Abstract
The Rift Valley fever virus (RVFV) encodes the structural proteins nucleoprotein (N), aminoterminal glycoprotein (Gn), carboxyterminal glycoprotein (Gc), and L protein, 78-kD, and the nonstructural proteins NSm and NSs. Using the baculovirus system, we expressed the full-length coding sequence of N, NSs, NSm, Gc, and the ectodomain of the coding sequence of the Gn glycoprotein derived from the virulent strain of RVFV ZH548. Western blot analysis using anti-His antibodies and monoclonal antibodies against Gn and N confirmed expression of the recombinant proteins, and in vitro biochemical analysis showed that the two glycoproteins, Gn and Gc, were expressed in glycosylated form. Immunoreactivity profiles of the recombinant proteins in western blot and in indirect enzyme-linked immunosorbent assay against a panel of antisera obtained from vaccinated or wild type (RVFV)-challenged sheep confirmed the results obtained with anti-His antibodies and demonstrated the suitability of the baculo-expressed antigens for diagnostic assays. In addition, these recombinant proteins could be valuable for the development of diagnostic methods that differentiate infected from vaccinated animals (DIVA).
Key Words: Rift Valley fever virus, Structural proteins, Nonstructural proteins, Immunoreactivity profiles, Sheep
Introduction
Rift Valley fever (RVF), caused by Rift Valley fever virus (RVFV), represents a potential threat to global human and animal health. The virus belongs to the genus Phlebovirus of the family Bunyaviridae and causes severe epidemics among ruminants and humans in Africa and the Arabian Peninsula (Jupp and Cornel 1988, Shoemaker et al. 2002). Although the RVFV natural infection cycle has been linked to the Aedes and Culex spp. mosquitos, other mosquito species, including those found in North America, can be infected and subsequently transmit the virus (McIntosh and Jupp 1981, Gad et al. 1987, Gargan et al. 1988, Jupp and Cornel 1988, Turell et al. 2010). The fact that a variety of mosquito species can act as vectors increases the likelihood of RVFV becoming endemic in areas outside of its traditional geographic range (Turell et al. 1988). The potential risk of spread of RVFV to temperate regions is exacerbated by global climate change, the almost ubiquitous presence of potential mosquito vectors, and international trade and travel. Currently, there are multiple challenges associated with efforts to combat RVF disease. Most important amongst them are the development and availability of an effective vaccine and pen-side diagnostic assays. In Africa, a live attenuated vaccine based on the Smithburn strain is available and provides lasting protective immunity, but it is abortigenic in pregnant livestock (Smithburn 1949, Hunter, et al. 2002, Botros et al. 2006), thus limiting its use. Killed or inactivated vaccines require multiple inoculations to be efficacious, which poses logistical challenges in developing countries where Rift Valley fever is endemic. The use of live-attenuated vaccines in nonendemic countries is less desirable due to concerns of introduction of live RVFV into a nonendemic region and the potential risk of reversion to virulence. Importantly, development of sensitive diagnostics, as well as effective vaccines, will require the identification and expression of sufficiently immunoreactive and potentially immunoprotective proteins that could be used in diagnostic assays or as candidates for vaccine development.
Like all members of the Bunyaviridae family, the RVFV genome comprises three single negative-stranded RNA strands, composed of large (L), medium (M), and small (S) segments (Elliott 1996, Giorgi 1996, Schmaljohn 1996). The L segment encodes the viral RNA-dependent RNA polymerase. The M segment encodes the structural glycoproteins, Gn and Gc, which are expressed as a polyprotein precursor that is processed by cellular proteases during maturation (Collett 1986, Suzich et al. 1990, Gerrard and Nichol 2007). It has been shown that these structural glycoproteins elicit production of virus-neutralizing antibodies important for protective immunity (Niklasson et al. 1985, Besselaar and Blackburn 1994, Bird et al. 2011, Papin et al. 2011, Piper et al. 2011). The M segment also encodes a nonstructural protein, NSm, which was shown to have an antiapoptotic function and a 78-kD protein whose function is not known yet (Anderson and Peters 1988, Won et al. 2007). The S segment encodes the nucleocapsid protein, N, and a nonstructural protein, NSs, which is recognized as a major virulence factor of the virus by counteracting host innate immunity (Vialat et al. 2000, Bouloy et al. 2001, Billecocq et al. 2004, Le May et al. 2004). In this study, we describe the expression of various RVFV proteins using the baculovirus system and profile their reaction pattern with antisera derived from RVFV vaccinated or challenged sheep.
