Abstract
Eyes gather information and color forms an extremely important component of the information, more so in the case of animals to forage and navigate within their immediate environment. By using the ONIOM (QM/MM) method, we report a comprehensive theoretical analysis of the structure and molecular mechanism of spectral tuning of monkey-red and green-sensitive visual pigments. We show that, interaction of retinal with three hydroxyl-bearing amino acids near the β-ionone ring part of the retinal in opsin, A164S, F261Y and A269T, increases the electron delocalization, decreases the BLA of the retinal and leads to variation in the wavelength of maximal absorbance in the red- and green-sensitive visual pigments. Based on the analysis, we propose the “OH-site” rule for seeing red and green. This rule is also shown to account for the spectral shifts obtained from hydroxyl-bearing amino acids near the Schiff base in different visual pigments: at site 292 (A292S, A292Y, and A292T) in bovine and at site 111 (Y111) in squid opsins. Therefore, the OH-site rule is shown to be site-specific and not pigment-specific and thus can be used for tracking spectral shifts in any visual pigment.
1. INTRODUCTION
Visual pigments found in the rod outer segments are called rhodopsins and those that are found in the cone outer segments are called cone pigments. Rhodopsin mediates the transformation of light into dim-light vision and cone pigments mediate the transformation of light into trichromatic color vision (blue, green and red) in humans and/or mammals.1 Irrespective of the difference in location and function, all visual pigments contain 11-cis-retinal covalently bound to an opsin apoprotein via protonated Schiff base linkage (PSB11) as the light-sensing chromophore.2 One of the fundamental characteristics used to distinguish different visual pigments from each other is the wavelength of maximal absorption (λmax). Based on the wavelength of their peak light sensitivity, visual pigments are classified into five sub-groups: Rhodopsin or RH1 pigments (~480–510 nm), RH2 pigments (~450–530 nm), SWS1 pigments (~360–440 nm), SWS2 pigments (~400–450 nm), and M/LWS pigments (~510–560 nm), respectively.3 Three visual pigments with each corresponding to a different λmax mediate human color vision: 414 nm for blue, 530 nm for green and 552 or 557 nm for red.4 Most mammals have dichromatic color vision, but hominoids and Old World monkeys have trichromatic vision for it helps them to detect yellow and red fruits against dapped foliage.5 Although considerable progress has been made in exploring the structure-function relationships responsible for dim-light vision,6 understanding color vision at a molecular level has remained difficult to date due to the absence of X-ray structures of cone pigments. Inspite of the difficulties, experimental mutagenesis studies have yielded valuable insights required for the identification of key molecular players mediating color vision.
Yokoyama et al. compared the partial amino acid sequences of red and green color pigments from five mammalian orders, and showed that green pigments evolved from its red-sensitive ancestral pigments. The authors proposed the famous “five-sites” rule for understanding the variance in λmax of different M/LWS pigments. However, they also realized the possibility of existence of other spectral tuning mechanisms outside the “five-sites” rule.7 According to the “five-sites” rule, amino acid changes at sites 164, 181, 261, 269 and 292 would account for the 50 nm blue shift between the red (560 nm) and green (~510–530 nm) visual pigments. Apparently, the amino acid substitutions refer to replacement of SHYTA with AYFAS (S164A, H181Y, Y261F, T269A, A292S) composition.7 A simpler way to look at the rule is that, substitution of Serine with Alanine (S→A) and Histidine with Tyrosine (H→Y) and Tyrosine with Phenylalanine (Y→F) and Threonine with Alanine (T→A) and Alanine with Serine (A→S) would lead to loss or gain of a hydroxyl-group by the visual pigment, which in turn may cause the change in the observed λmax.
One of the limitations of the rule is that, it is pigment-specific and thus can only be applied to the M/LWS pigments. Because all of the visual pigments contain the same PSB11 chromophore,8 any rule that helps us to understand the mechanism of color vision should be applicable to all visual pigments independent of its structural and/or its functional characteristics. However, to derive such a rule, one has to first decode the “five-sites” rule, for it already accounts for variation in λmax of majority of the M/LWS pigments. In this report, by studying the structure and spectral tuning mechanism of monkey red- and green-sensitive visual pigments, we present the “OH-site” rule that is shown to be site-specific and not pigment-specific.
According to the “OH-site” rule, if an amino acid contains an OH-group and is present near the β-ionone ring part of the retinal in opsin, it will stabilize the excited state relative to the ground state of the retinal and will induce a red shift in the λmax. In contrast, if an amino acid contains an OH-group and is present near the Schiff base site, it will stabilize the ground state relative to the excited state and will induce a blue shift in the λmax. In other words, interaction of retinal with dipolar residues near the β-ionone ring will increase the electron delocalization and decrease the bond length alternation (BLA) leading to red shifts, whereas, interaction of retinal with dipolar residues near the Schiff base site will decrease the electron delocalization and increase the BLA leading to blue shifts. We demonstrate this principle by using amino acid substitutions at sites 164, 261 and 269 (A164S, F261Y and A269T) in monkey M/LWS pigments, at site 292 (A292S, A292T, A292Y) in bovine RH1 pigment and at site 111 (Y111) in squid RH1 pigment. The findings allow us to prove that the “OH-site” rule is site-specific and not pigment-specific.
