Abstract
A critical question in tuberculosis control is why some strains of Mycobacterium tuberculosis are preferentially associated with multiple drug resistances. We demonstrate that M. tuberculosis strains from Lineage 2 (East Asian lineage and Beijing sublineage) acquire drug resistances in vitro more rapidly than M. tuberculosis strains from Lineage 4 (Euro-American lineage) and that this higher rate can be attributed to a higher mutation rate. Moreover, the in vitro mutation rate correlates well with the bacterial mutation rate in humans as determined by whole genome sequencing of clinical isolates. Finally, using a stochastic mathematical model, we demonstrate that the observed differences in mutation rate predict a substantially higher probability that patients infected with a drug susceptible Lineage 2 strain will harbor multidrug resistant bacteria at the time of diagnosis. These data suggest that interventions to prevent the emergence of drug resistant tuberculosis should target bacterial as well as treatment-related risk factors.
Recently, strains of Mycobacterium tuberculosis have emerged that are resistant to most or all effective antibiotics1-4. Given the low mutation rate of M. tuberculosis5,6 and its slow replication rate, it is unclear how the bacterium acquires resistance to multiple antibiotics, especially in the face of concurrent multiple drug treatment. The most commonly cited risk factors for treatment failure due to antibiotic resistance are patient noncompliance1-4,7-9, inappropriate drug regimens and dosing2,5,6,9,10 and primary infection with drug resistant strains11,12. The relative importance of bacterial determinants of treatment failure has been unclear. Recently, whole genome sequencing of M. tuberculosis isolates has revealed the importance of novel mutation in the emergence of drug resistance13-15. Sequencing of M. tuberculosis from patients failing antibiotic therapy revealed that new antibiotic resistance mutations can arise multiple times within a given individual15. Moreover, several studies suggest that certain strains of M. tuberculosis may be more frequently associated with multi-drug resistance (MDR) 16,17. Given that all drug resistances in M. tuberculosis occur through chromosomal mutation, these data suggest that the mutational capacity of the bacterium may be an important determinant of the likelihood of drug resistance.
M. tuberculosis forms phylogeographic lineages associated with particular human populations18-21. Though less genetically diverse than many other pathogens, there is both experimental and clinical evidence that M. tuberculosis strains from different lineages vary in their capacity to cause disease20-24 and acquire drug resistance11,12,16,20,25-28. Specifically, M. tuberculosis strains within Lineage 2 (the East Asian lineage, which includes the Beijing family of strains) have been epidemiologically associated with an increased risk of drug resistance in several cross-sectional studies in diverse locales, though not in all29. Strains from this lineage have polymorphisms in DNA replication, recombination, and repair genes as compared to Lineage 4 (the Euro-American lineage) strains, raising the possibility that they are more mutable than other M. tuberculosis strains30. However, these epidemiologic observations might also reflect social and programmatic factors (such as noncompliance and inappropriate dosing) correlating with the phylogeography of the Lineage 2 strains. Indeed, in vitro studies comparing the rate or frequency of drug resistance in the Lineage 2 and Lineage 4 strains have produced differing results31,32. Here, we sought to determine the rate at which M. tuberculosis strains of different lineages acquire drug resistance and the effect of strain based differences in mutation rate on the predicted de novo generation of MDR in patients with tuberculosis.
Results
Effect of mutation and genetic background on drug resistance in M. tuberculosis
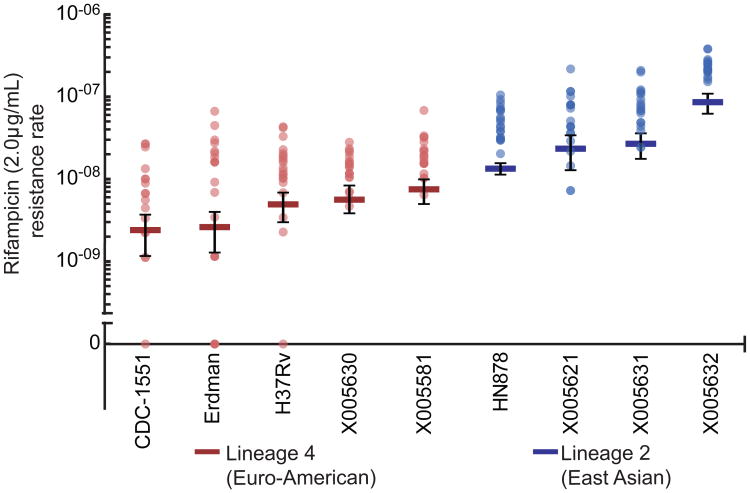
To measure the drug resistance rate of strains from different M. tuberculosis lineages, we performed Luria-Delbrück fluctuation analysis33,34 on a panel of laboratory and clinical isolates from both Lineage 2 and Lineage 4. All strains were fully drug susceptible, with MIC's at least 100 fold less than the drug concentrations at which we assessed drug resistance rates (MICrif<0.015μg/mL; MICINH<0.007 μg/mL, Supplemental Table 1). Within both lineages, there was some strain-to-strain variation in the rate at which rifampicin resistance was acquired (Figure 1). However, every strain from Lineage 2 acquired resistance to rifampicin (2μg/mL) at a significantly higher rate than every Lineage 4 strain, with a nearly 10-fold difference between the means of the two groups (Figure 1, Supplementary Table 1).
Figure 1. Lineage 2 strains more rapidly acquire rifampicin resistance.
Fluctuation analysis was used to determine the rifampicin (2μg/mL) resistance rate of clinical and laboratory strains from both Lineage 2 and Lineage 4. Strains from Lineage 4 are in red; strains from Lineage 2 are in blue. Circles represent mutation frequency (number of mutants per cell plated in a single culture), where darker circles represent multiple cultures with the same frequency. Bars represent the estimated mutation rate, with error bars representing the 95% confidence interval. Strains are displayed on the x-axis and the rifampicin resistance rate is displayed on the y-axis in log-scale. Values are listed in Supplementary Table 1.
The higher rate of rifampicin resistance could reflect three possible mechanisms: 1) differences in the ability to survive and mutate after exposure to antibiotic, 2) inherent differences in the number of rpoB mutations conferring rifampicin resistance (target size), or 3) differences in the basal mutation rate in the absence of selection. To test these hypotheses, we chose well-characterized representatives from Lineage 4 and Lineage 2 - CDC1551 and HN878, respectively - for further study.
