Abstract
Aggressive metastasis is the chief cause of the high morbidity and mortality associated with pancreatic cancer, yet the basis for its aggressive behavior remains elusive. Extracellular DNA (exDNA) is a recently discovered component of inflammatory tissue states. Here we report that exDNA is present on the surface of pancreatic cancer cells where it is critical for driving metastatic behavior. ExDNA was abundant on the surface and vicinity of cultured pancreatic cancer cells, but absent from normal pancreas cells. Strikingly, treatment of cancer cell cultures with DNase I to degrade DNA non-specifically reduced metastatic characters associated with matrix attachment, migration and invasion. We further assessed the role of exDNA in pancreatic cancer metastasis in vivo using an orthotopic xenograft model established by implantation of pancreatic cancer cells expressing firefly luciferase. Non-invasive bioluminescent imaging confirmed that DNase I treatment was sufficient to suppress tumor metastasis. Mechanistic investigations suggested the existence of a positive feedback loop in which exDNA promotes expression of the inflammatory chemokine CXCL8 which leads to higher production of exDNA by pancreatic cancer cells, with a significant reduction in CXCL8 levels achieved by DNase I treatment. Taken together, our results strongly suggest that exDNA contributes to the highly invasive and metastatic character of pancreatic cancer.
Keywords: extracellular DNA, DNase, CXCL8/IL-8, pancreatic cancer, metastasis, inflammation
Introduction
Pancreatic cancer is the fourth leading cause of cancer death in the United States and one of the most lethal malignancies, with a five-year survival rate less than 5% and a death/incidence ratio of approximately 0.99 (1). Eighty-90% of pancreatic cancer patients have already developed metastatic cancer at the time of diagnosis (2). The mechanisms underlying the development of metastases during pancreatic carcinogenesis are poorly understood (3). Understanding these mechanisms may provide novel approaches to pancreatic cancer treatment and/or prevention.
The development of metastasis is determined by both genetic alterations in tumor cells and by the surrounding microenvironment (4, 5). Inflammation has been shown to facilitate tumor metastasis, from dissemination of cancer cells at the primary tumor site to their implantation at secondary sites (6–8). The link between inflammation and pancreatic cancer development has been well recognized. Chronic pancreatitis/inflammation is among the few high risk factors of developing pancreatic cancer (1, 9–13).
Inflammation mediators including CXCL8 have been shown to induce the production of extracellular DNA (exDNA)-containing fibers by neutrophils (nicknamed neutrophil extracellular traps or NETs) (14). Extracellular DNA traps have been associated with several chronic inflammatory diseases, including cystic fibrosis, small-vessel vasculitis, and deep vein thrombosis (14–18). It has been recently reported that cytokine induced production of NETs contribute to cancer-associated thrombosis (19). Production of exDNA has been reported in a number of inflammatory cell types in addition to neutrophils, including macrophages, mast cells, platelets, dendritic cells, B- and T-lymphocytes. These inflammatory cells have all been found in the stroma of pancreatic cancer (20, 21) and have been implicated in angiogenesis and tumor metastasis by secreting cytokines and chemokines (22).
Others have shown significantly elevated production and secretion of the chemokine CXCL8 in the highly liver-metastatic pancreatic cancer cell line BxPC-3 in comparison with the immortalized normal human pancreatic ductal epithelium HPDE cell line (23). In this study, we asked whether normal or neoplastic pancreatic cells produced exDNA. We observed a large amount of exDNA on the surfaces and in the vicinity of the cultured BxPC-3 and MiaPaCa2 pancreatic cancer cell lines but not the normal pancreas HPDE cell line. We went on to assess the relationship between exDNA, CXCL8 and features of metastasis in both cell and tumor models of pancreatic cancer. This is the first report of the association of an abundant amount of surface exDNA with pancreatic cancer cells and its possible role in pancreatic cancer metastasis. This discovery may provide a new target and novel strategies for prevention, diagnosis, and treatment of this lethal cancer.
Materials and Methods
Cell culture and DNase I treatment of cells
We obtained MiaPaCa-2, BxPc3, and Panc-1 human pancreatic cancer cell lines from American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured at 37°C with 5% CO2 in RPMI 1640 medium (Mediatech, Inc., Herndon, VA), supplemented with 10% fetal bovine serum (Omega Scientific, Inc, Tarzana, CA); 2.5 mg/ml glucose; 1% L-glutamine; and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). Immortalized normal human pancreatic ductal epithelial (HPDE) cells and HPDE-KRASG12D cells expressing mutated KRAS, were kindly provided by Dr. Ming-Sound Tsao, University of Toronto, Canada (24). HPDE cells were cultured in keratinocyte serum-free medium supplemented with epidermal growth factor (EGF) and bovine pituitary extract (Invitrogen). DNase I treatment of 105 cells was at the concentration of 3 units/100µl. All the in vitro DNase I treatments lasted for 24 – 72 hours depending on different assays.
MTT cell viability and cell growth assay
Cell survival and growth was measured by MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) assay as previously described (25, 26). HPDE control cell line and pancreatic cancer cell lines BxPc3 and MiaPaCa-2 were examined with or without DNase I treatment (3 units/well/10,000 cells) 24 hours after cells were treated with DNase I.
Wound-healing assay
Cells were grown in 24-well plates in 500 µL medium/well until confluence was reached. A wound was made by scratching the cells with a 10-ul pipette tip in PBS, followed by replacement by culture media with and without DNase I (15 U/well for up to 3 days). The wounded monolayer was photographed overtime and cell migration was assessed by measuring gap sizes at multiple fields using ImageJ (National Institute of Mental Health, Bethesda, Maryland, USA).
Cell migration assay
Cell migration assays were conducted using a modified 24-well Boyden chamber. The top chamber (Transwell) with 8.0 µm pores on the filter membrane (BD Labware, Le Pont De Claix, France) was inserted into a 24-well plate (bottom chamber). Ten percent fetal bovine serum-containing medium was placed in the lower chambers to be used as a chemo-attractant. Cells (3×105) in a 300 µL volume of serum-free medium with or without DNase I were placed in the upper chambers and incubated at 37°C for 24 h. Migrated cells on the bottom surface of the filter were fixed, stained with Crystal Violet.
Crystal violet staining
Twenty four hours after culturing cells in the top chamber, medium in the transwell was siphoned off and the chamber was moved to the bottom chamber containing 4% paraformaldehyde to fix cells for 10 minutes. Top chamber was rinsed in PBS and inverted for staining. 50 µl of 5% Crystal Violet (Sigma-Aldrich, St. Louis, MO) in 25% methanol was applied onto the bottom of the filter of the top chamber and cells were stained for 10 minutes. Excess crystal violet was washed off by plunging the top chamber into distilled water in a beaker several times. Finish washing in a second beaker till water is clear. Cells on top side of filter (cell that did not migrate) were removed using a moist cotton swab. The filter was then air dried. Cells in 5–7 random fields were counted at 40× objective lens under an inversion microscope.
Cell invasion assay
The system setup for invasion assay using Boyden chamber was exactly the same as for cell migration assay, except that the Transwell filter was coated with 40 µL Matrigel (BD Bioscience, Bedford, MA) and cells were stained 48h instead of 24h after culture.
Fluorescent dye staining
Cells (1×105/well) were seeded on sterile cover slips that were placed in 6-well plate. Two days later, culture medium was aspirated and the cover slips were rinsed with PBS. Cells grew on the cover slips were then fixed in 4% paraformaldehyde for 10 min, followed by rinsing with water. Cells were stained with DAPI or Sytox Green by mounting the cover slip with the mounting medium containing DNA dye 4’, 6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA), or by mounting the cover slip on a regular glass slide with KPL mounting medium (Gaithersburg, MD) containing another DNA fluorescence dye Sytox Green (Molecular Probes), a non-living cell-permeant DNA binding dye. For staining exDNA induced by CXCL8, Sytox Green was added to cells cultured in a 24 well culture plate at 1 µM final concentration.
For staining cells in paraffin sections, tissue slides were deparaffinized with 100% xylene twice, 10 min each and hydrolyzed in 100% ethanol twice (5 min each), 95% ethanol twice (5 min each), 80% ethanol twice (5 min each), and water twice (5 min each). Then, DNA was stained by mounting the slides with the mounting medium containing DAPI (Vector Laboratories, Burlingame, CA).