Materials and Methods
Cloning and construction of recombinant bacmid
The full-length coding sequences of the RVFV nucleoprotein (N) and the nonstructural protein (NSs) were amplified by PCR using a proofreading DNA polymerase, Pfx50 (Life Technologies/Invitrogen, Carlsbad, CA), and primers (Table 1) designed from published sequences of RVFV strain ZH548 (accession no. DQ380151). Plasmid pET30 Ek/LIC containing the entire coding region of the S segment of ZH548 was used as template for the PCR. The ectodomain and full-length coding sequences of the virus structural glycoproteins Gne and Gc, respectively, as well as the nonstructural M protein NSm were synthesized according to the published sequences of the ZH548 strain (Genewiz, San Diego, CA). Amplicons were purified using a Qiagen gel or PCR purification kit (Qiagen, Valencia, CA). The purified products were cloned into a pFastBac/CT-TOPO vector (Life Technologies/Invitrogen, Carlsbad, CA). The TOPO cloning reactions were transformed into One Shot Mach1 T1 chemically competent Escherichia coli to produce the respective donor plasmids pRF-N, pRF-NSs, pRF-Gne, pRF-Gc, and pRF-NSm. The sizes and sequences of the inserts in the donor plasmids were confirmed by PCR using gene-specific primers (Table 1) that were confirmed by restriction enzyme analysis and DNA sequencing. Donor plasmids containing the gene of interest were purified from the E. coli transformants using a Qiagen Miniprep kit (Qiagen, Valencia, CA). The constructs were transformed into MAX Efficiency DH10Bac-competent E. coli to generate a recombinant bacmid by site-specific transpositioning. Recombinant bacmids were purified using HiPure Plasmid Miniprep kit. Transpositioning of the gene of interest into a recombinant bacmid was confirmed by PCR using M13F and M13R primers (Heidecker et al. 1980).
Table 1.
Primers Used for the Amplification and Cloning of Rift Valley Fever Virus Protein-Coding Sequences
| Primer | Orientation | Sequences | Target/description | Reference |
|---|---|---|---|---|
| JAR979 | Forward | CACCATGGACAACTATCAAGACCTTGCGATCC | N cds | Designed by this study |
| JAR980 | Reverse | GGCTGCTGTCTTGTAAGCCTGAGCG | N cds | Designed by this study |
| JAR981 | Forward | CACCATGGATTACTTTCCTGTGATATCTGTTGATTTG | NSs cds | Designed by this study |
| JAR982 | Reverse | ATCAACCTCAACAAATCCATCATCATCACTCTCC | NSs cds | Designed by this study |
| JAR975 | Forward | CACCATGATTGAAGGAGCTTGGGATTC | NSm cds | Designed by this study |
| JAR976 | Reverse | AGCAAAAACAACAGGTGCCAAAGC | NSm cds | Designed by this study |
| JAR996a | Forward | CACCATGACAGTCCTTCCAGCCTTAG | Gne or Gc cds | Designed by this study |
| JAR111 | Reverse | GGCACTGAGAGCAGTGTGACACTG | Gne cds | Designed by this study |
| JAR987 | Reverse | TGAGGCCTTCTTAGTGGCAGCAAG | Gc cds | Designed by this study |
| JAR990 | Forward | CCCAGTCACGACGTTGTAAAACG | M13 sequencing primer | Heidecker et al. 1980 |
| JAR991 | Reverse | AGCGGATAACAATTTCACACAGG | M13 sequencing primer | Heidecker et al. 1980 |
A signal peptide specific primer common for structural glycoproteins, Gne and Gc.
N, nucleoprotein; cds, coding sequence; NSs, nonstructural protein S; NSm, nonstructural protein m; Gne, ectodomain of Gn glycoprotein; Gc, Gc glycoprotein.
Signal peptide prediction and modification of Gn and Gc
The RVFV M segment, which encodes the envelope glycoproteins Gn and Gc, has at least four translation initiation sites within the single mRNA transcribed from the M segment. We hypothesized that sequences starting from one of the initiation codons (ATG) to the start of Gn may serve as a signal peptide and guide the translocation of the polyprotein from the cytoplasm to the endoplasmic reticulum (ER). Prediction of signal peptides and signal peptidase cleavage sites was performed using hidden Markov model and neural network model prediction programs at SignalP 3.0. A 54-nucleotide sequence beginning from the fifth ATG of the RVFV M segment, upstream of Gn coding sequence, was identified as a strong signal peptide with a single signal peptidase cleavage site. To ensure expression of glycosylated forms of the proteins, this signal sequence was fused to the amino terminus of Gne and Gc and amplified by PCR using primers shown in Table 1. The sequences were cloned into recombinant bacmids and expressed in a baculovirus expression system (Life Technologies/Invitrogen, Carlsbad, CA).
Expression and purification of recombinant RVFV proteins
To express recombinant RVFV proteins, highly purified recombinant bacmids were transfected, using Cellfectin II reagent (Life Technologies/Invitrogen Carlsbad, CA), into Spodoptera frugiperda, Sf9, cells (Life Technologies/Invitrogen) grown in Sf-900 II SFM medium (Life Technologies/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL–100 μg/mL penicillin–streptomycin. Protein expression was carried out using P2 or higher passage recombinant baculovirus stock (>107 pfu/mL) in T75 or T175 Biolite flasks (ThermoFisher Scientific, Dubuque, IA). To visualize expression of recombinant proteins, samples were subjected to western blot analysis by resolving on 12% Bis-Tris polyacrylamide gel and detected using anti-His-horseradish peroxidase (HRP) antibody. Purification of recombinant proteins was carried out using Ni-NTA superflow resin (Novagen, Rockland, MA) according to manufacturer's instructions. Recombinant proteins were eluted with an elution buffer containing 300 mM NaCl, 50 mM Na2PO4 (pH 8.0), and 250 mM imidazole and dialyzed overnight against storage buffer, phosphate-buffered saline (PBS; pH 7.4), and 5% glycerol. The purified proteins were stained with Coomassie Blue, and protein concentrations were determined using the bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL) at an absorbance of 562 nm, using bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, Mo) as the protein standard. Aliquots were stored at −80°C until used.