Differences in response to antibiotic
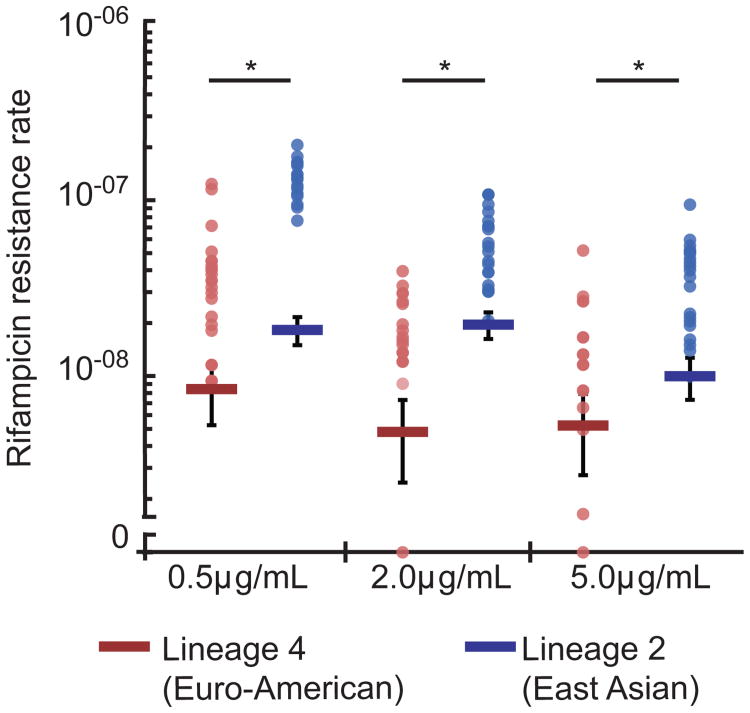
We first sought to determine whether Lineage 4 and Lineage 2 strains differed in their ability to acquire drug resistance after exposure to rifampicin. Fluctuation analysis assumes that all mutations occur prior to selection and that all mutants replicate as well as wild type33; however, if a strain is capable of surviving and mutating in the presence of drug, it will produce a greater number of drug-resistant mutants. Building on similar studies in Saccharomyces cerevisiae35, we reasoned that if mutations occur in the presence of a drug, then lowering the drug concentration might allow strains from both lineages to grow and acquire mutations post-exposure. Conversely, increasing the drug concentration might abrogate the ability of both strains to survive and acquire mutations in the presence of drug. However, we found that statistically significant increases in the rifampicin resistance rate for the Lineage 2 strain, HN878, relative to the Lineage 4 strain, CDC1551, were maintained over 10-fold variation in drug concentration (0.5μg/mL – 5μg/mL) (Figure 2, Supplementary Table 1).
Figure 2. Altering drug concentration does not alter the observation that Lineage 2 strains more rapidly acquire rifampicin resistance.
Fluctuation analysis was used to determine the rifampicin (0.5, 2, 5μg/mL) resistance mutation rate of representative strains from both Lineage 2 and Lineage 4 (HN878 and CDC1551, respectively). The Lineage 4 strain CDC1551 is in red, and the Lineage 2 strain HN878 is in blue. Circles represent mutation frequency (number of mutants per cell plated in a single culture), where darker circles represent multiple cultures with the same frequency. Bars represent the estimated mutation rate, with error bars representing the 95% confidence interval. Significance was determined by comparing strain pairs using the Wilcoxon rank-sum test. Strains are displayed on the x-axis and the rifampicin resistance rate is displayed on the y-axis in log-scale. Values are listed in Supplementary Table 1.
To extend these findings, the distribution of mutations observed in each fluctuation assay can be analyzed using tools developed by Lang et al35. This analysis takes advantage of the fact that mutations occurring in culture, prior to antibiotic exposure, result in a Luria-Delbrück distribution. While mutations arise according to a Poisson distribution, the subsequent outgrowth of mutants during broth culture generates a Luria-Delbrück distribution. In contrast, mutations occurring after plating on antibiotic will occur according to a Poisson distribution without the expansion in culture that creates a Luria-Delbrück distribution33,36,37. Thus, the appearance of any additional mutants resulting from acquisition of resistance after antibiotic exposure will generate a mixed distribution of the number of mutants, containing a Poisson-distributed number of post-plating mutants and a Luria-Delbrück-distributed number of mutants occurring prior to drug exposure.
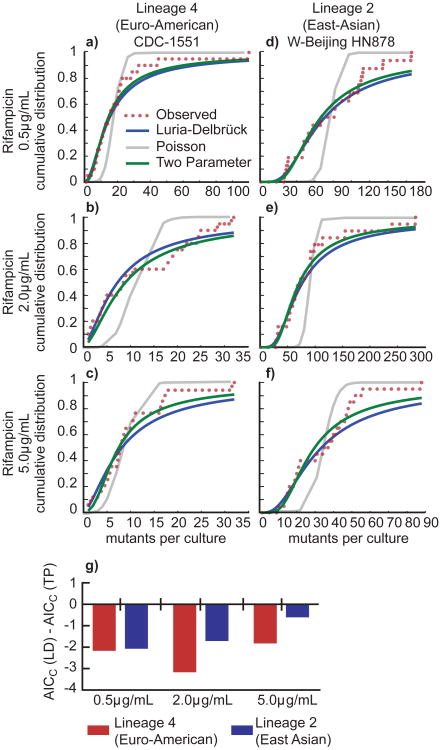
We therefore used a curve-fitting approach to determine whether the distribution of mutant frequencies in the two strains is better fit using a one-parameter Luria-Delbrück model, or a two-parameter Luria-Delbrück and Poisson mixture model (Figure 3a-f). The algorithm first fits the data to a Luria-Delbrück model alone, and then optimizes the fit with the addition of a Poisson model. A purely Poisson model was also fit to serve as a reference. We used the Akaike information criterion with correction for sample size (AICC) to identify instances where the Luria-Delbrück model alone provided the optimal fit over either the Poisson model or the two parameter mixture model38,39. The AICC quantifies the evidence in favor of using a more complex model (here, the two parameter mixture model) over a simpler (here, Luria-Delbrück) model, appropriately penalizing the fit of the more complex model by the increase in model complexity. As expected, the ΔAICc (AICC(Luria-Delbrück) - AICC(Poisson)) was highly negative in all conditions, confirming that the Luria-Delbrück model fits the distributions significantly better than the Poisson model alone (Supplementary Table 2). More revealing, the ΔAICc (AICC(Luria-Delbrück) - AICC(two parameter)) was also negative, indicating that there is insufficient evidence to support the inclusion of an additional Poisson component in the Luria-Delbrück distributions (Figure 3g, Supplementary Table 2). This suggests that post-exposure mutation is not responsible for the higher rifampicin resistance rate in HN878, the Lineage 2 (East Asian) strain.
Figure 3. The cumulative distribution of drug resistant mutants from both lineages indicates that mutations do not occur after exposure to antibiotic.
