartoshuk LM. 2004. Associations between taste genetics, oral sensation and alcohol intake. Physiol Behav. 82(2–3):435–445 [DOI] [PubMed] [Google Scholar]
- Essick GK, Chopra A, Guest S, McGlone F. 2003. Lingual tactile acuity, taste perception, and the density and diameter of fungiform papillae in female subjects. Physiol Behav. 80(2-3):289–302 [DOI] [PubMed] [Google Scholar]
- Friedl WA, Nachtigall PE, Moore PWB, Chun NKW, Haun JE, Hall RW, Richards JL. 1990. Taste teception in the Pacific bottlenose dolphin (Tursiops Truncatus Gilli) and the California sea Lion (Zalophus Californianus). In: Thomas JA, Kastelein RA, editors. Sensory abilities of cetaceans: laboratory and field evidence, NATO ASI series. New York: Plenum; p. 447–454 [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Drayna D. 2010. Association between common variation in genes encoding sweet taste signaling components and human sucrose perception. Chem Senses. 35(7):579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. 2009. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 19(15):1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glendinning JI, Bloom LD, Onishi M, Zheng KH, Damak S, Margolskee RF, Spector AC. 2005. Contribution of alpha-gustducin to taste-guided licking responses of mice. Chem Senses. 30(4):299–316 [DOI] [PubMed] [Google Scholar]
- Green BG, George P. 2004. ‘Thermal taste’ predicts higher responsiveness to chemical taste and flavor. Chem Senses. 29(7):617–628 [DOI] [PubMed] [Google Scholar]
- Green BG, Hayes JE. 2004. Individual differences in perception of bitterness from capsaicin, piperine and zingerone. Chem Senses. 29(1):53–60 [DOI] [PubMed] [Google Scholar]
- Guo SW, Reed DR. 2001. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 28(2):111–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Keast RS. 2011. Two decades of supertasting: where do we stand? Physiol Behav. 104(5):1072–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB. 2011. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults. Chem Senses. 36(3):311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Feeney EL, Allen AL. 2013. Do polymorphisms in chemosensory genes matter for human ingestive behavior? Food Qual. Pref. 30(2): 202–216 Available from: URL http://dx.doi.org/10.1016/j.foodqual.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevezi P, Moyer BD, Lu M, Gao N, White E, Echeverri F, Kalabat D, Soto H, Laita B, Li C, et al. 2009. Genome-wide analysis of gene expression in primate taste buds reveals links to diverse processes. PLoS One. 4(7):e6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Abe M, Asakura T, Imai H, Abe K. 2012. Expression analysis of taste signal transduction molecules in the fungiform and circumvallate papillae of the rhesus macaque, Macaca mulatta. PLoS One. 7(9):e45426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK. 2012. Major taste loss in carnivorous mammals. Proc Natl Acad Sci U S A. 109(13):4956–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppel G. 1991. Design and analysis: a researcher’s handbook. 3rd ed Englewood Cliffs (NJ): Prentice Hall [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 299(5610):1221–1225 [DOI] [PubMed] [Google Scholar]
- Kim UK, Wooding S, Riaz N, Jorde LB, Drayna D. 2006. Variation in the human TAS1R taste receptor genes. Chem Senses. 31(7):599–611 [DOI] [PubMed] [Google Scholar]
- Kurihara K, Kashiwayanagi M. 2000. Physiological studies on umami taste. J Nutr. 130(4S Suppl):931S–934S [DOI] [PubMed] [Google Scholar]
- Kusuhara Y, Yoshida R, Ohkuri T, Yasumatsu K, Voigt A, Hübner S, Maeda K, Boehm U, Meyerhof W, Ninomiya Y. 2013. Taste responses in mice lacking taste receptor subunit T1R1. J Physiol. 591(Pt 7):1967–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc Natl Acad Sci U S A. 99(7):4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugaz O, Pillias AM, Faurion A. 2002. A new specific ageusia: some humans cannot taste L-glutamate. Chem Senses. 27(2):105–115 [DOI] [PubMed] [Google Scholar]
- Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA. 2010. Individual differences in AMY1 gene copy number, salivary α-amylase levels, and the perception of oral starch. PLoS One. 5(10):e13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Jr, Reedy FE., Jr 1990. Variations in human taste bud density and taste intensity perception. Physiol Behav. 47(6):1213–1219 [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. 2002. An amino-acid taste receptor. Nature. 416(6877):199–202 [DOI] [PubMed] [Google Scholar]
- Pepino MY, Finkbeiner S, Beauchamp GK, Mennella JA. 2010. Obese women have lower monosodium glutamate taste sensitivity and prefer higher concentrations than do normal-weight women. Obesity (Silver Spring). 18(5):959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirastu N, Robino A, Lanzara C, Athanasakis E, Esposito L, Tepper BJ, Gasparini P. 2012. Genetics of food preferences: a first view from silk road populations. J Food Sci. 77(12):S413–S418 [DOI] [PubMed] [Google Scholar]
- Raliou M, Boucher Y, Wiencis A, Bézirard V, Pernollet JC, Trotier D, Faurion A, Montmayeur JP. 2009a. Tas1R1-Tas1R3 taste receptor variants in human fungiform papillae. Neurosci Lett. 451(3):217–221 [DOI] [PubMed] [Google Scholar]
- Raliou M, Grauso M, Hoffmann B, Schlegel-Le-Poupon C, Nespoulous C, Débat H, Belloir C, Wiencis A, Sigoillot M, Bano SP, et al. 2011. Human genetic polymorphisms in T1R1 and T1R3 taste receptor subunits affect their function. Chem Senses. 36(6):527–537 [DOI] [PubMed] [Google Scholar]
- Raliou M, Wiencis A, Pillias AM, Planchais A, Eloit C, Boucher Y, Trotier D, Montmayeur JP, Faurion A. 2009b. Nonsynonymous single nucleotide polymorphisms in human tas1r1, tas1r3, and mGluR1 and individual taste sensitivity to glutamate. Am J Clin Nutr. 90(3):789S–799S [DOI] [PubMed] [Google Scholar]
- Ratcliffe JM, Fenton MB, Galef BG. 2003. An exception to the rule: common vampire bats do not learn taste aversions. Ani Behav. 65:385–389 [Google Scholar]
- Rawal S, Wallace MR, Hayes JE, Bartoshuk LM, Langee T, Sholudko A, Duffy VB. 2009. TAS1R1-intronic SNP associations with liking for dietary sources of glutamate and orosensory intensity [abstract]. Thirty-first Annual Meeting of the Association for Chemoreception Sciences Sarasota (FL) 70 p 2009. Apr 22–26; Chem Senses. 34: A63. [Google Scholar]
- San Gabriel A, Uneyama H, Yoshie S, Torii K. 2005. Cloning and characterization of a novel mGluR1 variant from vallate papillae that functions as a receptor for L-glutamate stimuli. Chem Senses. 30 (Suppl 1):i25–i26 [DOI] [PubMed] [Google Scholar]
- Shigemura N, Shirosaki S, Sanematsu K, Yoshida R, Ninomiya Y. 2009. Genetic and molecular basis of individual differences in human umami taste perception. PLoS One. 4(8):e6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PB, Schuster B, Seo HS. 2010. Variation in umami taste perception in the German and Norwegian population. Eur J Clin Nutr. 64(10):1248–1250 [DOI] [PubMed] [Google Scholar]
- Tepper BJ. 2008. Nutritional implications of genetic taste variation: the role of PROP sensitivity and other taste phenotypes. Annu Rev Nutr. 28:367–388 [DOI] [PubMed] [Google Scholar]
- Thompson RD, Elias DJ, Shumake SA, Gaddis SE. 1982. Taste Preferences of the Common Vampire Bat (Desmodus-Rotundus). J Chem Ecol. 8:715–721 [DOI] [PubMed] [Google Scholar]
- Toyono T, Seta Y, Kataoka S, Kawano S, Shigemoto R, Toyoshima K. 2003. Expression of metabotropic glutamate receptor group I in rat gustatory papillae. Cell Tissue Res. 313(1):29–35 [DOI] [PubMed] [Google Scholar]
- Wooding S. 2006. Phenylthiocarbamide: a 75-year adventure in genetics and natural selection. Genetics. 172(4):2015–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. 2003. The receptors for mammalian sweet and umami taste. Cell. 115(3):255–266 [DOI] [PubMed] [Google Scholar]
- Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol Evol. 4(1):73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]