Immunofluorescence antibody assay
To confirm expression of recombinant RVFV proteins, we carried out an immunofluorescence antibody assay. Briefly, Sf9 cells were infected with recombinant baculovirus carrying RVFV N- or Gne-coding sequences. About 36 h later, cells were harvested and adhered to glass slides using a cytocentrifuge (Cytopro, Wescor, Logan, UT) according to manufacturer's instruction and fixed in acetone for 5 min at −20°C. Slides were blocked in PBS containing 1% BSA at 37°C for 45 min and then incubated for 30 min at 37°C with mouse monoclonal antibodies ID8 and 4D4 (provided by Dr. Connie W. Schmaljohn, the United States Army Medical Research Institute for Infectious Diseases, USAMRIID), against N and Gn proteins, respectively. Slides were rinsed for 10 min in 1× fluorescence antibody (FA) rinse buffer (2.85 g Na2CO3, 8.4 g NaHCO3, 2.125 g NaCl, distilled water to 1000 mL; pH 9 to 9.5) and the probed with anti-mouse fluorescein isothiocyantate (FITC)-conjugated secondary antibody for 30 min at 37°C (Sigma-Aldrich, St. Louis, MO). Slides were rinsed for 10 min in 1× FA rinse buffer and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, Molecular Probes®) containing mounting medium. Slides were examined under a fluorescent microscope (Nikon, Eclipse 90i®) at 100× magnification.
Tunicamycin assay
A baculovirus expression system was used to produce glycosylated recombinant proteins, a posttranslational modification shown to enhance antigenicity/immunogenicity (Gavrilov et al. 2011). To confirm that the modified Gne and Gc proteins were glycosylated, a tunicamycin glycosylation inhibition assay was performed. For this, six-well plates were seeded with 2×106 Sf9 cells per well and infected with recombinant baculoviruses expressing RVFV Gne or Gc at a multiplicity of infection (MOI) of 1. Immediately following infection, tunicamycin was added to each well at varying concentrations of 0.5 μg/mL, 1 μg/mL, 3 μg/mL, 6 μg/m, 8 μg/mL, and 10 μg/mL. Cells were harvested 5 days postinfection (pi) and resuspended in PBS (pH 7.4) containing 1× Complete protease inhibitor (Roche Diagnostics®, Indianapolis, IN). The samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in NuPAGE 12% Bis-Tris gels (Life Technologies/Invitrogen) and transferred on to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Amersham Hybond-P). The membranes were probed as described above using anti-His (carboxy-terminal)-HRP monoclonal antibody (Life Technologies/Invitrogen, Carlsbad, CA). Reactivity was detected using the AEC (3-amino-9-ethyl-carbazole) peroxidase substrate system (Abcam, Cambridge, MA).
Rift Valley fever virus anti-sera
Sheep (Rambouillet) were vaccinated subcutaneously with 106 plaque-forming units (pfu) of RVFV vaccine strain MP12. These studies were performed at Kansas State University (Biosecurity Research Institute [BRI] BSL-3Ag facility) or the Arthropod-Borne Animal Diseases Research Unit (ABADRU) large animal isolation building (LAIB) in Laramie, WY (day 28 sheep sera, P1–P6). Blood samples were collected from individual animals at specific time points (days) postvaccination (pv) for sera. Prevaccination blood samples were collected from each individual sheep for use as preimmune control sera. Day-28 pi antisera were obtained from previous sheep challenged at the CFIA BSL-3Ag facility in Winnipeg, Canada, with the wild-type RVFV ZH501 strain (sheep P7–P10). All sera were stored at −80°C, and sera from the CFIA facility were heat-inactivated with 0.5% Tween (56°C for 30 min) prior to removal from the BSL-3Ag laboratory. Institutional Animal Care and Use Committee (IACUC) protocols were approved by Kansas State University and University of Wyoming (ABADRU-LAIB).
Western blot analysis
Briefly, approximately 5 μg of each of recombinant RVFV protein was subjected to electrophoresis in 12% Bis-Tris polyacrylamide gel in 1×3-(N-morpholino)propanesulfonic acid (MOPS) running buffer (Life Technologies/Invitrogen). The proteins were transferred by electroblotting onto PVDF membranes according to standard protocols. The membrane was blocked in 0.1% Tween-20 in PBS (pH 7.4) containing 3% BSA at room temperature for 1 h. The blot was washed three times for 5 min each in 0.1% Tween-20 in PBS. All subsequent washing steps were carried out as described above. For analysis of recombinant protein expression, the membrane was incubated with anti-His(carboxyterminal)-HRP (Life Technologies/Invitrogen) diluted 1:5000 in blocking solution. Expression of recombinant proteins N and Gne was further confirmed using a primary antibody, mouse anti-N (R3-ID8) and mouse anti-Gn monoclonal antibody (4D4), respectively, at a dilution of 1:2000. For analysis of reactivity against sheep sera, the membrane was incubated with a 1:100 dilution of each test serum for 1 h at room temperature. After washing, the membrane was incubated for 1 h at room temperature with Protein G–HRP (Abcam, Cambridge, MA) diluted 1:25,000. After the final washing steps, specific reactivity was detected using AEC peroxidase substrate (Sigma-Aldrich, St. Louis, MO).