(a) Curve fitting analysis was performed to determine if the cumulative distribution of the fluctuation analysis data from Lineage 4 strain CDC-1551 plated on rifampicin, 0.5μg/mL better fit a one-parameter Luria-Delbrück (LD) model, a one parameter Poisson model (Poiss), or a two-parameter Luria-Delbrück and Poisson mixture model (TP). The number of mutants per culture is displayed on the x-axis, and the probability of observing x or fewer mutants per culture is shown on the y-axis. (b) Fitting as in (a) for Lineage 2 strain HN878 plated on rifampicin, 0.5μg/mL. (c) Fitting as in (a) for Lineage 4 strain CDC-1551 plated on rifampicin, 2μg/mL. (d) Fitting as in (a) for Lineage 2 strain HN878 plated on rifampicin, 2μg/mL. (e) Fitting as in (a) data for Lineage 4 strain CDC-1551 plated on rifampicin, 5μg/mL. (f) Fitting as in (a) data for Lineage 2 strain HN878 plated on rifampicin, 5μg/mL. (g) To determine which model best fit each data set, we determined the Akaike Information Criterion, corrected for small sample size (AICC). A smaller AICC represents a better fit, given a penalty for more parameters in a model. If the AICC(LD) is smaller than the AICC(TP), then the resulting value will be negative, reflecting a better fit for the LD model (see Supplementary Table 2).
This analytic approach also suggests that the difference in rifampicin resistance rates is not due to strain based differences in the fitness effects of the drug-resistance mutations33,37. If the drug resistant mutants in either strain suffered a strong fitness cost, the outgrowth of mutants in culture prior to selection would have been slower than drug susceptible cells, driving the Luria-Delbrück distribution back towards the underlying Poisson distribution of mutation. However, our data suggest that in both strains, drug-resistant mutants occur primarily according to a Luria-Delbrück distribution.
Differences in target size
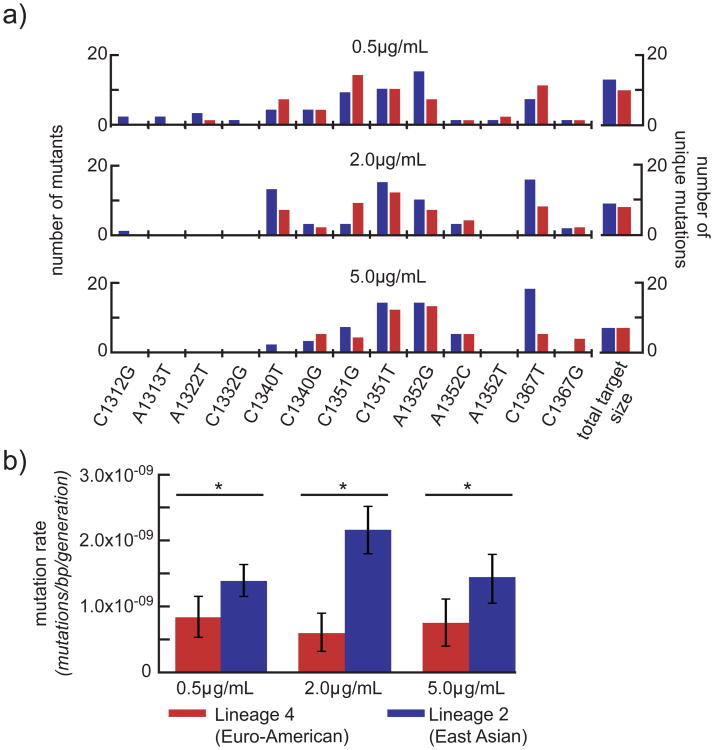
Rifampicin resistance is encoded by multiple mutations in the rifampicin resistance-determining region (RRDR) of rpoB. Strains in which there are a greater number of potential mutations in the RRDR that produce drug resistance would more rapidly acquire resistance to rifampicin. Therefore, we sought to determine whether M. tuberculosis strains of different lineages differ in the total number of mutations that confer rifampicin resistance, accounting for differences in the rifampicin resistance rates. We sequenced the RRDR of rpoB from 600 independent mutants (100 from each fluctuation assay in Figure 2) to determine the number of mutations conferring rifampicin resistance in both strains under each condition tested above (Figure 4a, Supplementary Table 3). Though there is potentially a discrepancy between drug resistance mutants found in vitro and in vivo40, all of the mutations that we identified correspond to mutations seen clinically41. For both strains, the target size became smaller as drug concentration increased, indicating that some rpoB mutations generate lower level rifampicin resistance. There were small differences in target size between the two strains at two of the three drug concentrations tested, such that the number of mutations conferring resistance at both 0.5 and 2μg/mL was higher in the Lineage 2 strain. These data suggest that target size differences between strains contribute to strain based differences in the acquisition of rifampicin resistance. However, correcting for target size to determine the per base pair mutation rate, the Lineage 2 strain, HN878, remained significantly higher than the Lineage 4 strain, CDC1551, at each of the drug concentrations tested (Figure 4b, Supplementary Table 4). Therefore, it is likely differences in basal mutation rate that lead to differences in the acquisition of rifampicin resistance.
Figure 4. Small differences in target size and differences in basal mutation rate are responsible for the observed differences drug resistance rate.
(a) The target size (the number of mutations conferring rifampicin resistance) of each strain under each condition was determined by sequencing the rifampicin resistance determining region of 100 isolates from each strain in each condition. Each mutation is shown on the x-axis, with coordinates representing position within rpoB (Rv0667). The number of mutants per strain uniquely formed within a culture is shown on the y-axis. The Lineage 4 strain, CDC1551, is shown in red; the Lineage 2 strain is shown in blue. On the right, the target size - the number of unique mutations conferring rifampicin resistance – is shown. (b) The per base pair mutation rate is determined by normalizing the drug resistance rate by target size. Drug concentration is shown on the x-axis, mutation rate per base pair is shown on a linear scale on the y-axis. Lineage 4 is shown in red; Lineage 2 is shown in blue. Significance was determined by comparing strain pairs using the Wilcoxon rank-sum test; error bars represent 95% confidence intervals. Values are found in Supplementary Table 4.