2. COMPUTATIONAL
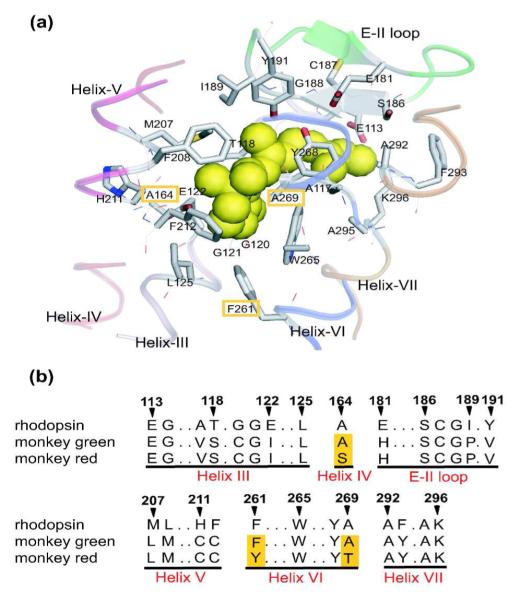
Although bovine and more specifically monkey RH1 pigments are made up of equal number of amino acids (348 AA), they are not identical.9 Compared to the amino acid composition of bovine opsin, 22 AA’s are different in monkey opsin (22/348) and they correspond to the sites K16A, A26Y, M49V, L112A, V173F, M183L, P194L, H195K, E196P, T198V, I213T, L216M, L266V, G270S, D282N, T297S, S298A, A299S, V300I, V318L, L321I and T335A, respectively. Compared to the monkey RH1 pigment, the red- and green-sensitive pigments are larger in size by 16 amino acids. Therefore, the counterion site E113 in RH1 pigment is equivalent to site E129 in M/LWS pigments.9 Throughout this report, the amino acid numbering will correspond to that of the RH1 pigment, rhodopsin. Both red- and green-sensitive opsins are composed of 364 AA’s and differ by only 15 AA’s. Although the difference in AA sequence is large between the RH1 and M/LWS opsins, if we take the amino acids surrounding the retinal binding site into consideration,10 the situation changes dramatically, more so in the case of red-sensitive opsin. That is, within the binding pocket, 15 AA’s (15/27) differ between RH1 and red-sensitive opsins and 12 AA’s (12/27) differ between RH1 and green-sensitive opsins. Therefore red- and green-sensitive opsins differ by only 3 AA’s at sites 164, 261 and 269 (see Figure 2) and the situation appears to be the same as in human green and red-sensitive pigments.11 Between RH1 and M/LWS opsins, there are four identical AA changes in helix III (sites A117V, T118S, G120C, E122I); one in helix IV at site A164S in red-opsin; three in the E-II loop at sites E181H, I189P, Y191V; four in helix V at sites M207L, F208M, H211C, F212C; two at sites F261Y, A269T in red-opsin; and one in helix VII at site F293Y.12
Figure 2.
a) The X-ray crystallographic structure of the chromophore-binding site of bovine rhodopsin (Protein Data Bank entry: 1U19[13]), which is viewed from the helix VI side. The upper and lower regions correspond to the extracellular and cytoplasmic sides, respectively. The retinal chromophore, which is bound to Lys296, is shown by green space-filling model. Side chains of the 27 amino acids around retinal are shown by stick drawings, though some residues behind the retinal are hidden. Ribbon drawings illustrate the secondary structures around the retinal. Corresponding amino acids in monkey green and red pigments are identical except for three amino acids highlighted in orange. b) Partial amino acid sequences of rhodopsin (bovine and monkey), monkey green, and monkey red. The amino acids are identical between bovine and monkey rhodopsins. The three amino acids that differ in monkey green and red are highlighted in orange columns. The residue numbers are based on the bovine rhodopsin sequence. This figure is taken from reference 12.