Immunofluorescence staining with DNA antibody
Cells (1×105/well) were seeded on sterile cover slips that were placed in 6-well plate. Two days later, culture medium was aspirated and the cover slips were rinsed with PBS. Cells grew on the cover slips were then fixed in 4% paraformaldehyde for 10 min. followed by cold methanol and acetone treatment (5 min./each) for cell fixation and permeabilization, then by PBS rinsing. Some cells were not treated for permeabilization after the fixation in paraformaldehyde. Next, cells were blocked with 5% bovine serum albumin (BSA) in PBS for 1 hour at room temperature, followed by rinsing with PBS three times, 3 min/each. Then cells were probed with mouse DNA antibody (1:100 dilution in 5%BSA/ PBS) either 1 hour at room temperature or at 4°C overnight. DNA antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). After the primary antibody reaction, cells were rinsed with PBS three times, followed by secondary antibody (1:800 dilution in 5%BSA/ PBS) reaction at room temperature for 40 min. The secondary antibody was Alexa Fluor 680- conjugated bovine antimouse (Life Technologies, NY). Following secondary antibody reaction, cover slips were rinsed in PBS and water separately, three time each solution. Finally, cover slips were mounted on regular glass slides either with KPL mounting medium, or with the mounting medium containing DAPI.
Immunofluorescence staining with integrin beta1 antibody and DNA antibody
Cells and cover slips were prepared the same way as above for staining with DNA antibody alone. Cells were not treated for permeabilization after the fixation. Here, 1:100 dilution of both mouse DNA and rat integrin beta 1 (a gift from Dr. Anne Cress) primary antibodies were mixed to probe cells. Alexa Fluor 680/488- conjugated bovine anti-mouse/anti-rat secondary antibodies for DNA and for beta 1 separately were mixed for secondary antibody reaction.
Immunohistochemistry (IHC) staining with DNA antibody
Tissue slides were deparaffinized and hydrolyzed as described in the “Fluorescent dye staining” section. After the hydration, antigen retriever step was followed. Slides were emerged in the antigen retriever solution (0.01M sodium citrate, 0.05% Tween 20, pH 6.0) and heated in a microwave. The solution was brought to boil and the power was turned off. Then heat was turned on again to bring the solution to boil. The power “on and off” was repeated for 10 min. to keep the solution to boil, but was kept from boiling out of the container. At the end of 10 min, the container was left on ice for 20 min. Then, slides were rinsed with PBS 3 times before they were subjected to immunoreaction. Procedures for blocking, primary antibody reaction, and secondary antibody reaction were the same as for the immunofluorescence stain as described above, except for using house radish peroxidase conjugated mouse secondary antibody (Santa Cruz, CA) instead of Alexa Fluor 680 conjugated secondary antibody. NovaRED substrate (Vector Laboratories, Burlingame, CA) was used for detection. After secondary antibody reaction, slides were rinsed and mounted the same as for immunofluorescence stain
Orthotopic xenograft mouse model
All procedures involving animal were approved by the University of Arizona institutional animal care and use committee, IACUC protocol #07029. Pancreatic cancer cell line MIA PaCa-2/LucE expressing firefly luciferase was used. 2 × 106 cells in 100ul Matrigel (Becton Dickenson) were injected into donor mice with severe combined immunodeficiency (SCID). After 4 days, Matrigel plugs were removed from the donor mice and implanted in the body-tail of the pancreases of receiving SCID mice at age of 6–8 weeks. Two groups of SCID mice were used. Group A: 10 SCID mice were injected with MIA PaCa-2/LucE cells and received an intraperitoneal (IP) injection of saline every other day as a control treatment. Group B: 14 SCID mice were injected with MIA PaCa-2/LucE cells together with DNase I at a concentration of 1 unit/mouse in subgroup 1 (n=7) and at a concentration of 50 units/mouse in subgroup 2 (n=7). Group B mice received IP injections of DNase I every other day at the same dosage as used in the first injection. Mice were purchased and maintained through the Experimental Mouse Shared Service in University of Arizona Cancer Center (EMSS/UACC). Mouse xenograft and DNase I application were carried out by specialists at the EMSS facility. In vivo tumor growth and spontaneous metastasis were monitored and recorded noninvasively every week by bioluminescence imaging (BLI) measuring the bioluminescence from tumor cells using an In Vivo Imaging System (AMI-1000, Spectral Instrument Imaging). At the end of the project, the mice were sacrificed and various organs were harvested for H&E staining and histology, and ex vivo bioluminescence analysis to confirm the tumor metastasis.
Histology of Pancreatic Xenografts
The pancreas, livers, spleens, diaphragms, and lungs were harvested, visually inspected, fixed in 10% formalin buffer, processed, embedded, sectioned, H&E and IHC stained with DNA antibody, and analyzed by a pathologist for the presence of tumors.
Cell adhesion assay
The 96-well microtiter plate was coated with Matrigel to assess cell adhesion to extracellular matrix as described before (27). A 150 µl cell suspension of MiaPaCa-2, BxPc3, and HPDE cells were added to the coated wells (1× 104/well) with or without the presence of 7 units DNase I/well and incubated at 37°C for 1 hr. After washing off un-attached cells, attached cells were stained with Crystal violet and counted under an inverting microscope.
Hanging drop assay
Cells were harvested and suspended in corresponding media as single cells. Drops (30-µl/drop) of media containing 20,000 cells/drop of HPDE or MiaPaCa-2 cells were pipetted onto the inner surface of a lid of a 24-well plate. The lid was placed upright on the plate so that the drops were hanging from the lid with cells suspended within them. The corresponding bottom wells contained media to maintain humidity. After culturing cells at 37°C overnight, hanging drops were transferred to microfuge tubes and were pipetted seven times up and down with a 200-µl standard tip, fixed with 2% paraformidehyde, and aliquots were spread on regular microscope glass slides covered with coverslips. Images of five random fields were taken and clusters containing ≥ 10 cells were counted.
Real-time PCR and CXCL8 ELISA
Cells (3×105 in 500 µl/well) were seeded in 24-well plate. In 24 hrs, media were changed and DNase I were added (15U/wells) to treatment wells. Cells without DNase I addition were controls. Two days later, culture media were collected for CXCL8 ILISA while cells were lysed for total RNA extraction and real-time PCR. Media were pulse centrifuged at 1000 rpm. Supernatant were transferred to new tube and stored at −80°C for later ELISA analysis. The secreted CXCL8 level was measured by using a CXCL8 ELISA assay kit (QuantiGlo Chemiluminescent Immunoassay; R&D systems). Total RNA extraction and real-time PCR were conducted as described before (26). CXCL8 forward primer sequence: ATGACTTCCAAGCTGGCCGTGGCT; and CXCL8 reverse primer sequence: TCTCAGCCCTCTTCAAAAACTTCTC.
Measurement of exDNA induced by CXCL8
100 µl of 10,000 cells/well were seeded in a 96-well plate together with 80 ng/ml of CXCL8 protein (BD Bioscience, San Jose, CA). Cells without CXCL8 treatment are control cells. After overnight incubation, 1 µM of Sytox Green was added to the cells to detect exDNA. The plates were read within 15 min in a Spectra MAX Genini fluorescence microplate reader with a filter setting of 485 nm (excitation)/538 nm (emission). For non-quantitive observation of exDNA after CXCL8 treatment, cells were seeded in a 24-well plate with or without 80 ng/ml CXCL8. After overnight incubation, 1 µM Sytox Green were added to the cells. Cells were observed under a Carl Zeiss fluorescent microscope. Images were captured using the AxioVisionE software.
Statistical analyses
All data are reported as mean ± standard deviation (SD). Statistical comparisons between treatment groups were done by the Student t test and by the Single factor ANOVA analysis. For all analyses, differences were considered significant at P < 0.05.