Resistance to other antibiotics
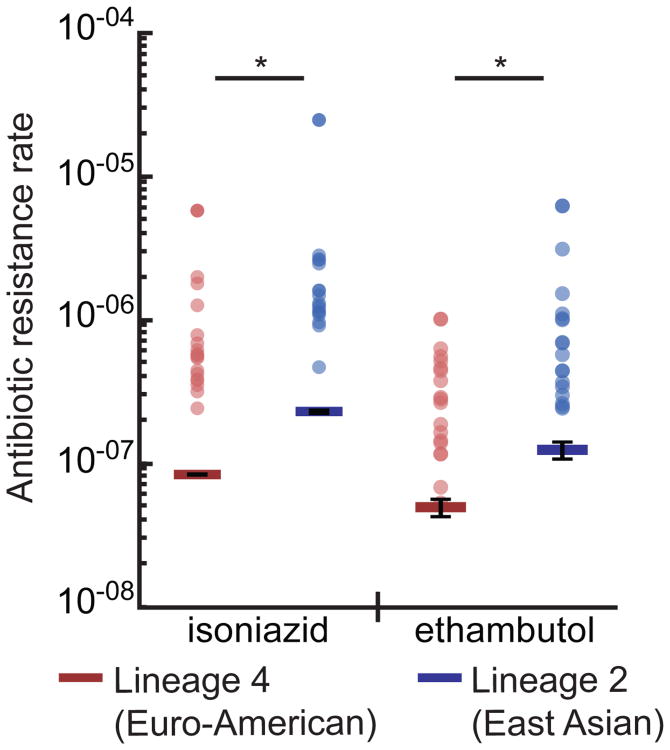
If the mutation rate of HN878 is higher than that of CDC1551, then the Lineage 2 M. tuberculosis strain should also acquire resistances to other antibiotics at a higher rate. We therefore assessed the rate at which HN878 and CDC1551 acquire resistance to ethambutol (5μg/mL) and isoniazid (1μg/mL). For both antibiotics, the rate of resistance was nearly 3-fold (2.51 and 2.75, respectively) higher in the Lineage 2 strain, HN878, consistent with the increased rate of rifampicin resistance (Figure 5, Supplementary Table 1). Taken together, these results suggest that M. tuberculosis strains from Lineage 2 have a higher basal mutation rate than strains from Lineage 4.
Figure 5. A representative Lineage 2 strain acquires isoniazid and ethambutol resistance at a higher rate.
Fluctuation analysis was used to determine the isoniazid (1μg/mL) and ethambutol (5μg/mL) resistance rate for the Lineage 4 strain, CDC1551, (shown in red) and the Lineage 2 strain, HN878 (shown in blue). Circles represent mutation frequency (number of mutants per cell in a single culture), where darker circles represent multiple cultures with the same frequency. Bars represent the estimated mutation rate, with error bars representing the 95% confidence interval. Significance was determined by non-overlapping 95% confidence interval. Values are listed in Supplementary Table 1.
In vitro mutation rates to predict in vivo resistance
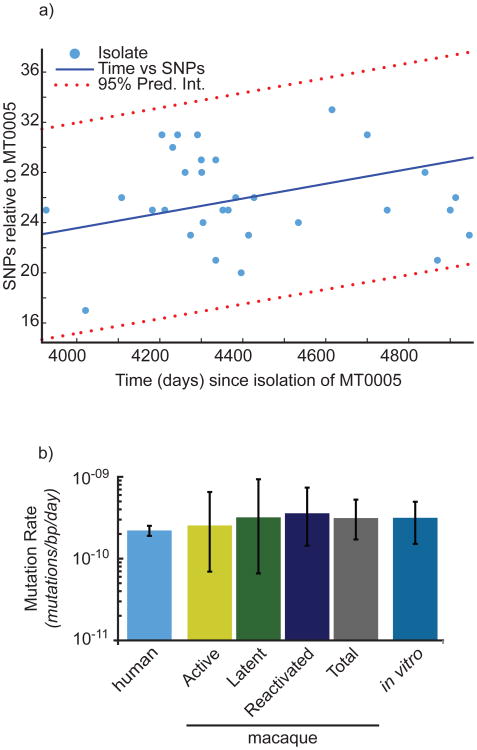
We then sought to understand how these in vitro measures of mutation translate to the in vivo environment. In our previous work, we determined that in nonhuman primates, M. tuberculosis mutates at a relatively fixed rate over time and this in vivo per-day mutation rate is well-approximated by the in vitro per-day mutation rate as measured by fluctuation analysis and adjusted for target size6. To determine if the in vitro mutation rate is similarly concordant with the mutation rate of M. tuberculosis during human infection, we analyzed the whole genome sequences of M. tuberculosis isolates derived from an outbreak of a Lineage 4 (Euro-American) strain in British Columbia, Canada42. We determined the number of SNPs in each strain relative to a historical isolate, identifying SNPs according to parameters that were experimentally validated through Sanger resequencing in our previous work (Figure 6a). By reconstructing the phylogeny of these strains through Bayesian Markov chain Monte Carlo analysis 43,44 (Supplementary Figure 1), and informing the phylogeny with dates for each isolate, we have estimated the base substitution rate (equivalent to the mutation rate under a neutral model of evolution45) in this outbreak. Strikingly, we found that the British Columbia strains acquired mutations at approximately the same rate over time as previously shown for M. tuberculosis strains isolated from macaques with active and latent disease irrespective of disease course (Figure 6b, Supplementary Table 5). In addition, the rate at which these strains acquired mutations in vivo was well approximated by the in vitro per-day mutation rate of the Lineage 4 strain (Erdman) used in the macaque infections as defined by fluctuation analysis. Finally, consistent with both our previous work6 and the work of others46, these data indicate that a molecular clock of 0.3-0.5 mutations/genome/year may be applicable to analysis of M. tuberculosis genetic diversity over the time scales assessed here.
Figure 6. Bayesian MCMC analysis reveals a mutation rate in humans similar to that estimated in strains from the macaque model and in vitro.
(a) The number of SNPs and the number of days separating the clinical isolate and MT0005 are plotted. SNPs located in repeat regions (PE_PGRSs, PPEs, and transposable elements) were excluded, consistent with our previous analysis6. The data are fit to a first order polynomial to illustrate the trend. (b) Estimates of mutation rate in human isolates were derived by reconstructing the phylogeny from the isolates represented in (a). Mutation rate is shown on the y-axis in log scale. Estimates of mutation rate from the macaque model and the infecting strain, Erdman (in vitro) were determined previously6. Error bars represent 95% confidence intervals.
A time-based model of mutation and drug-resistance predicts MDR before treatment
Given these data, we developed a stochastic simulation model of the evolution of drug resistance within a patient in order to assess the potential clinical impact of the observed differences in mutation rate between Lineage 2 and Lineage 4 strains. Our model of drug resistance utilizes a stochastic mutation parameter in which mutation occurs at a constant rate over time and we informed this parameter with the in vitro mutation rates for CDC1551 and HN878 as a proxy for their mutation rates in the human host (Supplementary Figure 2a, Supplementary Table 6). We used this model to simulate the emergence of MDR (defined here as resistance to both rifampicin and isoniazid) within an infected individual prior to diagnosis and treatment (Supplementary Figure 2b).