The QM/MM optimized structure of monkey RH1 pigment was prepared using the previously published wild-type bovine RH1 pigment as a template.14 All QM/MM calculations in this study were performed using the two-layer ONIOM (QM:MM) scheme,15 in which the QM part contains the full PSB11 and side-chain N–H moiety of Lys296 is treated at the B3LYP/6-31G(d) level. The MM part contains the full opsin, depicted as point charges using AMBER96 (348 residues in RH1 and 364 residues in M/LWS opsins) plus van der Waals interactions; the interface between the QM and MM regions was treated by the hydrogen link atom. The total energy of the system was obtained as EEE, the energy in the electronic embedding (EE) scheme, in which the QM:MM interaction is included in the QM calculation and therefore the QM wave function is polarized by the MM point charges.15 Throughout this study, we employed for geometry optimization the QM/MM-EE scheme and the calculations are carried out using the ONIOM (Our own N-layer Integrated molecular Orbital plus molecular Mechanics) method implemented in Gaussian09 program package.16 To calculate the absorption and circular dichroism (CD) spectra in the gas phase (QM) and in the protein (QM/MM) environments, ab initio multireference QM/MM calculations on the resulting structures at the spectroscopy oriented configuration interaction SORCI+Q method17 at 6-31G(d) level was applied using the ORCA 2.6.19 program package18 on top of the three-root (6e, 6o) complete active space self-consistent field (CASSCF) wave functions.19 The vertical excitation energies as well as oscillator and rotatory strengths to first (S1) and second (S2) excited states were calculated for all of the structures discussed in this study. Accuracy of this computational setup is within ±10 nm, and we showed previously that the present setup yielded results in good agreement with experimental measurements.20
3. RESULTS and DISCUSSION
3.1. The Chloride-Binding Site in M/LWS Pigments
Molecular determinants underlying the evolution of red and green color vision in mammals have been studied in the past. However, the mechanism by which the amino acid changes determine the spectral properties of red- and green-pigment continues to remain unclear. This could be due to lack of proper understanding of the retinal binding sites in M/LWS pigments that are characterized by the presence of a chloride (Cl−) binding site. In the absence of X-ray structures, identifying the possible location of Cl− binding site and understanding its role on the spectral tuning mechanism has proved to be difficult.
Mathies et al. showed that anions close to the Schiff base would stabilize the ground state more than the excited state and induce strong blue shifts in the λmax.21 Fager et al. showed that the Cl− binding site might protect the retinal from hydroxylamine attack22 and Klienschmidt et al. argued for the presence of an anion-binding site near the Schiff base end of retinal.23 In the absence of X-ray structures of rhodopsin at that time, it is possible, the site proposed by the authors may well correspond to that of the E113 counterion. Apparently, Wang et al. identified the location of Cl− binding site in M/LWS pigments. They showed His181 and Lys184 to be positively charged and strictly conserved in all LWS cone pigments and as a result serve as Cl− binding sites.24 Hirano et al. suggested that Cl− binding site interacts directly with the photoisomerized retinal (batho state) in chicken iodopsin.25
All these arguments were taken into consideration and a Cl− ion is introduced near the positively charged H181 and K184 residues. During the QM/MM geometry optimization, three water molecules including wat2a, which was originally H-bonded to the E181 residue move closer to the Cl− binding site and orient themselves towards the Cl− ion (Figure 3). This finding is in good agreement with the recent FTIR measurements that showed the presence of at least three internal water molecules near the Cl− binding site.26 Conserved water molecules are known to play a crucial role in the functioning of visual pigments.27 In the presence of H181, K184 and three water molecules, it is possible that the Cl− ion is weakly hydrated in M/LWS pigments. We suggest that the presence of weakly hydrated chloride-binding site may mediate the reaction process of monkey M/LWS pigment.28 Also, presence of positively charged histidine and/or lysine may well serve as a common structural motif for identifying Cl− binding sites in related photo-biological systems.29
Figure 3.
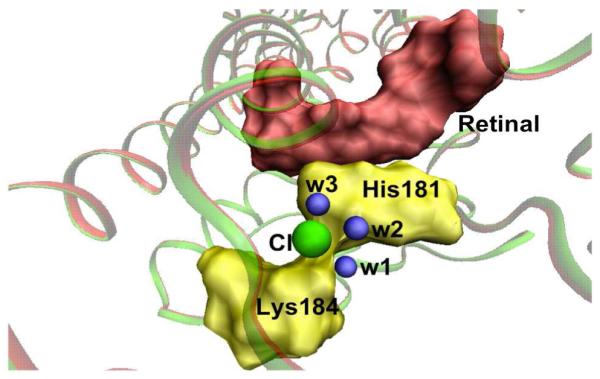
Chloride-binding site in the QM/MM optimized structure of monkey red- and green-sensitive visual pigments. Chloride ion is shown in green, His181 and Lys184 are given in yellow, water molecules in blue and retinal in red.