Results
Extracellular DNA (exDNA) on the surface of pancreatic cancer cells in vitro
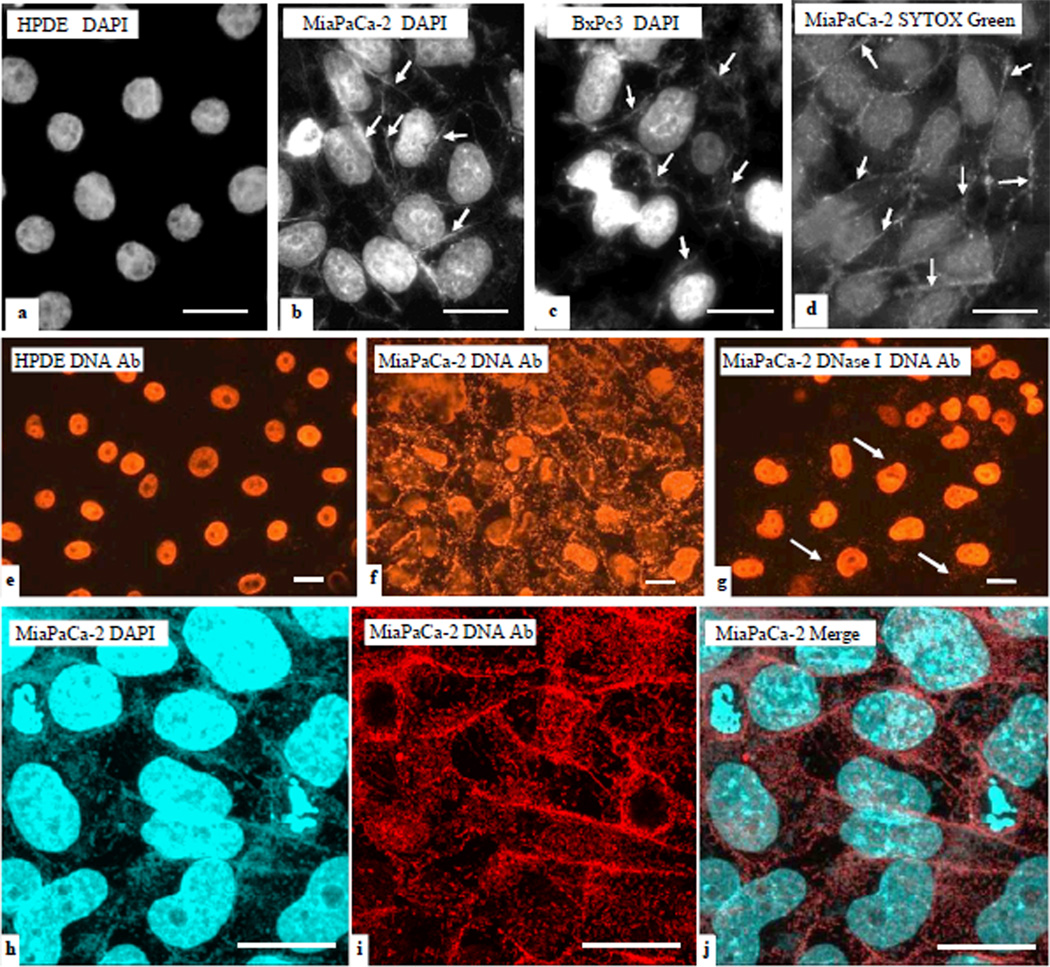
We detected exDNA on the surface and in the vicinity of cultured pancreatic cancer cell lines. We used fluorescent DNA dyes DAPI and SYTOX Green to stain cells cultured on cover slips. DAPI, or 4',6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to A-T rich regions in DNA. To validate the DAPI stain, we used another DNA fluorescent dye: Sytox Green. Unlike DAPI, this dye shows little base selectivity. ExDNA associated with cancer cell lines BxPc3 and MiaPaCa-2 was observed (Fig 1 b–d), but no exDNA was visible associated with the immortalized normal human pancreatic duct epithelial cell line HPDE (Fig. 1a). All cells were tested mycoplasma free.
Figure 1. exDNA associated with pancreatic cancer cells, but not with normal pancreas cells.
Immortalized normal human pancreatic duct epithelial (HPDE), and human pancreatic cancer cell lines BxPC3 and MiaPaCa-2 cells were cultured on cover slips, and then fixed and stained with fluorescent DNA dyes DAPI (blue in a–c) and SYTOX Green (green in d). exDNA appears as fibrous materials (arrows). Bar = 20 µm. Immunofluoerescent stain (IF) using DNA antibody was conducted on HPDE, MiaPaCa-2, and BxPc3 cells. e. DNA antibody reacts to nuclei DNA in HPDE cells. f. DNA antibody reacts to exDNA and nuclei DNA in MiaPaCa-2 cells. g. MiaPaCa-2 cells were subjected to DNase I treatment before IF. e–g, images were taken under the regular fluorescent microscope. h–j, MiaPaCa-2 cells were double stained with DAPI and DNA antibody and images were taken under the confocal microscope. DAPI stain was done after DNA antibody. Cells in h–j were not permeabilized before staining whereas cells in e–g were permeabilized. Bar = 20 µm
To confirm that the fluorescent dyes were detecting DNA molecules, we used a DNA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) to carry out the immunofluorescence assay (IFA) with a fluorochrome Alexa Fluor 594 conjugated secondary antibody. In agreement with the fluorescent dye staining, abundant exDNA associated with pancreatic cancer cell lines was observed (Fig. 1f) by IFA, whereas no exDNA but only nuclei DNA positively reacted to the DNA antibody in the control cell line HPDE (Fig. 1e). The IFA results were examined under both regular fluorescence microscope (Fig. 1e–g) and the confocal microscope (Fig. 1h–j). When MiaPaCa-2 cells were treated with DNase I before IFA, only trace amount of extracellular fluorescent materials were seen (Fig. 1g arrows). The exDNA associated with cancer cells does not seem to be produced by dying cells, because HPDE has a higher apoptosis level than BxPC3 cells but no associated exDNA (Fig. S1). We also used fluorescein diacetate to conduct a viability assay and showed that DNase I treatment did not affect cell viability (data not shown). Inactivated DNase I had no effect on exDNA (Fig. S2).
Extracellular DNA (exDNA) association with metastasized pancreatic cancer cells in situ
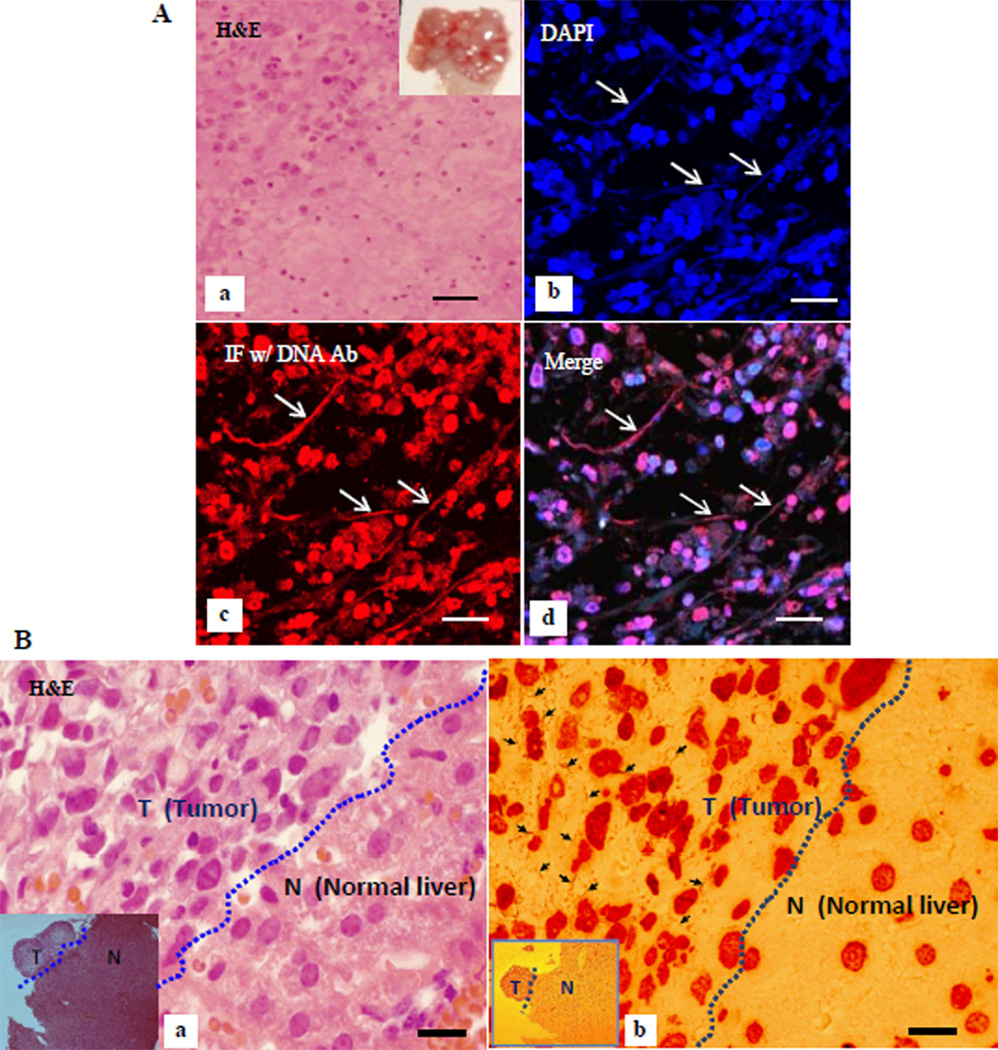
To examine the presence of exDNA in pancreatic cancer tissue in vivo, we conducted immunofluorescence assay (IFA) using the DNA antibody on mouse tissue sections. Figure 2A shows IFA of exDNA in metastatic pancreatic cancer in diaphragm of an orthotopic xenograft mouse model. Figure 2B shows exDNA detected by IHC in metastatic pancreatic cancer in liver. These results suggest exDNA may be related to pancreatic cancer metastasis.