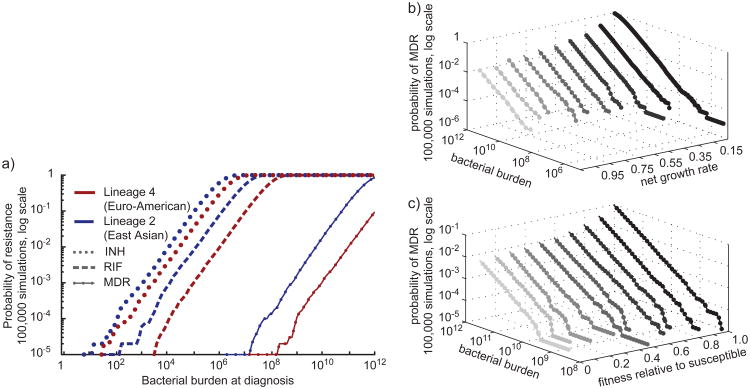
In the model, as a result of the differences in mutation rate, patients infected with the Lineage 2 strain, HN878, are at a significantly increased risk of MDR before treatment as compared to patients infected with the Lineage 4 strain, CDC1551 (Figure 7a). When all other parameters (birth, death, fitness and bacterial burden at the time of diagnosis) are kept equal, the difference in the probability of MDR before diagnosis and treatment is approximately 22-fold. We find similar results when using an alternative model of drug resistance developed by Colijn et al in which mutation is replication- rather than time-dependent (Supplementary Figure 2c)47. We assessed the sensitivity of our model to fluctuations in both growth rate and fitness (Figure 7b & c). Varying these parameters does not alter our principle conclusion that patients infected with Lineage 2 strains of M. tuberculosis are at a significantly higher risk for the de novo acquisition of MDR, reflecting the multiplicative effects of an increased risk of acquiring each individual drug resistance due to a higher basal mutation rate.
Figure 7. A stochastic simulation mathematical model predicts the emergence of MDR-TB before the onset of treatment.
(a) Estimates of the probability of observing MDR within a population were derived using a stochastic mathematical model of resistance in 200,000 simulations, 100,000 for each lineage. Model parameters are listed in Supplementary Table 6. Bacterial burden at diagnosis is shown on the x-axis, the probability of observing resistance is shown on the y-axis in log scale. Estimates for Lineage 4 are shown in red, Lineage 2 in blue. (b, c) To determine the sensitivity of our model to variations in growth rate and fitness, we varied each parameter (see Supplementary Table 6) and determined the probability of observing resistance (z-axis, log scale) at any given bacterial burden (y-axis, log scale) for a specified parameter set (x-axis).
Discussion
Here we demonstrate that strains from Lineage 2 of M. tuberculosis (the East Asian lineage, which includes the Beijing family of strains) acquire drug resistances in vitro more rapidly than strains from Lineage 4 (the Euro-American lineage). This is likely not the result of an enhanced ability of these strains to survive and mutate in the presence of drug, and we find no evidence of strong fitness effects that would explain the observed differences in drug resistance rates. Interestingly, we do find evidence that the genetic context of a given M. tuberculosis strain can impact the range of observed mutations conferring resistance to a single drug. In our analysis, the Lineage 2 strain, HN878, was permissive for a broader range of rpoB mutations than the Lineage 4 strain, CDC1551. However, the difference in target size is not sufficient to explain the observed difference in rifampicin resistance rates, suggesting that a basal difference in mutation before selection drives the accelerated rate of drug resistance in HN878. In support of this, we find that HN878 more rapidly acquires resistance to not only rifampicin, but also isoniazid and ethambutol.
We also find significant variation in the mutation rates of the other strains within each lineage. Previous analyses suggest that there is substantial genetic and phenotypic diversity among isolates from the various M. tuberculosis lineages48. We expect that differences in mutation rate and differences in target size both contribute to the two to thirty-five fold differences in rifampicin resistance rates that we have measured in these other strains. Further work will be required to establish the relative contribution of these factors to the drug resistance rate of each strain and to determine to what extent these findings can be generalized to strains from other M. tuberculosis lineages.
To establish the in vivo relevance of these findings, we sought to assess the concordance between mutation rates measured in vitro and in vivo. Strikingly, we found that the mutation rate of M. tuberculosis in vitro is very close to the mutation rate – assessed as mutation per unit time – in isolates from a human transmission chain. Thus, M. tuberculosis acquires mutations at a similar rate over time in people as it does in an actively growing culture. This is consistent with our previous findings that the in vitro mutation rate over time was similar to the rate of mutation over time in M. tuberculosis isolated from macaques with latent and active disease. Taken together, these findings define a molecular clock for M. tuberculosis that may be used in future evolutionary and epidemiologic studies, though care must be taken to identify and compensate for potential sources of variation in rate49,50.
We propose two possible explanations for the finding that M. tuberculosis acquires mutations at the same rate over time in vitro, in macaques, and across a human transmission chain. First, there may be a population of M. tuberculosis replicating in vivo at a rate similar to the replication rate in vitro. These same bacteria may be over-represented in culturable clinical isolates, suggesting that they may be more likely to cause disease and be transmitted. Alternatively, it is possible that the mutation rate of M. tuberculosis is driven by a time dependent rather than a replication dependent factor. For example, the replicative error rate in M. tuberculosis could be very low relative to time and mutations may occur both in vitro and in vivo largely through DNA damage from endogenous metabolic processes or exogenous stressors.
We expect that these models may be resolved in part by elucidating the molecular basis of strain-based differences in mutation rate. The differences in mutation rate between clinical M. tuberculosis strains measured here are more modest than the differences that distinguish clinical isolates of other bacteria such as Pseudomonas aeruginosa or Escherichia coli51-53. Clinical isolates of these pathogens may become orders of magnitude more mutable than wild type strains through the loss of mismatch repair51. However, mycobacteria, like all other actinomycetes, lack mismatch repair entirely54,55, and the molecular basis of replicative fidelity in mycobacteria remains unclear. While mutations in DNA replication and repair genes are enriched in some M. tuberculosis strains from Lineage 230, no single point mutation has been found to accelerate the basal mutation rate of M. tuberculosis in isogenic strains. Importantly, Lineage 2 strains also differ in important metabolic pathways from Lineage 4 strains56,57. Thus, it is possible that genetic differences outside DNA replication and repair contribute to the differences in mutation rate that we have measured.
We have used the observation that M. tuberculosis mutates at a constant rate per unit time to develop a predictive model of the evolution of MDR in vivo. Our model demonstrates that it is possible to see multi-drug resistance evolve before the onset of treatment, consistent with a prior replication-dependent model47. Moreover, differences in mutation rate have approximately multiplicative effects for each mutation required, leading to stark differences in the de novo generation of MDR. Indeed, we parameterized our model with data from HN878, which has only modestly elevated acquisition rates of rifampicin and isoniazid resistance. Strain X005632 has a 35-fold higher rate of rifampicin resistance as compared to CDC1551. With a similarly elevated rate of isoniazid resistance, the risk of MDR in an individual infected with X005632 would be nearly three orders of magnitude higher than for a patient infected with CDC1551. The predicted differences in the occurrence of at least one MDR bacterium within infected patients may not be directly proportional to the clinical risk of drug resistant tuberculosis because individual bacteria may not have equal capacity to cause disease. However, the magnitude of these differences suggests that strain based variation in drug resistance rates is an important risk factor for the development of drug resistant disease. Indeed, just as strains with higher mutation rates have increased risk of drug resistance mutations and mutations that facilitate acquisition of drug resistance (i.e. efflux pumps58-60), these strains have increased capacity to acquire compensatory mutations61 that may enhance their fitness in vivo.