3.2. The Retinal Geometry in M/LWS pigments
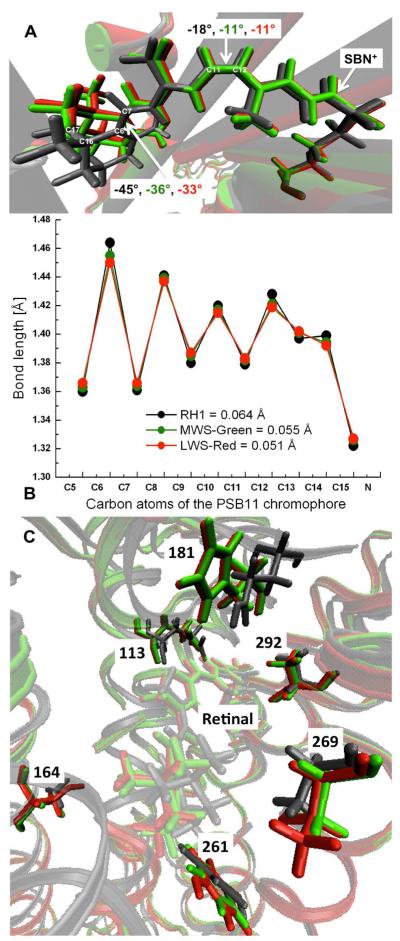
Although the chromophore is the same, its geometry is found to be slightly different in RH1 and M/LWS pigments. As one end of the retinal tether is held fixed by the β-ionone ring and other end by the Schiff base lysine linkage (Figure 4), amino acid replacements near the polyene chain can significantly alter this architecture. To start with, orientation of the β-ionone ring relative to the polyene chain is altered in all the three pigments. Compared to the geometry of retinal in RH1 opsin, C6—C7 twist angle is reduced by 9° in MWS (from - 45° to -36°) and by 11° in LWS (from −45° to −33°) pigments. As a result, the retinal relaxes leading to a small displacement of the β-ionone ring in M/LWS pigments. Residues Ile189 and Met207 are close to the methyl groups attached to C16 and C17 carbon atoms of the retinal and it is possible that amino acid substitutions at sites 189, 207, 211, and 261 may have created the space required for the slight displacement of β-ionone ring. At the other end of the retinal, negative pretwist of the isomerizing C11—C12 bond is reduced from −18° in RH1 to −11° in M/LWS pigments. The decrease in distortion leads to an increase in length of the retinal conjugation (from C5 to SBN+ moiety), from 11.26 Å in RH1 to 11.69 Å in MWS and to 11.82 Å in LWS pigments. As a consequence, the retinal is found to be lot more relaxed and less distorted in the M/LWS pigments.
Figure 4.
a) Overlay of the QM/MM optimized retinal geometries in RH1 (gray), MWS (green), and LWS (red) opsins. Dihedral angles of the C5—C6 and C11—C12 double bonds of the retinal are also given. b) Bond lengths along the retinal polyene chain are plotted and the corresponding bond length alternation (BLA) values, which is defined as the average of the bond lengths of single bonds minus that of double bonds (C5 to SBN+ moiety) are given in parentheses. c) Overlay of the retinal binding sites in RH1 (gray), in MWS (green), and in LWS (red) pigments and some key residues responsible for spectral tuning, 113, 181, 164, 261, 269, and 292 are also shown.
The QM/MM calculations also reveal significant change in the BLA of retinal polyene chain in M/LWS pigments (Figure 4b). In particular, decrease in C6—C7 and C11—C12 twist angles is accompanied with the decrease and increase of single and double bond lengths. In particular, the effect of BLA is more pronounced near C13=C14—C15=N+ region (Figure 4b), which leads to a slight increase in the retinal-counterion (SBN+---E113) distance in M/LWS pigments. As a consequence, the BLA of retinal decreases from 0.064 Å in RH1 to 0.055 Å in MWS-green and to 0.051 Å in LWS-red pigments in agreement with resonance Raman spectra analysis that also argued for the decrease in BLA of retinal in human green and red visual pigments.30 In the absence of X-ray structures, the present QM/MM analysis will serve as a good reference point for studying the retinal architecture in M/LWS cone pigments.
3.3. The Retinal Binding Pocket in M/LWS Pigments
Although the same set of structural elements is retained in all the three pigments, residues constituting the “five-sites” rule are found to undergo a slight displacement in M/LWS pigments (Figure 4c). The most prominent change is seen in the vicinity of C11 and C12 atoms due to the substitution of Glu with His at site 181. In the presence of H181, hydrogen bonds connecting Glu181, wat2a, and backbone atoms of S186 in the EII loop in RH1 opsin13 are broken and the displaced wat2a moves towards the new chloride-binding site involving H181 and K184. In rhodopsin, the closest atom to the retinal isomerizing region is the peptide oxygen of C187 in the EII loop.31 It is suggested that the peptide oxygen of C187 assists in the large dislocation of the C12 atom by repulsive interaction during photoisomerization.32 Because the peptide contact between C187 and G188 and the disulfide bond between C187 and C110 are found to remain intact in M/LWS pigments, it is possible that C187 also assists in the isomerization of retinal in M/LWS pigments. In RH1 pigment the retinal polyene chain is held close to the helices III and VI. Due to the amino acid substitutions in helix III (A117V, T118S, G120C) and in helix V (M207L, F208M, H211C, F212C), the stiffness in the polyene chain is released and the pocket becomes wider. This could be one of the reasons for the presence of several internal water molecules in M/LWS pigments.26 Residue W265 in helix VI is highly conserved in all the three pigments and continues to remain in van der Waals contact with the β-ionone ring of retinal.13
3.4. Variance in the Wavelength of Maximal Absorbance
Molecular basis of variations in the λmax of cone photopigments has been widely studied in the past. Neitz et al. compared the amino acid sequences of eight new world monkeys and concluded that three amino acid substitutions at positions 164, 261, and 269 produce the ~30 nm difference (from 530 to 562 nm) underlying the human red-green color vision.33 Yokoyama et al. studied the convergent evolution of red- and green-like visual pigments in blind cave fish and found the involvement of these three residues for spectral tuning.34 Henderson et al. also argued in favor of these experimental findings by comparing the structure of rhodopsin and color pigments with that of bacteriorhodopsin.35 Merbs et al. identified a role for hydroxyl-bearing amino acids in color vision.36 Chan et al. introduced hydroxyl-bearing amino acids at these three sites in rhodopsin and showed that they indeed cause red shifts.37
Asenjo et al. performed an extensive mutagenesis study and found that in addition to these three sites, amino acid susbtitutions at four more sites 116, 230, 233, and 309 are involved in spectral tuning.38 According to this finding seven amino acid changes are required for converting green- to red-sensitive pigment. By using the parsimony and Bayersian methods, Yokoyama et al. challenged this finding and showed ancestors of Old World and New World monkeys to be only red-sensitive as they lacked the green gene7,39 Therefore, amino acid substitutions should be introduced in the ancestral pigment and only three and not seven amino acid changes should mediate color vision.