Figure 2. exDNA detected in vivo in mouse tissues.
A, pancreatic cancer metastasized to the diaphragm in a xenografted mouse. a, H&E of a tumor growing diaphragm (inset) section. b–d, double staining of a tumor growing diaphragm section with DAPI and imunofluorescent stain (IF) with DNA antibody, imaged under a confocal microscope. b, DAPI stain, c, IF with DNA antibody, and d, merge image of b and d. Arrows point to exDNA. Bar = 40um. B, exDNA detected in the metastasized tumor. IHC using DNA antibody revealed abundant exDNA (b, arrows) in a pancreatic cancer metastasized to liver tissue harvested from an orthotopically xenografted mouse. a, the corresponding H&E of IHC (b). No exDNA were observed in the normal liver tissue. Insets are lower magnification image using a 5× objective lens (T-tumor, L-liver). Bar = 20 µm.
ExDNA does not affect cell growth in vitro
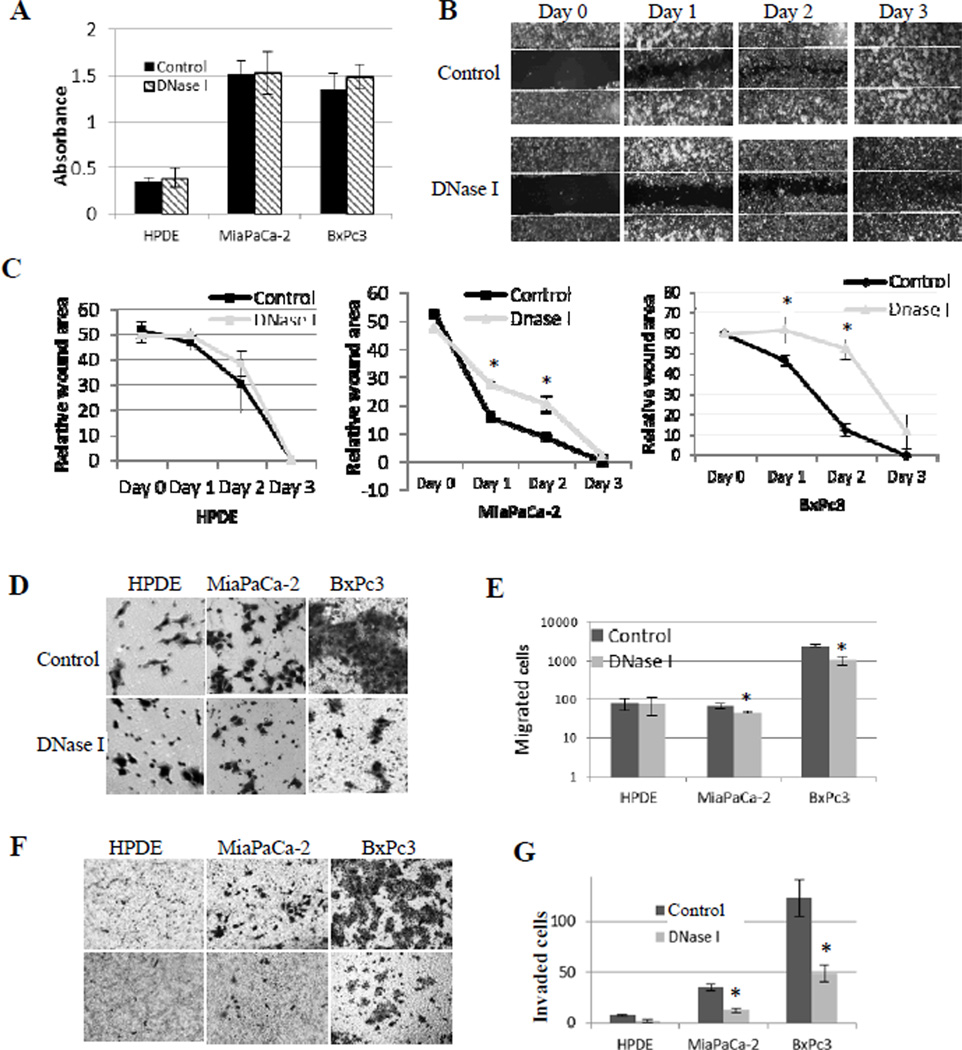
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) colorimetric assay is an established method of determining cell viability and cell growth (25). To assess if exDNA affect cell proliferation, we used MTT assay to measure the cell growth of different cell lines with and without DNase I treatment. Our results show that cell growth and cell viability were not affected by DNase I treatment (Fig. 3A). The degradation of cell surface exDNA by DNase I is shown in Fig. 1g by arrows. These data indicate the presence or absence of cell surface exDNA does not affect cell viability and growth. Also these results demonstrate that at the concentration we used in our experiments DNase I can digest exDNA but does not affect cell viability and cell growth, which is important for using DNase I to study cancer cell metastasis in vitro.
Figure 3. exDNA affects cell migration and invasion, but not cell growth in pancreatic cancer cell lines.
A, Cell proliferation was assayed by MTT method and the absorbance was read at 595 nm. Absorbance value positively correlated with cell numbers. Three triplicates sample in each plate per experiment. The results were reported as mean value of three experiments. B, Wounded areas were measured for 3 days after scratching the monolayer of cells in three cell lines. Images are representative of the wound-healing by MiaPaCa-2 cells. C, DNase I delays the wound healing in cancer cell lines, but not the HPDE normal cell line. Charts are plots of wounded areas of three cell lines measured through ImageJ of three random areas in three days. Data from three repeated experiments are presented as means ± SD, (*, p < 0.05, n=9). D, representative photographs of transwell-migration assay. Cells were seeded in transwells in the absence (control) and presence of DNase I (15 unites/well); after 24 hours, cells migrated to the basal side of the transwell membranes were stained with 0.5% Crystal Violet and 7 random views were counted. E, bar graph shows migrated cell numbers in normal pancreas HPDE cell line and two cancer cell lines. Data from three repeated experiments are presented as means ± SD, (*, p < 0.05, n=21). F, cell invasion setting is the same as for cell migration in D, except for transwell membranes were coated with Matrigel. G, bar graph shows numbers of cells invading through Matrigel in normal pancreas HPDE cell line and two cancer cell lines. Data from three repeated experiments are presented as means ± SD, (*, p < 0.01, n=21).