While our model focused on the evolution of multidrug resistance (rifampicin and isoniazid resistance), the findings are applicable to the evolution of resistances to any antibiotic. New antibiotics and novel regimens of new and current antibiotics are being developed, combining drugs for which resistance is not yet prevalent. The expectation is that with proper implementation, a novel regimen will treat patients infected with MDR M. tuberculosis and prevent the emergence of resistance to new drugs62,63. However, both clinical and high resolution sequencing evidence suggests patients fail therapy with strains that are resistant to only a subset of antibiotics administered14,15,64. Thus, even in the context of a novel regimen, resistance to these new antibiotics may be difficult to avoid especially in the context of infection by strains from Lineage 2.
Consistent with epidemiologic data suggesting that severe disease at diagnosis is associated with the acquisition of MDR in new cases65,66, our model also predicts that bacterial burden is a critical determinant of the probability of drug resistance. In the face of substantial capacity for mutation and resistance, early and active case detection with novel, sensitive point of care diagnostics remains our best hope of curbing the drug resistance epidemic. Smear microscopy is the most common primary diagnostic for M. tuberculosis around the world but is orders of magnitude less sensitive than both culture and molecular diagnostics67,68. If, as our model suggests, higher bacterial burden at diagnosis results in increased risk for the evolution of MDR M. tuberculosis, then improving diagnostic sensitivity will not only curtail ongoing transmission of disease as previously suggested69,70, but also limit the de novo emergence of drug resistance. The risk of drug resistance appears to be even higher in the setting of infection with Lineage 2 strains of M. tuberculosis. Taken together, these data emphasize that M. tuberculosis strains differ in their propensity for acquiring drug resistance and suggest that these biological factors should be considered in efforts to limit the emergence of novel resistances to both existing antibiotics and new treatment regimens.
Methods
Culture and MIC determination of clinical isolates
Clinical Strains were identified as previously described71. Strains were grown in broth culture of 7H9 supplemented with 10% Middlebrook OADC, 0.0005% tween 80, and 0.005% glycerol. To determine MIC, strains were grown to log phase (OD 0.5 to 0.8), and diluted to an OD of 0.006. Wells were inoculated with 50μL of culture and then supplemented with 50μL of media containing the appropriate concentration of drug. All concentrations were tested in triplicate, and two sets of triplicate control wells were established for each strain. After 6 days, 10μL of Alamar Blue (Life Technologies) was added to each well of one triplicate set of controls, where control wells were not supplemented with antibiotic. If control wells converted from blue to pink after 24 hours, 10μL of Alamar Blue was added to each triplicate set of experimental wells as well as a new set of control wells. All wells were examined after 24 hours. Any wells that remained blue represented inhibition of growth. The following concentrations were tested: 0.5, 0.25, 0.125, 0.06125, 0.03063, 0.01531μg/mL of rifampicin (Sigma); 0.25, 0.125, 0.06125, 0.03063, 0.01531, 0.00766μg/mL of isoniazid (Sigma). The MIC was defined as the lowest concentration of drug that prevented color change from blue to pink.
Fluctuation analysis
Fluctuation analysis was performed as previously described6. Briefly, for a single strain of M. tuberculosis, starter cultures were inoculated from freezer stocks of optical density (OD) 1.0 culture. Once at an OD of 0.7 to 1.1, approximately 300,000 cells were used to inoculate 120mL of Middlebrook 7H9 supplemented with 10% Middlebrook OADC, 0.0005% tween 80, and 0.005% glycerol, giving a total cell count of 10,000 cells per 4mL culture. This volume was immediately divided to start 24 cultures of 4mL each in 30mL square PETG culture bottles (Nalgene, Rochester NY). Cultures were grown at 37°C with shaking for 11 to 14 days, until reaching an OD of 1.0. Once at an OD of 1.0, 20 cultures were transferred to 15mL conical tubes and spun at 4000 RPM for 10 minutes at 4°C. Cultures were then resuspended in 250-500μL of 7H9/OADC/tween/glycerol and spotted onto 7H10/OADC/tween/glycerol plates supplemented with 0.5, 2, or 5μg/mL rifampicin (Sigma, R3501), 1μg/mL isoniazid (Sigma, I3377), or 5μg/mL ethambutol (MP Biomedicals, 157949). Once spread using sterile glass beads (4mm diameter), plates were allowed to dry and subsequently incubated at 37°C for 28 days. Cell counts were determined by serial dilution of 4 cultures for each strain. The drug resistance rate was determined by calculating m (the estimated number of mutations per culture) based on the number of mutants (r) observed on each plate using the Ma, Sarkar, Sandri (mss) method as previously described33,35. Dividing m by Nt, the number of cells plated for each culture, gives an estimated drug resistance rate. 95% confidence intervals were estimated using equations (24) and (25) as described in Roshe and Foster33,36. For comparing pairs of fluctuation analysis data (Figure 2), the nonparametric two-sided Wilcoxon rank sum test (also known as the Mann-Whitney U-test) was performed using the ranksum command in Matlab with alpha set to 0.05, comparing the frequency of drug resistant mutants in each culture.
Fluctuation analysis data analysis
To estimate the extent to which the data met the assumptions of Luria-Delbrück fluctuation analysis, we performed a curve fitting analysis as described by Lang and Murray35. Briefly, data were fit using either a one-parameter model consistent with the Luria-Delbrück model, or a two-parameter model containing an additional parameter describing a Poisson distribution. The fit of each model was assessed using the least-squares methodology described by Lang and Murray, with AICC calculated as described previously39. A lower AICC reflects a better fit given a penalty for increasing the number of parameters, and a negative ΔAICC (ΔAICC = AICC (one-parameter) – AICC (two parameter)) indicates the one parameter model is a better approximation of the data.
Determination of target size
The number of rpoB mutations conferring resistance to 0.5, 2, and 5μg/mL of rifampicin was determined by isolating 100 colonies, five from each fluctuation analysis culture, into 100μL Middlebrook 7H9 supplemented with 10% Middlebrook OADC, 0.0005% tween 80, and 0.005% glycerol. Cultures were grown overnight at 37°C, and then heat-inactivated at 85°C for 2 hours. Heat-inactivated culture was then used as template for PCR and sequencing using primers previously described72. Sequences were analyzed for mutation relative to the reference sequence H37Rv, and totaled. For each culture, duplicate mutations were only counted once. The absolute number of unique mutations observed across cultures for a given condition was used to determine target size for each strain under each condition.