Indirect enzyme-linked immunosorbent assay
A 96-well plate (Nunc, Maxisorp®) was used for the indirect enzyme-linked immunosorbent assay (ELISA). Each well was coated overnight at 4°C with 100 ng of recombinant protein in 100 μL of Dulbecco's coating buffer (pH 7.4) (Life Technologies/Invitrogen). Plates were blocked for 15 min at 37°C with PBS (pH 7.4) containing 0.1% Tween and 1% skim milk. After washing three times with 0.1% Tween-20 in PBS, a volume of 200 μL of test serum, diluted 1:200 in the blocking solution, was added and incubated at 37°C for 1 h. All subsequent washing steps were carried out three times as indicated above. Each serum sample was tested in duplicate. Each test included a positive control, obtained from sheep challenged with a virulent strain of RVFV (ZH501), and negative controls obtained from the respective sheep prior to vaccination.
After washing, plates were incubated with Protein G–HRP (Abcam, Cambridge, MA), diluted 1: 50,000 in blocking solution, at 37°C for 1 h. Protein G has high binding affinity to immunoglobulin (IgG) from sheep, goats, horses, and rabbits, with little or no binding affinity to IgM. After washing, 100 μL of substrate buffer containing 0.1 mg/mL 3,3′,5,5′-tetramethylbenzidine (TMB) (Thermo Scientific, Rockford, IL) and hydrogen peroxide (H2O2) was added and the plates were incubated in the dark for 25 min. The reaction was stopped with 2 M H2SO4, and optical densities (OD) were measured at 450 nm using a microplate reader (Fluostar Omega, BMG Labtech, Cary, NC). For each ELISA, the cutoff OD value was determined by addition of 2 standard deviations (SD) to the mean OD value of serum obtained from prevaccinated/noninfected sheep.
Statistical analysis
Data were analyzed using a t-test of independent samples with equal or unequal variances. To reduce the effect of variable individual host immune response on reactivity, we calculated geometric means of OD values and analyzed the differences for statistical significance.
Results
Expression of RVFV recombinant proteins
Using recombinant baculovirus, RVFV structural (Gc, Gn, and N) and nonstructural (NSs and NSm) proteins, each containing a hexahistidine tag at their carboxyl terminus, were expressed in a eukaryotic expression system using Sf9 cells. To ensure translocation in the ER and glycosylation of the structural glycoproteins Gne and Gc, a signal peptide was fused upstream of the amino terminus of both proteins. Infection of Sf9 cells with recombinant baculovirus carrying the full-length Gn coding sequence resulted in no or a low amount of Gn protein. To minimize interactions of the full-length Gn with cellular membranes, only the ectodomain of the protein Gne, without the transmembrane and cytosolic regions, was expressed. The proteins were purified and detected by western blot using anti-His-HRP monoclonal antibody (Fig. 1A). Recombinant proteins of the expected molecular weights were expressed (Gc, 60 kD; Gne, 54 kD; N, 30 kD; NSs, 33 kD; NSm, 17 kD). Expression of N and Gn proteins was further confirmed by immunoreaction with anti-N or anti-Gn mouse monoclonal antibodies (Fig. 1B). Coomassie-stained gels of the purified proteins are shown in Figure 1C. Immunofluorescent antibody assay using the mouse monoclonal antibodies against the Gn and N proteins further confirmed the expression of the proteins in Sf9 cells (Fig. 2). A specific green immunofluorescent signal was observed around the periphery of the nucleus of Gne-expressing cells, whereas N appeared to show a more diffuse staining (Fig.2).
FIG. 1.
Baculovirus-expressed Rift Valley fever virus (RVFV) proteins. Proteins were purified and detected using anti-His(C-term)HRP monoclonal antibody. Gne, 54 kD; Gc, 60 kD; NSs, 34 kD; N, 31 kD; NSm, 14 kD; M, marker (A). Monoclonal antibodies against N and Gn, IDE8 and 4D4, respectively, were used to confirm expression of the respective proteins (B). (C) Coomassie Blue staining of the purified proteins. N, nucleoprotein; NSs, nonstructural protein S segment; NSm, nonstructural protein m segment; Gne, ectodomain of amino-terminal glycoprotein; Gc, carboxy-terminal glycoprotein; M, molecular weight marker.
FIG. 2.
Immunofluorescence antibody assay confirms expression of the recombinant proteins Gne and N in Sf9 cells. Monoclonal antibodies 4D4 and ID8 were used to detect expression of Gne and N, respectively. A specific green fluorescent signal around the nucleus of Gne-expressing cells is seen indicating that the recombinant protein is within the cell. Anti-Gn control, noninfected Sf9 cells stained with Gn monoclonal antibody (4D4); anti-N control, noninfected Sf9 cells stained with N monoclonal antibody (ID8) show negative staining. The cell nucleus is stained in blue color.
Baculovirus-expressed Gn and Gc proteins are N-glycosylated
It is known that the RVFV Gn and Gc glycoproteins carry one and four putative N-glycosylation sites, respectively (Gerrard and Nichol 2007). To characterize glycosylation patterns of Gne and Gc, biochemical inhibition of N-glycosylation by treatment of cells with tunicamycin, a potent inhibitor of bacterial and eukaryote N-acetylglucosamine transferases, was carried out. Treatment of Sf9 cells infected with either recombinant Gc or Gne baculoviruses with varying concentrations of tunicamycin (0.5–10 μg) resulted in inhibition of glycosylation of the proteins demonstrated by a lower molecular weight of the reactive proteins when compared to nontreated controls. The shift was more obvious for Gc (Fig. 3A), known to have four putative glycosylation sites when compared to Gne (showing marginal shift), known to have only one glycosylation site (Fig. 3B). As a control, baculovirus-expressed sheep prion protein (PrP) expressed in Sf9 cells was used. Treatment with tunicamycin resulted in significant inhibition of N-glycosylation (Fig. 3C).