ExDNA affects pancreatic cancer cells’ metastatic potential in vitro
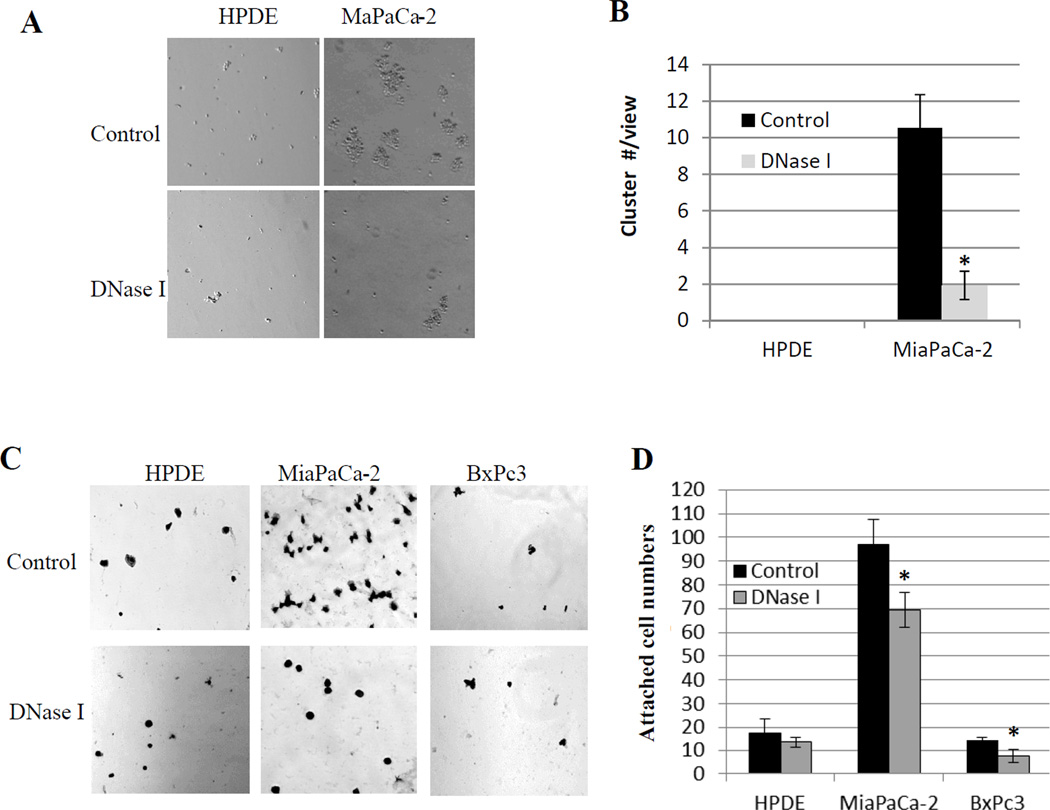
We used wound healing and transwell cell migration assays to assess the effect of exDNA on cell migration. We used transwell with Matrigel coated membrane to examine cell invasion. At the concentration of 10 units/5×105 cells, DNase I treatment did not affect cell viability, but significantly reduced the rate of pancreatic cancer cell migration (Fig. 3B–E) and invasion (Fig. 3F, G). The same DNase I treatment did not affect cell migration and invasion of the normal pancreas cells (HPDE). In addition to cell migration and invasion, cancer cell adhesion to each other and to cell matrix is another important characteristic correlated with their metastatic potential (28). We used hanging-drop assay to examine the cell-cell adhesion ability. Fig. 4A and B show that exDNA increased MiaPaCa-2 cancer cell aggregation ability, which may help them to survive circulating system during the metastasis and promote metastasis by increase cancer cell arrest in microvasculature (29). To assess the effect of exDNA on cell’s attachment ability to extracellular matrix, we carried out the cell attachment assay on Matrigel that consists of assortment of extracellular matrix proteins and is used in vitro as basement membrane (30). Fig. 4C and D showed that exDNA aids pancreatic cancer cell attachment to Matrigel, which may facilitate the invasion of metastatic cancer cells through the basement membrane in vivo. Furthermore, we examined the effect of DNase I on integrin beta-1, an integrin protein important for cell adhesion and metastasis, to see if DNase I affect other surface proteins. Figure S3 showed DNase I did not abolish beta-1 on cell surface.
Figure 4. DNase I inhibits pancreatic cancer cells aggregate to each other and attach to Matrigel.
A, Hanging droplet assay shows cell’s ability to aggregate to form clusters. MiaPaCa-2 cells form clusters, which was inhibited by DNase I treatment. B, clusters with ≥ 10 cells were counted in five random views in three repeated experiments. Bar chart shows the mean cluster numbers, *, p<0.01. C, Cell attachment assay where 1×104 cells of normal pancreas HPDE cell line and pancreatic cancer cell lines MiaPaCa-2 and BxPc3 were seeded in each well of the Matrigel-coated 96 well plate with and without the presence of DNase I (15U/well). One hour after incubate cells at 37°C, unattached cells were washed off and attached cells were stained with 0.5% Crystal Violet and 5 random views were counted under an inverted microscope. D, bar chart shows attached cell numbers presented as means ± SD, *, p < 0.01 as determined by the Student unpaired t test.
DNase I inhibits pancreatic tumor cells metastasis in orthotopic xenograft mouse model
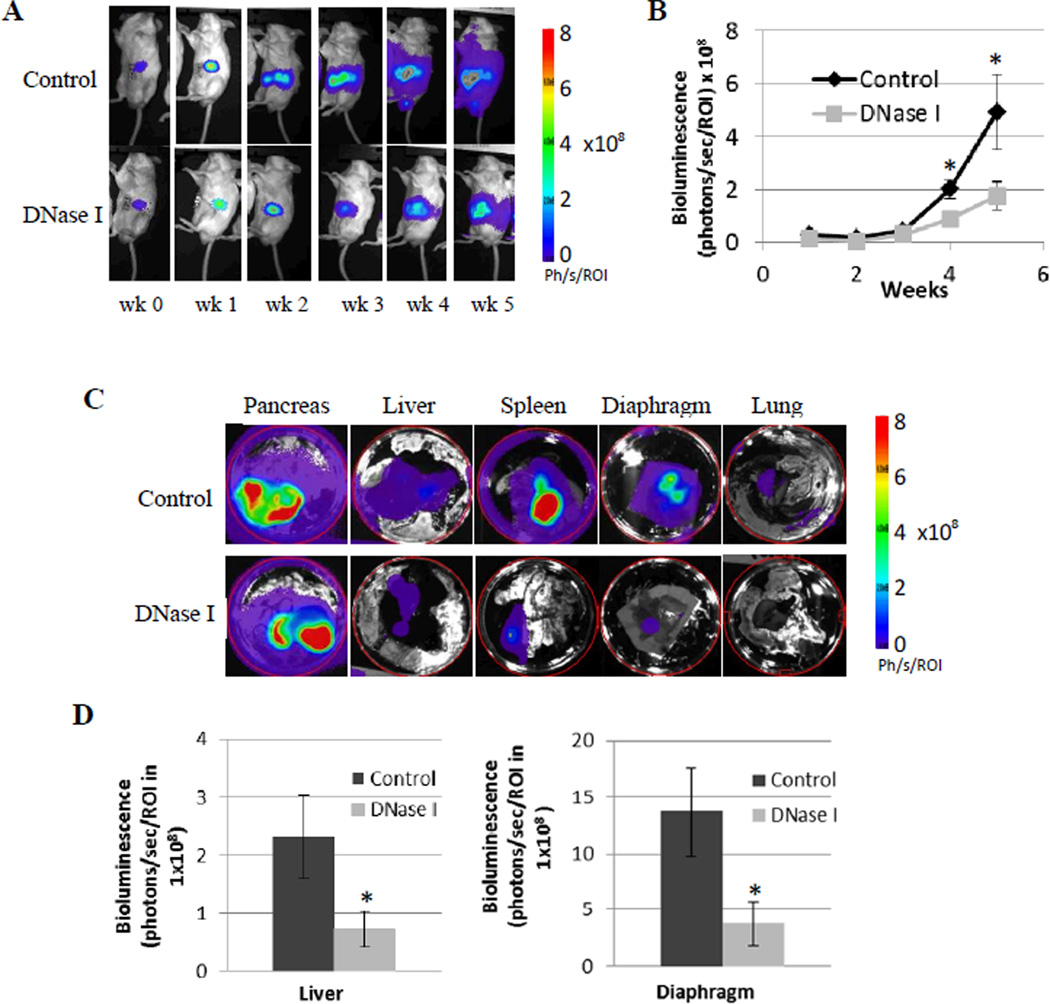
We established an orthotopic pancreatic cancer mouse model, using cancer cells genetically modified to express luciferase. This cell line was established by the EMSS/UACC and has the unique feature that it spontaneously metastasizes. Using an In Vivo Imaging System, a bioluminescence technology, we were able to noninvasively monitor the orthotopic tumor growth/metastasis in the whole mouse and to confirm the distant metastasis ex vivo after organs were harvested at the end of the experiment. Bioluminescent imaging (BLI) allowed us to quantitatively measure the tumor development, as bioluminescence intensity measured in photons/second/Region of interest (ph/s/ROI) is proportional to tumor burden. Fig. 5A and 5B show that tumor burden was significantly inhibited by DNase I treatment at weeks 4 and 5 after tumor implantation.
Figure 5. Bioluminescence imaging (BLI) monitoring of tumor growth and metastasis.
A, representative bioluminescence images of xenografts mice in control and DNase I treated group over 5 weeks of the experiment. Bioluminescence is from cancer cells genetically modified to express luciferase. Mice were imaged after injection of D-luciferin. B, chart shows intensity of bioluminescence measured as photons/second/Region of interest (ph/s/ROI) that positively correlated with tumor burden. The data are presented as means ± SD (n = 7), *, p < 0.05 as determined by the Student t test. C, Ex vivo detection of distant tumor metastasis. At the end of the experiment, organs were harvested and BLI were taken from different organs. The circle around each organ is the defined area that is the same for every image to quantify the BLI. D, DNase I treatment significantly inhibited pancreatic cancer metastasizing to liver and diaphragm. Data are presented as means ± SD (n = 7), *, p < 0.05 as determined by the Student t test and the single factor ANOVA analysis.