To determine the variance in λmax of the M/LWS pigments, it is important to understand the difference in response between the excited and ground states to the external perturbation from the protein environment. Analyses of the data in Table 1 show that, the gas phase PSB11 (QM-only) at QM/MM optimized geometry absorbs at 619 nm in RH1 and at 623 nm in M/LWS pigments. The counterion, Glu113, induces a strong blue shift of ~120 nm in all the three pigments and shifts the λmax from gasphase to protein environments.40-42 The counterion also plays a critical role in regulating the phototransduction of short-wavelength sensitive cone pigments.43
Table 1.
The calculated SORCI+Q first vertical excited state absorption wavelengths (λ) in nm, oscillator (f) and rotatory (R) strengths in au and difference in the ground- (S0) and excited state (S1) dipole moments (Δμ) of the PSB11 chromophore in the gas-phase (QM-only) and protein (QM/MM) environments of monkey RH1, monkey green- and red-sensitive visual pigments at QM/MM optimized geometries. The experimental values for monkey RH1, green- and red-sensitive pigments are taken from reference 33 and values for bovine RH1 and A292S mutant pigments are taken from reference 52.
| Monkey Visual Pigments |
First Vertical Excited State (S0→S1) Properties |
||||||
|---|---|---|---|---|---|---|---|
| Gas phase | Protein | ||||||
|
| |||||||
| λ | f | λ | f | R | Δ μ | Exp | |
| RH1-Dim/Light | 619 | 1.2 | 493 | 1.4 | 0.1 | 12.1 | 498 |
| MWS-Green | 623 | 1.3 | 521 | 1.5 | 0.3 | 12.9 | 530 |
| LWS-Red | 623 | 1.4 | 544 | 1.5 | 0.3 | 13.2 | 562 |
In the presence of electrostatic and polarization effects, the retinal absorbs at 493 nm in RH1, 521 nm in MWS and 544 nm in LWS pigments. The red shift separating RH1 and M/LWS pigments can be correlated to the increase in polarization of the retinal by M/LWS pigments. This property is evident in the calculated difference in dipole moment (Δμ) between the ground (S0) and excited (S1) states of the retinal, which increases from 12.1 D in RH1 to 12.9 D in MWS and to 13.2 D in LWS pigments. The calculated oscillator strength (f) increases only slightly (from 1.4 to 1.5), but the rotatory strength increases by three-fold (from 0.1 to 0.3 au) in M/LWS pigments. This finding correlates well with the experimental low-temperature circular dichroism measurements of chicken iodopsin (cone pigments), which also displayed an increase in rotatory strength compared to chicken rhodopsin.44 The spectral red shifts of 28 nm (calc.) vs. 32 nm (exptl.) separating RH1 and MWS-green pigments and 21 nm (calc.) vs. 30 nm (exptl.) between green and red pigments are also accounted for in the QM/MM calculations.