Estimate of mutation rate from human isolates
To determine the per base, per day mutation rate in human isolates, phylogenies were created using the concatenated SNP sequences reported by Gardy et al from a clonal outbreak of a Lineage 4 (Euro-American) strain in British Columbia, Canada42 using BEAST v.1.7.2 43,44 to perform Bayesian MCMC analysis. Prior to phylogenetic analysis, SNPs located in repeat regions (PE_PGRSs, PPEs, and transposable elements) were excluded, consistent with our previous experimentally validated analysis of SNPs from whole genome sequencing to estimate mutation rate6. Concatenated SNP sequences were compiled and prepared using BEAUti v1.7.2 to select analysis parameters and construct the xml input file. Concatenated sequences were converted to NEXUS format, and loaded into BEAUti where time was noted for each isolate. Time was defined in days based on time elapsed from symptom onset relative to isolation of the historical isolate, MT0005 (1995). A GTR substitution model was used with empirically determined base frequencies. Default priors were used for 10,000,000 chains. Output was analyzed in Tracer v1.5, and all parameters produced an effective sample size of 200 or greater. Phylogenetic tree construction was completed using TreeAnnotator v1.7.2 with a posterior probability limit of 0.5 and a burnin of 1000 trees, leaving 9001 potential trees for construction. Tree visualization was completed using FigTree v1.3.1 and the tree was rooted on MT0005, with the most likely tree depicted in Supplementary Figure 1.
Mathematical simulation of drug resistance
We developed a compartmental, partially stochastic mathematical model of the evolution of drug resistance within an individual according to the following set of equations:
| (1) |
| (2) |
| (3) |
| (4) |
Where:
| (5) |
| (6) |
| (7) |
| (8) |
Here, NS(t), NR(t), NH(t), NMDR(t) are the population sizes of the susceptible, rifampicin resistant, isoniazid resistant, and MDR populations at time (t), respectively. The parameters b and dA represent the birth and rates of bacteria in each population. The subscripted mX·Y parameters are the number of bacteria transitioning from population NY to population NX. m is a stochastically determined parameter, determined by a random Poisson variable where lambda is determined by the mutation rate to resistance times the population size of the susceptible population. These equations were parameterized with the values displayed in Supplementary Table 6. All simulations were run in Matlab (Natick, MA). For all simulations and for both mutation parameter sets (μH−W & μR−W, μH−CDC & μR−CDC), simulations of the evolution of drug resistance were run 100,000 times to determine the probability of observing drug resistance with a given set of parameters. To determine the effect of varying birthrate, 10 simulations of 200,000 simulated patients (100,000 per simulated strain) each were run with b = (0.20:1.10 in increments of 0.10), giving a net birth rate of 0.05:0.95. To determine the effect of varying the fitness of drug resistance mutants, 10 simulations of 200,000 patients each (100,000 per simulated strain) were run with crH = crR = (0.0 : 0.90 in increments of 0.10). For all simulations, bacterial burden was allowed to increase to 1012 bacteria within a patient, and the probability of observing rifampicin resistance, isoniazid resistance, and MDR was determined by dividing the number of simulated patients with at least one resistant bacterium by the total number of simulated patients. To determine the probability of observing resistance in a model where mutation is determined by replication dynamics, we used equation 2 from Colijn et al (2011)47 with the parameters listed in Supplementary Figure 2c.
Supplementary Material
Acknowledgments
This work was supported by a New Innovator's Award, DP2 0D001378, from the Director's Office of the National Institute of Health to S.M.F., by a subcontract from NIAID U19 AI076217 to S.M.F. and M.B.M., by the US National Institutes of Health Models of Infectious Disease Agent Study program, through cooperative agreement 1 U54 GM088558 (M.L.), by the Howard Hughes Medical Institute, Physician Scientist Early Career Award (S.M.F.), and by the Doris Duke Charitable Foundation, Clinical Scientist Development Award (S.M.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Accession codes: Whole genome sequencing of human isolates was performed previously42, and reads were deposited in the National Center for Biotechnology Information's Sequence Read Archive under accession number SRA020129.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Velayati AA, et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest. 2009;136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 2.Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally Drug-Resistant Tuberculosis in India. Clin Infect Dis. 2011 doi: 10.1093/cid/cir889. [DOI] [PubMed] [Google Scholar]
- 3.Migliori GB, De Iaco G, Besozzi G, Centis R, Cirillo DM. First tuberculosis cases in Italy resistant to all tested drugs. Euro Surveill. 2007;12:E070517.1. doi: 10.2807/esw.12.20.03194-en. [DOI] [PubMed] [Google Scholar]
- 4.Gandhi NR, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 2010;375:1830–1843. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 5.David HL. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Applied microbiology. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ford CB, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011 doi: 10.1038/ng.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford WZ, et al. The changing epidemiology of acquired drug-resistant tuberculosis in San Francisco, USA. The Lancet. 1996;348:928–931. doi: 10.1016/S0140-6736(96)03027-9. [DOI] [PubMed] [Google Scholar]
- 8.Goble M, et al. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993;328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 9.Pablos-Méndez A, Knirsch CA, Barr RG, Lerner BH, Frieden TR. Nonadherence in tuberculosis treatment: predictors and consequences in New York City. Am J Med. 1997;102:164–170. doi: 10.1016/s0002-9343(96)00402-0. [DOI] [PubMed] [Google Scholar]
- 10.Udwadia ZF, Pinto LM, Uplekar MW. Tuberculosis management by private practitioners in Mumbai, India: has anything changed in two decades? PLoS ONE. 2010;5:e12023. doi: 10.1371/journal.pone.0012023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson R, et al. Drug-resistant tuberculosis epidemic in the Western Cape driven by a virulent Beijing genotype strain. Int J Tuberc Lung Dis. 2010;14:119–121. [PubMed] [Google Scholar]
- 12.Streicher EM, et al. Genotypic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from rural districts of the Western Cape Province of South Africa. Journal of Clinical Microbiology. 2004;42:891–894. doi: 10.1128/JCM.42.2.891-894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casali N, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–745. doi: 10.1101/gr.128678.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioerger TR, et al. The non-clonality of drug resistance in Beijing-genotype isolates of Mycobacterium tuberculosis from the Western Cape of South Africa. BMC Genomics. 2010;11:670. doi: 10.1186/1471-2164-11-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun G, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J INFECT DIS. 2012 doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerging Infect Dis. 2006;12:736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- 18.Hershberg R, et al. High Functional Diversity in Mycobacterium tuberculosis Driven by Genetic Drift and Human Demography. Plos Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glynn JR, Whiteley J, Bifani PJ, Kremer K, Van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerging Infect Dis. 2002;8:843–849. doi: 10.3201/eid0808.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato-Maeda M, et al. Beijing Sublineages of Mycobacterium tuberculosis Differ in Pathogenicity in the Guinea Pig. Clin Vaccine Immunol. 2012;19:1227–1237. doi: 10.1128/CVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drobniewski F, et al. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA. 2005;293:2726–2731. doi: 10.1001/jama.293.22.2726. [DOI] [PubMed] [Google Scholar]