FIG. 3.
In vitro glycosylation assay of Rift Valley fever glycoproteins, Gc and Gne. (A) Treatment of Gc-recombinant baculovirus-infected Sf9 cells with varying concentrations of tunicamycin (0.5 μg/mL to 10 μg/mL) resulted in inhibition of glycosylation shown by a shift in electrophoretic migration. (B) Similar treatment of Gne (8 μg/mL and 10 μg/mL) resulted in marginal molecular weight shift (compare with nontreated, nt), since the protein has one putative N-glycosylation site; Gc has four putative N-glycosylation sites. Treatment of baculovirus expressed sheep prion protein (PrP) with varying concentrations of tunicamycin (1 μg/mL to 10 μg/mL) resulted in inhibition of glycosylation of the protein(C). nt, nontreated controls; m, molecular weight marker.
Western blot analysis
We examined antibody reactivity against the recombinant proteins N, NSs, NSm, Gne, and Gc using sera collected at different time points pv (days 3, 10, and 28) from MP12 vaccinated sheep. Recombinant N was reactive with day-3 pv sera, and both N and NSs proteins showed antibody reactivity with day-10 pv and day-28 pv sera, with N showing consistently stronger reactivity than NSs (Fig. 4). Reactivity of the structural glycoproteins Gne and Gc showed that Gne was reactive with 1 of 2 serum samples on day 3 pv. Thereafter, it remained consistently reactive with all sera obtained on day 10 and 28 pv (Table 2). The Gc protein showed no reactivity at day 3 pv and weak reactivity with day 10 pv sera, where only 1 of the 4 serum samples tested was positive; it was 90% reactive with day 28 pv sera. To examine reactivity of RVFV proteins further, we performed immunoblot analysis using day-28 pi sera obtained from sheep infected with the wild-type strain ZH501 of RVFV. Recombinant N and Gne proteins showed consistent and strong reactivity against all sera (4/4); NSs and Gc were also reactive against all sera (4/4) but with relative lower signal intensities. We did not detect specific reactivity with the NSm protein, either with the MP12-vaccinated or ZH501-infected sheep sera.
FIG. 4.
Western blot showing a sample of immunoreactivity of purified baculovirus-expressed proteins with RVFV antisera from sheep. Reactivity shows that the proteins were expressed in the correct conformation. Gne, ectodomain of amino-terminal glycoprotein; N, nucleoprotein; NSs, nonstructural protein S segment; Gc, carboxy-terminal glycoprotein; CL, noninfected cell lysate; m, molecular weight marker; pv, postvaccination; pi, postinfection.
Table 2.
Reactivity of Baculovirus-Expressed Rift Valley Fever Virus Proteins with Sheep Antisera Determined by Western Blot
| |
MP12 vaccinated sheep sera: days pv |
|
||
|---|---|---|---|---|
| Recombinant proteins | Day 3 | Day 10 (nsp/total sample tested) | Day 28 | Wild type (ZH501) challenged sheep sera: day 28 pi(nsp/total sample tested) |
| Gne | 1/2 | 4/4 | 10/10 | 4/4 |
| Gc | 0/2 | 1/4 | 9/10 | 4/4 |
| N | 2/2 | 4/4 | 10/10 | 4/4 |
| NSs | 0/2 | 4/4 | 9/10 | 4/4 |
| NSm | 0/2 | 0/4 | 0/10 | 0/4 |
pv, postvaccination; pi, postinfection; nsp, number of samples tested positive; Gne, ectodomain of Gn glycoprotein; Gc, Gc glycoprotein; N, nucleoprotein; NSs, nonstructural protein S; NSm, nonstructural protein m.
Indirect ELISA
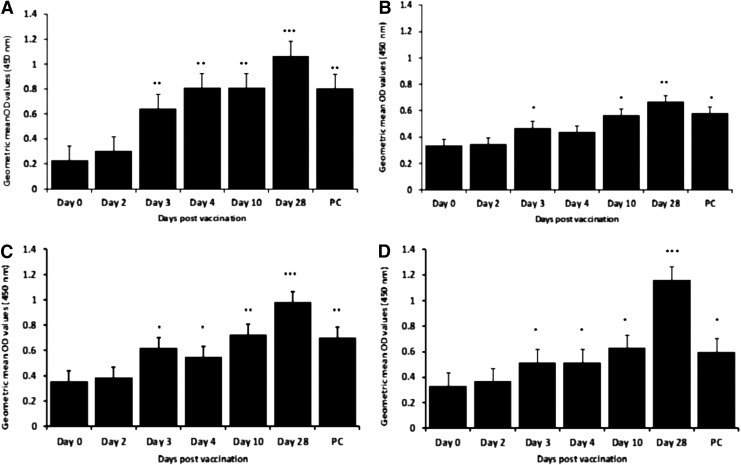
An indirect ELISA was developed to assess antibody reactivity of the recombinant proteins against sera obtained from sheep vaccinated with RVFV MP12 vaccine strain or challenged with wild-type ZH501 RVFV strain. For MP12-vaccinated sheep sera, the recombinant proteins N, NSs, Gc, and Gne showed time-dependent increase in reactivity, shown by an increase in OD values (Fig. 5A–D). A consistent strong reactivity for all proteins was observed with day-28 sera. There was particularly strong antibody reactivity with N and to a lesser extent with Gne on day 3 pv (Fig. 5A, C). The level of reactivity with NSs was also strong but had comparatively lower OD values (Fig. 5B). Reactivity with Gc was high on day 10 pv and was strongest on day 28 (Fig. 5D). For the wild type exposure-derived sheep sera, the recombinant proteins, N, NSs, Gne, and Gc were reactive with the day 28 pi (data not shown). In contrast, NSm showed low reactivity manifested by relatively low OD values with sera from both MP12 and wild-type infected sheep (Fig. 5E). Sera from three sheep (infected with the wild-type ZH501, P7 and P10, and MP-12-immunized sheep, P5) showed significant reactivity (p<0.05) in comparison with the negative control (day 0 serum). The positive sera were obtained from sheep on day 28 pi with the wild-type RVFV ZH501.