To determine the origin of spectral shift we turned off the charges of the three hydroxyl-bearing amino acids in LWS opsin, because as mentioned before, the MWS-green pigment evolved from its red-sensitive LWS ancestral pigment. Some of the earlier theoretical studies on color vision have over-looked this fact45-48 and as a result, neither the molecular basis of spectral tuning nor the evolutionary mechanisms of visual pigments was elucidated in detail. By turning off charges, we find S164, Y261 and T269 to contribute 6, 9, and 6 nm individually and 21 nm cumulatively to the spectral shift. The cumulative shift is additive and is in good agreement with the experimental assignment of 6, 9 and 15 nm for S164A, Y261F and T269A mutants in human red and green pigments.33 Although the Cl− ion induces a blue shift of ~20 nm, the overall effect of the chloride binding site (Cl, H181, and K184) is a weak red shift of ~10 nm47,49 and correlates well with the chloride depletion induced blue shift obtained in the gecko cone pigments.50 The findings also support the argument that, secondary interactions involving the binding site residues are as important as the first-order chromophore protein interactions in mediating the wavelength maximum in cone pigments.51
So, what is the mechanism by which these three hydroxyl-bearing amino acids induce red shifts? Does it mean all sites with hydroxyl-bearing amino acids induce a red shift? In other words, will increase in number of hydroxyl-bearing residues cause an increase in the λmax of visual pigments? To unravel the mechanism and to answer these questions on theoretical grounds, we introduced the same set of hydroxyl-bearing amino acids (serine, tyrosine and threonine) into bovine rhodopsin (RH1 pigment) at site 292. Note that, site 292 is near the Schiff base region and is occupied by alanine in bovine opsin. It is also one of the “five-sites” found to be consistently involved in the spectral tuning mechanisms of many M/LWS pigments. Experimental studies have shown that substitution of alanine with serine (A292S) induces a blue shift of 11 nm in rhodopsin52 and 8 nm in blue cone visual pigments.53 Furthermore, A292S mutant yields a blue shift of ~10 to 17 nm in the green-absorbing Drosophila pigment.54 There are exceptions where, the effect of A292S is negligible and/or gets manifested only in the presence of other mutants.55
By using the OH-site rule, we demonstrate that not only serine, but also tyrosine and threonine at site 292 can cause blue shifts. That is, replacement of A with S, Y or T at site 292 (A292S, A292Y, A292T) will stabilize the ground state, decrease the electron delocalization and increase the BLA of retinal leading to blue shifts of 11, 9, 14 nm in the λmax. This principle can also be applied in the case of invertebrate squid rhodopsin where, site 111 near the Schiff base region is occupied by tyrosine (Y111) and induces a blue shift of 10 nm.56 Note that, site 111 corresponds to site 113 in bovine rhodopsin and is occupied by the primary counterion, E113. Therefore, we suggest that interaction of retinal with dipolar residues near the β-ionone ring site will (i) increase the electron delocalization; (ii) decrease the BLA of the retinal, and (iii) lead to a red shift in the λmax. Whereas, interaction of retinal with dipolar residues near the Schiff base site will (i) decrease the electron delocalization; (ii) increase the BLA of the retinal, and (iii) lead to a blue shift in the λmax.
4. Conclusion
Visual pigment provides the decisive crossing point for interaction between an organism and its environment. In the case of vertebrates; visual pigments exhibit a remarkable level of diversity, which reflects their adaptive reaction to various color environments. On one hand, amino acid replacements are known to play a crucial role in mediating the variance in the λmax of color pigments in vertebrates.57 On the other hand, defects in color vision are known to arise due to difference in variation in the spectral positioning of the M/LWS pigments.58 Notice that, M/LWS pigments are not only present in animals with red-green color vision, but also in those with red-green color blindness.59 Therefore, a critical first step toward understanding the origin of color blindness is to determine the molecular basis of variance in the λmax of red and green pigments.
In this report, we have presented a QM/MM analysis of the structure and spectral tuning mechanism that determines the variance in λmax of monkey red- and green-sensitive visual pigments. The analysis allows us to propose the “OH-site” rule where, three hydroxyl-bearing amino acid substitutions, A164S, F261Y and A269T, are shown to account for 30 nm spectral shift, which separates the red- and green-sensitive visual pigments. The rule is also shown to account for the spectral shifts arising from OH-sites in bovine and squid visual pigments. Therefore, we suggest that the “OH-site” rule is site-specific and not pigment-specific and thus can be applied to any visual pigment.
Supplementary Material
Figure 1.
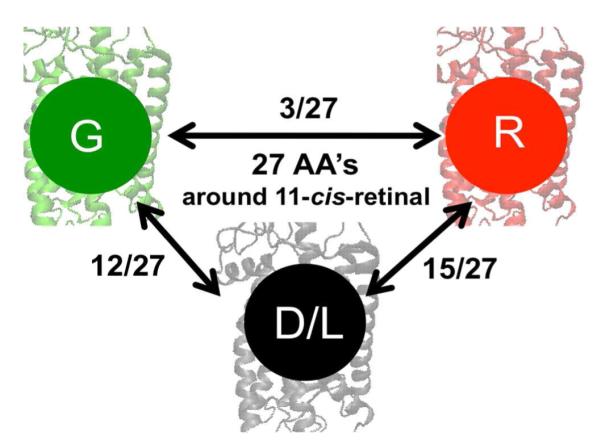
Schematic representation of the amino acid (AA) differences around 11-cis-retinal that separates dim-light (D/L) from red (R) and green (G) visual pigments.