- 23.Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discovery Today: Disease Mechanisms. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong BC, et al. Progression to Active Tuberculosis, but Not Transmission, Varies by Mycobacterium tuberculosisLineage in The Gambia. J INFECT DIS. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang Y, et al. Spoligotyping and Drug Resistance Analysis of Mycobacterium tuberculosis Strains from National Survey in China. PLoS ONE. 2012;7:e32976. doi: 10.1371/journal.pone.0032976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vadwai V, Shetty A, Supply P, Rodrigues C. Evaluation of 24-locus MIRU-VNTR in extrapulmonary specimens: Study from a tertiary centre in Mumbai. Tuberculosis. 2012:1–9. doi: 10.1016/j.tube.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Huang HY, et al. Mixed Infection with Beijing and Non-Beijing Strains and Drug Resistance Pattern of Mycobacterium tuberculosis. Journal of Clinical Microbiology. 2010;48:4474–4480. doi: 10.1128/JCM.00930-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anh DD, et al. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerging Infect Dis. 2000;6:302–305. doi: 10.3201/eid0603.000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taype CA, et al. Genetic diversity, population structure and drug resistance of Mycobacterium tuberculosis in Peru. Infection, Genetics and Evolution. 2012;12:577–585. doi: 10.1016/j.meegid.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Mestre O, et al. Phylogeny of Mycobacterium tuberculosis Beijing strains constructed from polymorphisms in genes involved in DNA replication, recombination and repair. PLoS ONE. 2011;6:e16020. doi: 10.1371/journal.pone.0016020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Steenwinkel JEM, et al. Drug Susceptibility of Mycobacterium tuberculosisBeijing Genotype and Association with MDR TB. Emerging Infect Dis. 2012;18:660–663. doi: 10.3201/eid1804.110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werngren J. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. Journal of Clinical Microbiology. 2003 doi: 10.1128/JCM.41.4.1520-1524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luria SE, Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart FM. Fluctuation tests: how reliable are the estimates of mutation rates? Genetics. 1994;137:1139–1146. doi: 10.1093/genetics/137.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart FM, Gordon DM, Levin BR. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics. 1990;124:175–185. doi: 10.1093/genetics/124.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 39.Burnham KP. Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociological Methods & Research. 2004;33:261–304. [Google Scholar]
- 40.Bergval IL, Schuitema ARJ, Klatser PR, Anthony RM. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. J Antimicrob Chemother. 2009;64:515–523. doi: 10.1093/jac/dkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandgren A, et al. Tuberculosis Drug Resistance Mutation Database. Plos Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 43.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M, Ota T. On the rate of molecular evolution. Journal of Molecular Evolution. 1971;1:1–17. doi: 10.1007/BF01659390. [DOI] [PubMed] [Google Scholar]
- 46.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2012 doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colijn C, Cohen T, Ganesh A, Murray MB. Spontaneous Emergence of Multiple Drug Resistance in Tuberculosis before and during Therapy. PLoS ONE. 2011;6:e18327. doi: 10.1371/journal.pone.0018327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portevin D, Gagneux S, Comas I, Young DB. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- 50.Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 51.Hall LMC, Henderson-Begg SK. Hypermutable bacteria isolated from humans--a critical analysis. Microbiology (Reading, Engl) 2006;152:2505–2514. doi: 10.1099/mic.0.29079-0. [DOI] [PubMed] [Google Scholar]
- 52.Blázquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 53.Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 54.Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 55.Springer B, et al. Lack of mismatch correction facilitates genome evolution in mycobacteria. Mol Microbiol. 2004;53:1601–1609. doi: 10.1111/j.1365-2958.2004.04231.x. [DOI] [PubMed] [Google Scholar]
- 56.Fallow A, Domenech P, Reed MB. Strains of the East Asian (W/Beijing) lineage of Mycobacterium tuberculosis are DosS/DosT-DosR two-component regulatory system natural mutants. J Bacteriol. 2010;192:2228–2238. doi: 10.1128/JB.01597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huet G, et al. A lipid profile typifies the Beijing strains of Mycobacterium tuberculosis: identification of a mutation responsible for a modification of the structures of phthiocerol dimycocerosates and phenolic glycolipids. Journal of Biological Chemistry. 2009;284:27101–27113. doi: 10.1074/jbc.M109.041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmalstieg AM, et al. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrobial Agents and Chemotherapy. 2012;56:4806–4815. doi: 10.1128/AAC.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Louw GE, et al. Rifampicin Reduces Susceptibility to Ofloxacin in Rifampicin Resistant Mycobacterium tuberculosis through Efflux. Am J Respir Crit Care Med. 2011 doi: 10.1164/rccm.201011-1924OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams KN, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Comas I, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2011;44:106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams K, et al. Sterilizing Activities of Novel Combinations Lacking First- and Second-Line Drugs in a Murine Model of Tuberculosis. Antimicrobial Agents and Chemotherapy. 2012;56:3114–3120. doi: 10.1128/AAC.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lienhardt C, et al. New Drugs for the Treatment of Tuberculosis: Needs, Challenges, Promise, and Prospects for the Future. J INFECT DIS. 2012;205:S241–S249. doi: 10.1093/infdis/jis034. [DOI] [PubMed] [Google Scholar]
- 64.Menzies D, et al. Standardized treatment of active tuberculosis in patients with previous treatment and/or with mono-resistance to isoniazid: a systematic review and meta-analysis. Plos Med. 2009;6:e1000150. doi: 10.1371/journal.pmed.1000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gelmanova IY, et al. Barriers to successful tuberculosis treatment in Tomsk, Russian Federation: non-adherence, default and the acquisition of multidrug resistance. Bull World Health Organ. 2007;85:703–711. doi: 10.2471/BLT.06.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seung KJ, et al. The effect of initial drug resistance on treatment response and acquired drug resistance during standardized short-course chemotherapy for tuberculosis. Clin Infect Dis. 2004;39:1321–1328. doi: 10.1086/425005. [DOI] [PubMed] [Google Scholar]
- 67.Helb D, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. Journal of Clinical Microbiology. 2010;48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weyer K. Laboratory services in tuberculosis control Part II: microscopy. World Health Organization; Geneva, Switzerland: 1998. [Google Scholar]
- 69.Dowdy DW, Basu S, Andrews JR. Is Passive Diagnosis Enough? The Impact of Subclinical Disease on Diagnostic Strategies for Tuberculosis. Am J Respir Crit Care Med. 2012 doi: 10.1164/rccm.201207-1217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaw JB, Wynn-Williams N. Infectivity of pulmonary tuberculosis in relation to sputum status. Am Rev Tuberc. 1954;69:724–732. doi: 10.1164/art.1954.69.5.724. [DOI] [PubMed] [Google Scholar]
- 71.Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci USA. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gagneux S. The Competitive Cost of Antibiotic Resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.