FIG. 5.
Reactivity of recombinant RVFV proteins, nucleoprotein, N (A), nonstructural protein, NSs (B), glycoprotein, Gne (C), glycoprotein, Gc (D), and nonstructural protein NSm (E) with antisera from MP12 vaccinated sheep. Day 0, prevaccination sera; PC, positive control sera derived from sheep challenged with RVFV wild-type, ZH501; P1–P6, day-28 sera from sheep vaccinated with MP12 RVFV strain (Laramie, WY); P7–P10, day-28 sera from sheep infected with the wild-type virus (ZH501). Asterisks (*) denote level of statistical significance and show that differences in optical density (OD) values of sera tested for each of the time-points was significantly different (p<0.05) from day 0 (prevaccination) sera. Cutoff OD value for each enzyme-linked immunosorbent assay (ELISA) was determined by addition of 2 standard deviations to the mean OD value of serum obtained from prevaccinated/noninfected sheep. N, 0.320; NSs, 0.358; Gn, 0.395; Gc, 0.387; NSm, 0.208.
Discussion
There is increasing demand for sensitive and safe diagnostic tests and efficacious vaccines for zoonotic pathogens, including RVFV, to protect human and animal health. The recent spread of RVFV beyond its traditional endemic boundaries in Africa to the Arabian Peninsula (Jupp and Cornel 1988, Ikegami and Makino 2009, Abdo-Salem et al. 2011) has resulted in increased interest for RVFV vaccines, rapid diagnostics, and associated immunoreagents. At present, diagnosis of RVFV infection is achieved using various techniques including virus isolation (Shope and Sather 1979, Anderson et al. 1989), antigen detection (Niklasson et al. 1983, Meegan et al. 1987, Drolet et al. 2012), nucleic acid amplification techniques (Ibrahim et al. 1997, Garcia et al. 2001, Drosten et al. 2002, Drolet et al. 2012), and detection of RVFV-specific antibodies (Niklasson et al. 1984, Swanepoel et al. 1986, Meegan et al. 1987, Paweska et al. 2003).
The use of virus isolation is not user-friendly, takes an extended period of time, and requires suitable biosafety protocols or measures; antigen or nucleic acid detection in the serum or blood of animals or humans only works in cases of host viremia, which in the case of RVFV infection is a narrow viremic window, lasting on average about 3 days in serum (Morrill et al. 1990, Nfon et al. 2012, Smith et al. 2012) or longer in whole blood (Grolla et al. 2012). The classical methods for detection of antibodies to RVFV include various forms of virus neutralization and hemagglutination-inhibition tests. Disadvantages of these techniques include health risk to laboratory personnel (Smithburn 1949), as well as restrictions to high biocontainment laboratories for their use outside RVF endemic areas (Barnard 1979).
On the other hand, application of ELISA to detect IgG antibody to RVFV relied largely on the use of inactivated whole virus lysate, which is also associated with potential health risks (Swanepoel et al. 1986a, Swanepoel et al. 1986b, Meegan et al. 1987, Paweska et al. 1995). The nucleocapsid (N) protein is the most abundant and highly immunogenic component of the RVF virion and has been used for development of diagnostic assays for detection of RVFV-specific antibodies in human and animal sera (Fafetine et al. 2007, Jansen van Vuren et al. 2007, Paweska et al. 2007, Paweska et al. 2008). Although the N protein is shown to be highly conserved among members of the Bunyaviridae family (Schwarz, et al. 1996, Magurano and Nicoletti 1999), a previous indirect ELISA based on the recombinant protein did not show cross-reactivity with other African phleboviruses that could hamper the reliability of using this protein in assays for serodiagnosis of RVFV infection (Paweska et al. 2007). However, the N protein did show serological cross-reactivity with an unidentified agent among some sera from US and Canadian sheep (Wilson et al., personal communication).
Recombinant proteins provide a safe platform for development of improved diagnostics and, in some cases, for subunit vaccines. In this study, we have expressed a panel of RVFV proteins (N, NSs, NSm, Gne, and Gc) using the baculovirus expression system and evaluated their potential as diagnostic targets. One of the advantages of using the baculovirus expression system is the ability to scale up expression; it also allows posttranslational protein modification, which in the case of glycoproteins, enhances antigenicity and immunogenicity. Sequence modifications made to Gn and Gc resulted in enhanced expression, and analysis of Gne expression by immunofluorescence showed that the protein was present within the cell, manifested by detection of green fluorescence around the cell nucleus, in the periphery of the plasma membrane of infected cells (Fig. 2).