ACKNOWLEDGMENTS
The critical comments of Prof. T. Okada and Prof. S. Yokoyama are gratefully acknowledged. The work is supported in part at Emory by a grant from the National Institutes of Health (R01EY016400) and at Kyoto by a Core Research for Evolutional Science and Technology (CREST) Grant on High Performance Computing for Multiscale and Multiphysics Phenomena JST and at Nagoya by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology to H.K (20108014, 22247024).
Footnotes
SUPPORTING INFORMATION Cartesian coordinates of the QM/MM optimized retinal geometries of monkey RH1, green- and red-sensitive visual pigments are available in the Supporting Information.
Notes The authors declare no competing financial interest.
REFERENCES
- (1).Birge RR. Annu. Rev. Phys. Chem. 1990;41:683–733. doi: 10.1146/annurev.pc.41.100190.003343. [DOI] [PubMed] [Google Scholar]
- (2).Shichida Y, Imai H. Cell. Mol. Lif. Sci. 1998;54:1299–1315. doi: 10.1007/s000180050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Yokoyama S. Annu. Rev. Genomics Hum. Genet. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- (4).Merbs SL, Nathans J. Nature. 1992;356:433–435. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- (5).Nei M, Zhang J, Yokoyama S. Mol. Biol. Evol. 1997;14:611–618. doi: 10.1093/oxfordjournals.molbev.a025800. [DOI] [PubMed] [Google Scholar]
- (6).Mathies RA, Lugtenburg J. In: Molecular Mechanisms of Visual Transduction. Stavenga DG, DeGrip WJ, Pugh EN Jr., editors. Elsevier; Amsterdam, The Netherlands: 2000. p. 55. [Google Scholar]
- (7).Yokoyama S, Radlwimmer F. Mol. Biol. Evol. 1998;15:560–567. doi: 10.1093/oxfordjournals.molbev.a025956. [DOI] [PubMed] [Google Scholar]
- (8).Sekharan S, Morokuma K. J. Am. Chem. Soc. 2011;133:19052–19055. doi: 10.1021/ja208789h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Oprian DD, Asenjo AB, Lee N, Pelletier SL. Biochemistry. 1991;30:11367–11372. doi: 10.1021/bi00112a002. [DOI] [PubMed] [Google Scholar]
- (10).Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- (11).Nathans J, Thomas D, Hogness DS. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- (12).Katayama K, Furutani Y, Imai H, Kandori H. Angew. Chem. Int. Ed. 2010;49:891–894. doi: 10.1002/anie.200903837. [DOI] [PubMed] [Google Scholar]
- (13).Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- (14).Altun A, Yokoyama S, Morokuma K. J. Phys. Chem. B. 2008;112:6814–6827. doi: 10.1021/jp709730b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. J. Comput. Chem. 2003;24:760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- (16).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, J. A. Montgomery J, Peralta JE, Ogliaro F, Bearpark M, J. J. Heyd EB, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision A.02. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- (17).Neese FA. J. Chem. Phys. 2003;119:9428–9443. [Google Scholar]
- (18).Neese F. ORCA-an ab initio, DFT and semiempirical electronic structure package. v. 2.6 2007. [Google Scholar]
- (19).Roos BO, Taylor PR. Chem. Phys. 1998;48:157–173. [Google Scholar]
- (20).Sekharan S, Altun A, Morokuma K. Annu. Rep. Comp. Chem. 2011;Vol. 7:215–231. [Google Scholar]
- (21).Mathies R, Stryer L. Proc. Natl. Acad. Sci. U.S.A. 1976;73:2169–2173. doi: 10.1073/pnas.73.7.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fager LY, Fager RS. Exp. Eye. Res. 1979;29:401–408. doi: 10.1016/0014-4835(79)90056-3. [DOI] [PubMed] [Google Scholar]
- (23).Kleinschmidt J, Harosi FI. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9181–9185. doi: 10.1073/pnas.89.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang Z, Asenjo AB, Oprian DD. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- (25).Hirano T, Imai H, Kandori H, Shichida Y. Biochemistry. 2001;40:1385–1392. doi: 10.1021/bi001332j. [DOI] [PubMed] [Google Scholar]
- (26).Katayama K, Furutani Y, Imai H, Kandori H. Biochemistry. 2012;51:1126–1133. doi: 10.1021/bi201676y. [DOI] [PubMed] [Google Scholar]
- 27).Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc. Natl. Acad. Sci. USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sato K, Yamashita T, Imamoto Y, Shichida Y. Biochemistry. 2012;51:4300–4308. doi: 10.1021/bi3000885. [DOI] [PubMed] [Google Scholar]