At the end of the mouse experiment, the In Vivo Imaging System allowed us to detect luciferase expressing tumor cells in each harvested organs quickly and quantitatively. Fig. 5C, D show that DNase I treatment significantly inhibited tumor metastasis from pancreas to liver and to the diaphragm. Although all pancreases developed high level of pancreatic cancer reflected by the highest level of the bioluminescence in BLI, there was no difference in the primary pancreatic tumor growth between control mouse group and DNase I treated mouse group.
We recorded tumor growth and metastasis in different organs by gross examination. Table 1 shows visible tumor in mice organs from control group and from DNase I treated group. Positive (+) indicates that tumor (s) was/were visible, whereas negative (−) indicates that no visible tumor on the organ examined. Although not as sensitive as bioluminescence imaging, data from gross examination in Table 1 also showed DNase I inhibited orthotopic pancreatic tumor metastasizing to different organs.
Table 1.
Harvested organs with visible tumor (tm +) or without visible tumor (tm −) in each mouse (ms #)
| Pancreas | Spleen | Liver | Mesentery | Diaphragm tumor coverage % |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control group |
DNase I treated |
Control group |
DNase I treated |
Control group |
DNase I treated |
Control group |
DNase I treated |
Control group |
DNase I treated |
||||||||||
| ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm | ms # | tm |
| 1 | + | 1 | + | 1 | + | 1 | + | 1 | + | 1 | + | 1 | + | 1 | − | 1 | 100 | 1 | 10 |
| 2 | + | 2 | + | 2 | − | 2 | − | 2 | + | 2 | − | 2 | − | 2 | − | 2 | 10 | 2 | 1 |
| 3 | + | 3 | + | 3 | + | 3 | − | 3 | − | 3 | + | 3 | − | 3 | − | 3 | 1 | 3 | 10 |
| 4 | + | 4 | + | 4 | − | 4 | − | 4 | − | 4 | − | 4 | − | 4 | − | 4 | 0 | 4 | 0 |
| 5 | + | 5 | + | 5 | + | 5 | − | 5 | + | 5 | + | 5 | + | 5 | − | 5 | 90 | 5 | 30 |
| 6 | + | 6 | + | 6 | + | 6 | − | 6 | + | 6 | − | 6 | + | 6 | − | 6 | 35 | 6 | 1 |
| 7 | + | 7 | + | 7 | − | 7 | − | 7 | − | 7 | + | 7 | − | 7 | − | 7 | 5 | 7 | 30 |
| 8 | + | 8 | + | 8 | + | 8 | − | 8 | 45 | ||||||||||
| 100% + | 100% + | 63% + | 14% + | 63% + | 57% + | 38% + | 0% + | Avg 35.8 | Avg 11.7 | ||||||||||
ExDNA affects CXCL8 production in pancreatic cancer cell lines
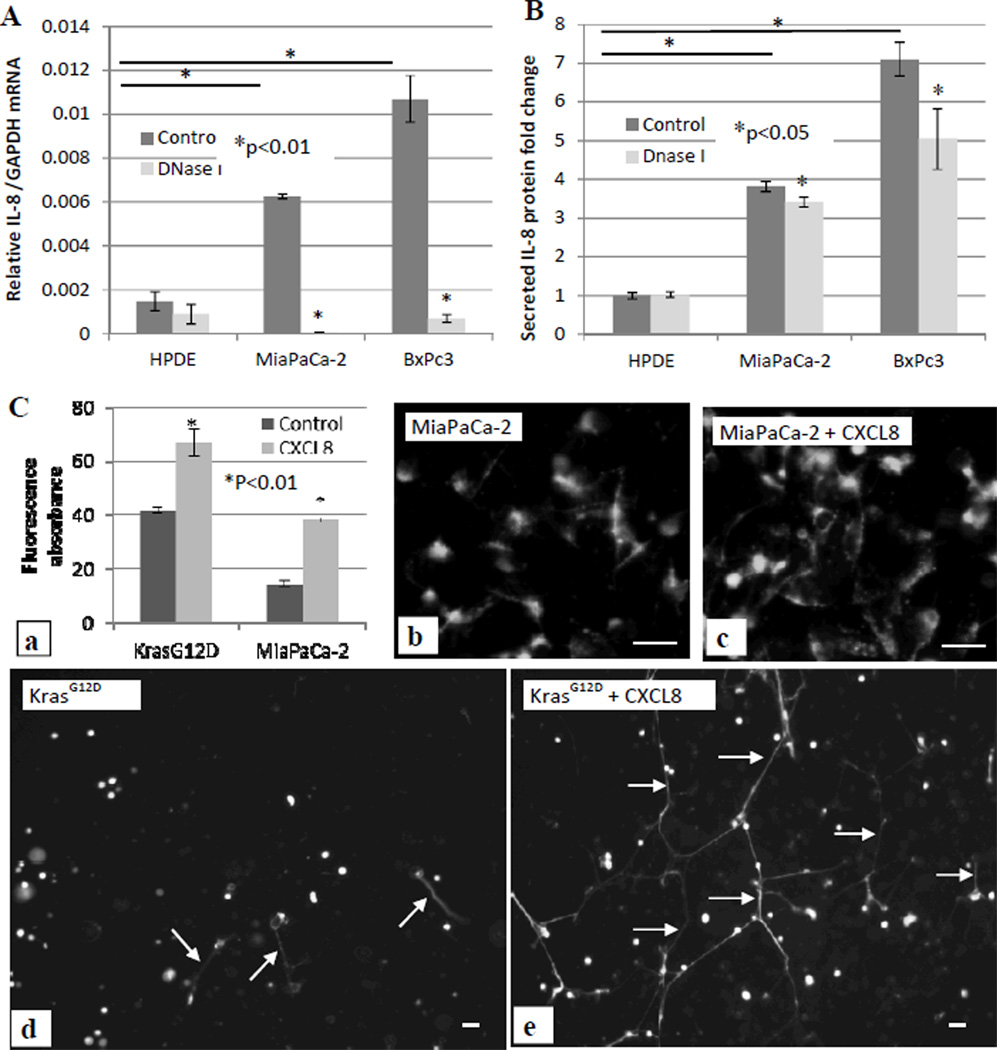
To elucidate the mechanism of exDNA in cancer metastasis, we examined the effect of DNase I treatment on cell production of cytokines with a Human Cytokine ELISA Plate Array I (Signosis, Sunnyvale, CA) followed manufacture’s instruction. The ELISA array test showed only CXCL8, one of the major inflammatory mediator, was affected by the DNase I treatment (data not shown). We then focused on CXCL8 for further examination. After treating cells with DNase I for 48 hours, we measured secreted CXCL8 protein level in the culture media and CXCL8 mRNA level in cells. Figure 6 shows both the transcription level of CXCL8 mRNA (Fig. 6A) and secreted CXCL8 protein level (Fig. 6B) are much higher in BxPc3 and MiaPaCa-2 cells compared with in HPDE cells, and the DNase I treatment significantly reduced CXCL8 mRNA (Fig. 6A) and secreted CXCL8 protein levels (Fig. 6B) in two cancer cell lines, but not in the normal HPDE cell line. The high CXCL8 level correlates with the high amount of cell surface exDNA in these cancer cells.
Figure 6. Feedback loop between exDNA and CXCL8.
Normal pancreatic HPDE cells, pancreatic cancer BxPc3 cells and MiaPaCa-2 cells were treated with DNase I. 500µl of 105 cells/well were seeded in absence and presence of DNase I (3 units/100µl) in a 24 well plate. 48 hrs after treatment, cells were harvested for total RNA isolation and qPCR to measure IL-8 mRNA. Relative fold changes of IL-8 mRNA level were normalized to GAPDH mRNA (A), and media were collected for ELISA assay to examine secreted IL-8 protein level. Fold changes of IL-8 protein level were against the level of IL-8 secreted from HPDE cells (B). C, MiaPaCa-2 and HPDE-KRASG12D cells were treated with 80 ng/ml exogenous CXCL8 for 18 hrs, then cells were stained with Sytox Green. exDNA were quantitatively measured (a) and were microscopically observed (b–e). Data from three repeated experiments are presented as means ± SD.
CXCL8 elevates exDNA production in cancer cell lines
To further evaluate whether there is a feedback loop of CXCL8 on exDNA production, we then examined if CXCL8 can increase exDNA production in pancreatic cancer cells as well as HPDE cells with mutated KRAS gene. We added CXCL8 to MiaPaCa-2, and HPDE-KRASG12D cells cultured in 96-well and 24-well plates. Figure 6C showed CXCL8 treatment significantly elevated the exDNA production quantitatively (Fig. 6C a) and microscopically in both cell lines (Fig. 6C b–e). DNase I treatment also reduced exDNA associated with HPDE-KRASG12D cells the same as in pancreatic cancer cells (Fig. S4).