Indeed, other studies reported specific targeting of recombinant RVFV glycoproteins to the plasma membrane of insect cells using the baculovirus expression system (Takehara et al. 1990, Liu et al. 2008) or localization of the proteins on the cell surface when expressed using alphavirus replicons (Filone et al. 2006). To ensure that Gne and Gc, are translocated into the ER and glycosylated, we added a signal peptide sequence carrying a signal peptidase cleavage site upstream of the 5′ end of the coding sequence. This ensured correct protein trafficking through the lumen of the ER, where glycosylation enzymes and signal peptidase are located (Gerrard and Nichol 2002, Gerrard and Nichol 2007). Biochemical characterization of glycosylation of Gne and Gc showed that the glycoproteins were N-glycosylated. Treatment with tunicamycin, a potent inhibitor of bacterial and eukaryote N-acetylglucosamine transferases, resulted in inhibition of glycosylation as detected by electrophoretic migration of the proteins (Fig. 3A, B), strongly indicating that Gne and Gc were subjected to posttranslational N-glycosylation.
Analysis of antibody reactivity of the recombinant proteins with RVFV antisera by western blot and indirect ELISA showed the proteins were immunoreactive, a strong indication of correct structural conformation. In western blot analyses (Fig. 4) and ELISA (Fig. 5) using MP12 vaccinated and wild-type–challenged sera, the RVFV nucleocapsid protein was the most reactive. As the most abundant and highly immunogenic structural component of RVFV virion, the N protein has been considered the best choice for the development of immunoreagents for antigen detection assays (Pepin et al. 2010).
The early immune response and strong antibody reactivity against N protein with MP12-vaccinated sera might suggest a possible role of N protein in protective immunity to RVFV infection. Indeed, vaccination with N antigen has been shown to protect mice from wild-type RVFV challenge (Wallace et al. 2006, Liu et al. 2008, Lorenzo et al. 2008, Lagerqvist et al. 2009, Jansen van Vuren et al. 2010, Lorenzo et al. 2010). Remarkably, the glycoprotein Gne also showed an early-response reactivity (day 3 pv) in western blot and ELISA with MP12-vaccinated sera and was detected by day-28 pi sera from all ZH501-infected sheep. This would qualify Gne, in addition to the N-protein, as a good choice as a diagnostic antigen. Moreover, the early and specific antibody response to Gne found with MP12-vaccinated sheep makes this protein a good candidate for vaccine development. Importantly, the RVFV glycoproteins Gne and Gc have been shown to contain epitopes that induce production of neutralizing antibodies, a correlate of protective immunity against RVFV infection (Niklasson et al. 1985, Morrill et al. 1987, Hubbard et al. 1991, Morrill et al. 1997).
Therefore, future studies should aim at establishing a possible correlation between Gne reactivity and the presence of neutralizing antibodies. Also, baculovirus-expressed recombinant Gne alone or in combination with recombinant N could improve the sensitivity and specificity of herd screening for RVFV infections in areas where cross-reactivity with other bunyaviruses/phleboviruses may be an issue. Recombinant Gc also showed strong reactivity with sheep sera, but seemed to be less sensitive when compared with Gne (Fig. 5). However, the induction of a strong antibody response to Gc appears later in MP12-vaccinated animals compared to N and Gn. For improved RVFV detection, molecular tools for detection of RVFV nucleic acids should be used in parallel with antibody detection to identify early-infected ruminants.
The recombinant NSs protein showed early-onset reactivity with MP12 sera, but had an overall lower reactivity than the N protein. This may be attributed to the typically low antibody titers raised against the NSs protein after RVFV infections (Pepin et al. 2010). The RVFV NSs protein is responsible for general suppression of the host transcriptional machinery, including type I interferons (Le May et al. 2004, Le May et al. 2008). In contrast to NSs, the other nonstructural protein, NSm, showed poor reactivity with sera from MP12 vaccinated and wild-type–challenged animals in western blot. Further assessment of its reactivity in an ELISA test showed little or no reactivity with MP12-vaccinated sera, demonstrated by extremely low OD values (Fig. 5E). Sera from 3 sheep (2 wild type–infected, P7 and P10, and 1 MP12-vaccinated, P5), at day 28 pi showed relatively high reactivity when compared to day-0 sera (Fig. 5E). Interestingly, in a virulent RVFV challenge experiment using ELISA, Bird et al. (2011) reported seropositivity for NSs antibodies in all 3 control sheep, whereas only 2 of the 3 animals were seropositive for NSm; this was the first time immunoreactivity against NSm had been detected in a natural host species (Bird et al. 2011). This might indicate that NSm is less immunogenic when compared to NSs or is expressed at lower levels resulting in a lower antibody response in mammalian hosts.
The development of a diagnostic tool that differentiates infected from vaccinated animals (DIVA) is very much needed in regions nonendemic for RVFV. The apparent poor immunogenicity of RVFV NSm in ruminant hosts reported in this paper and also shown previously (Bird et al. 2011) makes the use of NSm as a target for developing a DIVA diagnostic test questionable. In this study, NSs-specific antibodies were consistently detected in RVFV-infected/vaccinated sheep, which suggests that the NSs protein may serve as a candidate for the development of a DIVA diagnostic assay. Previous investigators have demonstrated variability of detection of antibodies to NSs (Fernandez et al. 2012). Therefore, further optimization and standardization of the use of this protein in diagnostic assays may be required for this antigen to be a reliable DIVA marker.