- (29).Rivalta I, Amin M, Luber S, Vassiliev S, Pokhrel R, Umena Y, Kawakami K, Shen JR, Kamiya N, Bruce D, Brudvig GW, Gunner MR, Batista VS. Biochemistry. 2011;50:6312–6317. doi: 10.1021/bi200685w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kochendoerfer GG, Wang Z, Oprian DD, Mathies RA. Biochemistry. 1997;36:6577–6587. doi: 10.1021/bi970322o. [DOI] [PubMed] [Google Scholar]
- (31).Sekharan S, Sugihara M, Weingart O, Okada T, Buss V. J. Am. Chem. Soc. 2007;129:1052–1054. doi: 10.1021/ja066970p. [DOI] [PubMed] [Google Scholar]
- (32).Liu RSH, Hammond GS. Acc. Chem. Res. 2005;38:396–403. doi: 10.1021/ar040246z. [DOI] [PubMed] [Google Scholar]
- (33).Neitz M, Neitz J, Jacobs GH. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- (34).Yokoyama R, Yokoyama S. Proc. Natl. Acad. Sci. U.S.A. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Henderson R, Schertler GFX. Phil. Trans. Roy. Soc. (Lond.) B. 1990;326:379–389. doi: 10.1098/rstb.1990.0019. [DOI] [PubMed] [Google Scholar]
- (36).Merbs SL, Nathans J. Photochem. Photobiol. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. [DOI] [PubMed] [Google Scholar]
- (37).Chan T, Lee M, Sakmar TP. J. Biol. Chem. 1992;267:9478–9480. [PubMed] [Google Scholar]
- (38).Asenjo AB, Rim J, Oprian D. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- (39).Yokoyama S, Yang H, Starmer WT. Genetics. 2008;179:2037–2043. doi: 10.1534/genetics.108.090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sekharan S, Sugihara M, Buss V. Angew. Chem., Int. Ed. 2007;46:269–271. doi: 10.1002/anie.200603306. [DOI] [PubMed] [Google Scholar]
- (41).Gascon JA, Batista VS. Biophys. J. 2004;87:2931–2941. doi: 10.1529/biophysj.104.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wanko M, Hoffmann M, Strodel P, Kosolowski A, Thiel W, Neese F, Frauhenheim T, Elstner M. J. Phys. Chem. B. 2005;109:3606–3615. doi: 10.1021/jp0463060. [DOI] [PubMed] [Google Scholar]
- (43).Babu KR, Dukkipati A, Birge RR, Knox BE. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- (44).Okada T, Matsuda T, Kandori H, Fukada Y, Yoshizawa T, Shichida Y. Biochemistry. 1994;33:4940–4946. doi: 10.1021/bi00182a024. [DOI] [PubMed] [Google Scholar]
- (45).Trabanino RJ, Vaidehi N, Goddard WA., III J. Phys. Chem. B. 2006;110:17230–17239. doi: 10.1021/jp057144q. [DOI] [PubMed] [Google Scholar]
- (46).Coto PB, Strambi A, Ferre N, Olivucci M. Proc. Natl. Acad. Sci. USA. 2006;103:17154–17159. doi: 10.1073/pnas.0604048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Fujimoto K, Hasegawa J, Nakatsuji N. Chem. Phys. Lett. 2008;462:318–320. [Google Scholar]
- (48).Rajamani R, Lin YL, Gao J. J. Comp. Chem. 2011;312:855–865. [Google Scholar]
- (49).Imamoto Y, Hirano T, Imai H, Kandori H, Maeda A, Yoshizawa T, Groesbeek M, Lugtenburg J, Shichida Y. Biochemistry. 1998;38:11749–11754. doi: 10.1021/bi990587y. [DOI] [PubMed] [Google Scholar]
- (50).Lewis JW, Liang J, Ebrey TG, Sheves M, Livnah N, Kuwata O, Jäger S, Kliger DS. Biochemistry. 1997;36:14593–14600. doi: 10.1021/bi9712908. [DOI] [PubMed] [Google Scholar]
- (51).Kusnetzow A, Dukkipati A, Babu KR, Singh D, Vought RW, Knox BE, Birge BB. Biochemistry. 2001;40:7382–7844. doi: 10.1021/bi010387y. [DOI] [PubMed] [Google Scholar]
- (52).Janz JM, Farrens DL. Biochemistry. 2001;40:7219–7227. doi: 10.1021/bi002937i. [DOI] [PubMed] [Google Scholar]
- (53).Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. J. Biol. Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- (54).Salcedo E, Farrell DM, Zheng L, Phistry M, Bagg EE, Britt SG. J. Biol. Chem. 2009;284:5717–5722. doi: 10.1074/jbc.M807368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Yokoyama S, Tada T, Zhang H, Britt L. Proc. Natl. Acad. Sci. USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sekharan S, Altun A, Morokuma K. Chem.—Eur. J. 2010;16:1744–1749. doi: 10.1002/chem.200903194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Yokoyama S. Gene. 2002;300:69–78. doi: 10.1016/s0378-1119(02)00845-4. [DOI] [PubMed] [Google Scholar]
- (58).Neitz M, Neitz J. Arch. Opthalmol. 2000;118:691–700. doi: 10.1001/archopht.118.5.691. [DOI] [PubMed] [Google Scholar]
- (59).Yokoyama S, Takenaka N. Mol. Biol. Evol. 2005;22:968–975. doi: 10.1093/molbev/msi080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.