Discussion
Our study provides the first evidence that exDNA is associated with pancreatic cancer cells in culture and in tissue sections, but not with normal pancreas cells. Our data indicates that cancer cell related exDNA is involved in cell metastatic potential in vitro and that DNase I treatment significantly decreased cancer metastasis in the orthotopic xenograft pancreatic cancer mouse model. Our data are consistent with studies dating to the 1960s, when reports appeared implicating exDNA in cancer development (29, 31, 32). This area of research fell dormant until recent reports of the suppressive effect of DNase I on the metastasis of pulmonary and liver cancers in murine tumor models (33). Studies identifying NETs and other extracellular DNA traps have provided a mechanistic rationale for exDNA in carcinogenesis (14–16, 18, 19, 34).
The exDNA in our cell culture experiments is found in pancreatic cancer cell cultures and is not dependent on microenvironmental factors. Thus, the results of our cell culture experiments are likely affected by extracellular traps made up of pancreatic cellular exDNA. The results from mouse models could involve traps containing tumor and/or stromal exDNA. Our current findings do not address the pathway(s) for producing exDNA in either tumor or stromal cells. Apoptosis, necrosis, inflammatory cell lysis, and active cell secretion all can release exDNA (35). Our apoptosis assessment however does not suggest exDNA is associated with cell apoptosis. Figure S1 shows significant higher level of caspase activity in HPDE cells, but we did not observe cell surface exDNA in this cell line. Micronuclei or microparticles are another possible source of exDNA. These are small, extranuclear bodies that are formed during mitosis from lagging chromosomes, often related to DNA damages. DNA in micronuclei/ microparticles is ultimately eliminated from cells (36). Levels of micronuclei in the blood are elevated in a wide variety of diseases (37). It has recently been shown that micronuclei in peripheral lymphocytes was associated with pancreatic cancer (38). We have frequently observed micronuclei in pancreatic cancer cells; micronuclei were often found connected with exDNA (Fig S5). Further examination of micronuclei would be important for tracing the origin and the fate of exDNA, further for understanding its function.
Cancer cell surface-associated DNA has been reported in other types of cancer cells (39–41). We found exDNA on the surfaces and in the vicinity of cultured pancreatic cancer cell lines, also observed abundant exDNA associated with metastasized pancreatic cancer cells in the tissue section from the xenograft mouse model (Fig. 2). We cross-examined the presence of pancreatic cancer cell associated exDNA using DNA fluorescent dyes, immunofluoresent, and immunohistochemical stainings using DNA antibody both in cultured cells and mouse tissue sections. The extracellular nature of exDNA was clearly demonstrated by immunofluorescent assays using DNA antibody on permeabilized (Fig. 1f) or non-permeabilized cells (Fig. 1i). DNA antibody reacted with both nuclei DNA and exDNA (Fig. 1f) in the permeabilized cells, whereas it reacted only with exDNA in the non-permeabilized cells (Fig. 1i). The fact that we observed abundant cell surface-bound exDNA in pancreatic cancer cell lines, but not in normal pancreas cell line and the early studies of using DNase to control cancer metastasis prompted us to test the hypothesis that exDNA associated with pancreatic cancer cells plays a role in pancreatic cancer metastasis.
A significant higher levels of CXCL8 mRNA and secreted CXCL8 protein were detected in pancreatic cancer cell lines compared with that in normal HPDE cells (Fig. 6), which is in agreement with earlier studies (23, 42). Our study showed that the levels of CXCL8 mRNA and secreted CXCL8 protein correspond to the level of exDNA in each cell lines, and DNase I treatment can reduce CXCL8 mRNA and secreted CXCL8 protein levels while degrading exDNA. The correlation between CXCL8 and exDNA has never been shown in cancer cells, although this relationship has been reported in inflammatory cells (14, 18). Our current study suggests that exDNA could very well be another mediator molecule common to cancer and inflammation. It has been shown that the production and secretion of cytokines in the inflammatory cells can be induced by bacterial DNA and by DNA-containing immune complex (43). In inflammatory neutrophils, production of exDNA in the form of NET (neutrophil extracellular trap) has been reported to be induced by CXCL8 (14, 18). A possible exDNA and CXCL8 feedback loop may exist in inflammatory cells. Our results indicated for the first time an exDNA and CXCL8 feedback loop in pancreatic cancer cells.
The elevated production of CXCL8 has been recently shown in transformed HPDE cells expressing mutated K-RAS. The increased production of cytokine CXCL8 was shown to promote invasion and angiogenesis of cancers including pancreatic cancer cells (44–47). Our results showed the presence of exDNA in HPDE-KRASG12D cells, and exogenous CXCL8 treatment increased exDNA production significantly in this cell line as in other pancreatic cancer cell line. Our study also showed DNase I treatment reduced cell migration and invasion abilities in HPDE-KRASG12D cells (Fig. S4c).
Results from our in vitro experiments demonstrated that exDNA plays a novel role in cancer cell metastasis potential, including cell migration, cell invasion, and cell adhesion to each other and to extracellular matrix. These are all vital steps in cancer metastasis (27, 48, 49). These findings provide a mechanistic explanation and insight into previous reports on the correlation between cell surface exDNA and circulating exDNA and cancer development (35, 39, 40, 50). The role of exDNA in pancreatic cancer metastasis was further delimited by the MTT assay, which demonstrated DNase I treatment did not affect the viability and proliferation of cancer cells. The study by Sugiharal et al. (29) also showed that exDNA had no effect on primary tumor growth when tumor was treated with DNase I.
Here we showed for the first time that DNase I inhibits pancreatic cancer metastasis. Our in vivo mouse model validated the role of exDNA in pancreatic cancer metastasis. Tumor cell metastasis is confirmed by ex vivo BLI measurement in different organs harvested at the end of the experiment. Higher concentration of DNase I treatment gave rise to similar result as that from the lower concentration DNase I treatment (data not shown). Our mouse data is in agreement with other mouse model studies showing that DNase I inhibits metastasis of other types of cancer (36, 39, 40). These findings unveiled that exDNA could be a potential novel hallmark in pancreatic cancer metastasis, which may lead to new drug discoveries and cancer detection methods.
Supplementary Material
Acknowledgments
We thank Dr. Wenxin Zheng for his critical reading of our manuscript and giving helpful suggestions. We thank Dr. Haiyan Cui for her help with statistical analysis. We thank Experimental Mouse Shared Service in University of Arizona Cancer Center (EMSS/UACC) for their service of the mouse model, and the University of Arizona TACMASS core facility for tissue sectioning and H&E services. This work was supported by NCI/NIH grant CA158895, Arizona Foundation Faculty Seed Grant, NIH University of Arizona Cancer Center Support Grant CA23074, and NIH GI SPORE CA095060.
Abbreviations
- exDNA
extracellular DNA
- IHC
immunohistochemistry
- IFA
immunofluorescence assay
- NET
neutrophil extracellular trap
- HPDE
human pancreatic ductal epithelial
- SCID
severe combined immunodeficiency
- DAPI
4’, 6-diamidino-2-phenylindole
- BLI
bioluminescence imaging
- GAPDH
glyceraldehydes-3-phosphate dehydrogenase
Footnotes
Author contribution: Fushi Wen, Jiaqi Shi designed research; Fushi Wen performed research, analyzed data and wrote the paper; Alex Shen and Andrew Choi helped with experiments; Eugene W. Gerner edited the paper.
Conflict of interest: Authors declare no conflict.
References
- 1.Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nature Reviews Gastroenterology & Hepatology. 2009;6:699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 2.Barkin JS, Goldstein JA. Diagnostic and therapeutic approach to pancreatic cancer. Biomedicine & Pharmacotherapy. 2000;54:400–409. doi: 10.1016/S0753-3322(01)80008-1. [DOI] [PubMed] [Google Scholar]
- 3.Tuveson DA, Neoptolemos JP. Understanding Metastasis in Pancreatic Cancer: A Call for New Clinical Approaches. Cell. 2012;148:21–23. doi: 10.1016/j.cell.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Kopfstein L, Christofori G. Metastasis: cell-autonomous mechanisms versus contributions by the tumor microenvironment. Cellular And Molecular Life Sciences. 2006;63:449–468. doi: 10.1007/s00018-005-5296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nature Reviews Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 6.Mantovani A. CANCER Inflaming metastasis. Nature. 2009;457:36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 7.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 8.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 9.Uomo I, Miraglia S, Pastorello M. Inflammation and pancreatic ductal adenocarcinoma: a potential scenario for novel drug targets. Jop J Pancreas. 2010;11:199–202. [PubMed] [Google Scholar]
- 10.Farrow B, Evers BM. Inflammation and the development of pancreatic cancer. Surg Oncol. 2002;10:153–169. doi: 10.1016/s0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 11.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlson BM, Ekbom A, Josefsson S, McLaughlin JK, Fraumeni JF, Nyren O. The risk of pancreatic cancer following pancreatitis: An association due to confounding? Gastroenterology. 1997;113:587–592. doi: 10.1053/gast.1997.v113.pm9247480. [DOI] [PubMed] [Google Scholar]
- 13.Ekbom A, McLaughlin JK, Nyren O. Pancreatitis And The Risk Of Pancreatic-Cancer. New England Journal Of Medicine. 1993;329:1502. doi: 10.1056/NEJM199311113292016. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs TA, Brill A, Duerschmied D, et al. Extracellular DNA traps promote thrombosis. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessenbrock K, Krumbholz M, Schonermarck U, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nature Medicine. 2009;15:623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bass JIF, Russo DM, Gabelloni ML, et al. Extracellular DNA: A Major Proinflammatory Component of Pseudomonas aeruginosa Biofilms. Journal Of Immunology. 2010;184:6386–6395. doi: 10.4049/jimmunol.0901640. [DOI] [PubMed] [Google Scholar]
- 18.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil Function: From Mechanisms to Disease. In: Paul WE, editor. Annual Review of Immunology. Vol 30. 2012. pp. 459–489. [DOI] [PubMed] [Google Scholar]
- 19.Demers M, Krause DS, Schatzberg D, et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proceedings of the National Academy of Sciences. 2012;109:13076–13081. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esposito I, Menicagli M, Funel N, et al. Inflammatory cells contribute to the generation of an angiogenic phenotype in pancreatic ductal adenocarcinoma. Journal Of Clinical Pathology. 2004;57:630–636. doi: 10.1136/jcp.2003.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erkan M, Reiser-Erkan C, Michalski CW JK. Tumor microenvironment and progression of pancreatic cancer. Exp Oncol. 2010;32:128–131. [PubMed] [Google Scholar]
- 22.Waugh DJJ, Wilson C. The Interleukin-8 Pathway in Cancer. Clinical Cancer Research. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 23.Matsuo Y, Sawai H, Funahashi H, et al. Enhanced angiogenesis due to inflammatory cytokines from pancreatic cancer cell lines and relation to metastatic potential. Pancreas. 2004;28:344–352. doi: 10.1097/00006676-200404000-00025. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Furukawa T, Kobari M, Tsao MS. Comparative phenotypic studies of duct epithelial cell lines derived from normal human pancreas and pancreatic carcinoma. American Journal of Pathology. 1998;153:263–269. doi: 10.1016/S0002-9440(10)65567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sylvester P. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol. 2011;716:157–168. doi: 10.1007/978-1-61779-012-6_9. [DOI] [PubMed] [Google Scholar]
- 26.Wen F, Zhou R, Shen A, Choi A, Uribe D, J S. The Tumor Suppressive Role of eIF3f and Its Function in Translation Inhibition and rRNA Degradation. PLoS ONE. 2012;7:e34194. doi: 10.1371/journal.pone.0034194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, Liu W, Guo X, et al. IRX1 influences peritoneal spreading and metastasis via inhibiting BDKRB2-dependent neovascularization on gastric cancer. Oncogene. 2011;30:4498–4508. doi: 10.1038/onc.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deer EL, Gonzalez-Hernandez J, Coursen JD, et al. Phenotype and Genotype of Pancreatic Cancer Cell Lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugihara S, Yamamoto T, Tanaka H, Kambara T, Hiraoka T, Miyauchi Y. Deoxyribonuclease Treatment Prevents Blood-Borne Liver Metastasis Of Cutaneously Transplanted Tumor-Cells In Mice. British Journal Of Cancer. 1993;67:66–70. doi: 10.1038/bjc.1993.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes CS, Postovit LM, Lajoie GA. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 31.Delamirande G. Action Of Deoxyribonuclease And Ribonuclease On Growth Of Ehrlich Ascites Carcinoma In Mice. Nature. 1961;192:52. doi: 10.1038/192052a0. [DOI] [PubMed] [Google Scholar]
- 32.Salganik RI, Martynova RP, Matienko NA, Ronichevskaya GM. Effect of Deoxyribonuclease on the Course of Lymphatic Leukaemia in AKR Mice. Nature. 1967;214:100. doi: 10.1038/214100a0. [DOI] [PubMed] [Google Scholar]
- 33.Patutina O, Mironova N, Ryabchikova E, et al. Inhibition of metastasis development by daily administration of ultralow doses of RNase A and DNase I. Biochimie. 2011;93:689–696. doi: 10.1016/j.biochi.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Wen FS, White GJ, VanEtten HD, Xiong ZG, Hawes MC. Extracellular DNA Is Required for Root Tip Resistance to Fungal Infection. Plant Physiology. 2009;151:820–829. doi: 10.1104/pp.109.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids - A promising, non-invasive tool for early detection of several human diseases. Febs Letters. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu N. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis. 2011;26:119–123. doi: 10.1093/mutage/geq053. [DOI] [PubMed] [Google Scholar]
- 37.Pisetsky DS, Gauley J, Ullal AJ. Microparticles as a source of extracellular DNA. Immunologic Research. 2011;49:227–234. doi: 10.1007/s12026-010-8184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang P, Li YN, Li DH. Micronuclei levels in peripheral blood lymphocytes as a potential biomarker for pancreatic cancer risk. Carcinogenesis. 2011;32:210–215. doi: 10.1093/carcin/bgq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aggarwal SK, Wagner RW, McAllister PK, Rosenberg B. Cell-Surface-Associated Nucleic-Acid In Tumorigenic Cells Made Visible With Platinum-Pyrimidine Complexes By Electron-Microscopy. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1975;72:928–932. doi: 10.1073/pnas.72.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juckett DA, Rosenberg B. Actions Of Cis-Diamminedichloroplatinum On Cell-Surface Nucleic-Acids In Cancer-Cells As Determined By Cell Electrophoresis Techniques. Cancer Research. 1982;42:3565–3573. [PubMed] [Google Scholar]
- 41.Russell JL, Golub ES. Leukemia In Akr Mice - Defined Suppressor Cell-Population Expressing Membrane-Associated Dna. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1978;75:6211–6214. doi: 10.1073/pnas.75.12.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuo Y, Ochi N, Sawai H, et al. CXCL8/IL-8 and CXCL12/SDF-1 alpha co-operatively promote invasiveness and angiogenesis in pancreatic cancer. International Journal Of Cancer. 2009;124:853–861. doi: 10.1002/ijc.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian J, Avalos AM, Mao SY, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nature Immunology. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo Y, Campbell PM, Brekken RA, et al. K-Ras Promotes Angiogenesis Mediated by Immortalized Human Pancreatic Epithelial Cells through Mitogen-Activated Protein Kinase Signaling Pathways. Molecular Cancer Research. 2009;7:799–808. doi: 10.1158/1541-7786.MCR-08-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rial NS, Lazennec G, Prasad AR, Krouse RS, Lance P, Gerner EW. Regulation of deoxycholate induction of CXCL8 by the adenomatous polyposis coli gene in colorectal cancer. International Journal Of Cancer. 2009;124:2270–2280. doi: 10.1002/ijc.24226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizukami Y, Jo WS, Duerr EM, et al. Induction of interleukin-8 preserves the angiogenic response in HIF-1 alpha-deficient colon cancer cells. Nature Medicine. 2005;11:992–997. doi: 10.1038/nm1294. [DOI] [PubMed] [Google Scholar]
- 47.Arenberg DA, Kunkel SL, Polverini PJ, Glass M, Burdick MD, Strieter RM. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. Journal Of Clinical Investigation. 1996;97:2792–2802. doi: 10.1172/JCI118734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Talmadge JE, Fidler IJ. AACR Centennial Series: The Biology of Cancer Metastasis: Historical Perspective. Cancer Research. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefebure B, Charbonnier F, Di Fiore F, et al. Prognostic Value of Circulating Mutant DNA in Unresectable Metastatic Colorectal Cancer. Annals Of Surgery. 2010;251:275–280. doi: 10.1097/SLA.0b013e3181c35c87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.