Research efforts aimed at the development and validation of a new generation of safe and accurate diagnostic immunoreagents and assays based on RVFV recombinant antigens are critical (Pepin et al. 2010). This is the first study to carry out expression of several RVFV proteins and assess their reactivity against a panel of immune sera from natural hosts. Analysis of antibody reactivity revealed that the proteins were expressed in the correct conformation in a baculovirus expression system. Addition of a fusion signal peptide sequence to the structural glycoproteins Gne and Gc ensured processing of the proteins through the cellular glycosylation pathway. We have shown that baculovirus-expressed N, Gne, Gc, and NSs could be used as potential serodiagnostic targets for monitoring host humoral immune response to infection and/or vaccination. The immunoreactivity toward NSm was rather weak making this RVFV antigen a less desirable diagnostic target. The general immunoreactivity profile suggests that N and Gne would represent desirable targets for development of highly sensitive serodiagnostic assays. In addition, future vaccine formulations will require DIVA compatibility with companion diagnostic DIVA tests. We have shown that baculovirus-expressed N and NSs assays, depending on the vaccine composition, can be used for such companion assays.
Acknowledgments
The authors thank Dr. Connie W. Schmaljohn, the United States Army Medical Research Institute for Infectious Diseases (USAMRIID), for providing the RVFV M segment plasmid and the monoclonal antibodies to Gn and N proteins. We thank Dr. Friedman Weber, University of Marburg, for providing the RVFV S segment plasmid. We thank Dr. Bhupinder Bawa, Kansas State University, for his overall support and Mark Minihan of Kansas State University Biosecurity Research Institute for the animal care. This work was funded by grants of the Department of Homeland Security Center of Excellence for Emerging and Zoonotic Animal Diseases (CEEZAD), Grant No. 2010-ST061-AG0001, the Kansas Bioscience Authority and the USDA Agricultural Research Service project 5430-050-005-00D.
Author Disclosure Statement
No competing financial interests exist.
References
- Abdo-Salem S. Waret-Szkuta A. Roger F. Olive MM, et al. Risk assessment of the introduction of Rift Valley fever from the Horn of Africa to Yemen via legal trade of small ruminants. Trop Anim Health Prod. 2011;43:471–480. doi: 10.1007/s11250-010-9719-7. [DOI] [PubMed] [Google Scholar]
- Anderson GWJ. Peters CJ. Viral determinants of virulence of Rift Valley fever (RVF) in rats. Microb Pathog. 1988;5:241–250. doi: 10.1016/0882-4010(88)90096-4. [DOI] [PubMed] [Google Scholar]
- Anderson GWJ. Saluzzo JF. Ksiazek TG. Smith JF, et al. Comparison of in vitro and in vivo systems for propagation of Rift Valley fever virus from clinical specimens. Res.Virol. 1989;140:129–138. doi: 10.1016/s0923-2516(89)80090-1. [DOI] [PubMed] [Google Scholar]
- Barnard BJH. Rift Valley fever vaccine antibody and immune response in cattle to a live and an inactivated vaccine. J S Afr Vet Assoc. 1979;50:155–157. [PubMed] [Google Scholar]
- Besselaar TG. Blackburn NK. The effect of neutralizing monoclonal antibodies on early events in Rift Valley fever virus infectivity. Res Virol. 1994;145:13–19. doi: 10.1016/s0923-2516(07)80002-1. [DOI] [PubMed] [Google Scholar]
- Billecocq A. Spiegel M. Vialat P. Kohl A, et al. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol. 2004;78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH. Maartens LH. Campbell S. Erasmus BJ, et al. Rift Valley Fever virus vaccine lacking the NSs and NSm genes is safe, nonteratogenic, and confers protection from viremia, pyrexia, and abortion following challenge in adult and pregnant sheep. J Virol. 2011;85:12901–12909. doi: 10.1128/JVI.06046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botros B. Omar A. Elian K. Mohamed G, et al. Adverse effect of non-indigenous cattle of European breeds to live-attenuated Smithburn Rift Valley fever vaccine. J Med Virol. 2006;78:787–791. doi: 10.1002/jmv.20624. [DOI] [PubMed] [Google Scholar]
- Bouloy M. Janzen C. Vialat P. Khun H, et al. Genetic evidence for an Iiterferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J Virol. 2001;75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett MS. Messenger RNA of the M segment RNA of Rift Valley fever virus. Virology. 1986;151:151–156. doi: 10.1016/0042-6822(86)90114-5. [DOI] [PubMed] [Google Scholar]
- Drolet BS. Weingartl HM. Jiang J. Neufeld J, et al. Development and evaluation of one-step rRT-PCR and immunohistochemical methods for detection of Rift Valley fever virus in biosafety level 2 diagnostic laboratories. J Virol Methods. 2012;179:373–382. doi: 10.1016/j.jviromet.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Drosten C. Gottig S. Schilling S. Asper M, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RMe. The Bunyaviridae. Plenum Press; New York: 1996. [Google Scholar]
- Fafetine J. Tijhaar E. Paweska J. Neves L, et al. Cloning and expression of Rift Valley fever virus nucleocapsid (N) protein and evaluation of a N-protein based indirect ELISA for the detection of specific IgG and IgM antibodies in domestic ruminants. Vet Microbiol. 2007;121:29–38. doi: 10.1016/j.vetmic.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Fernandez JC. Billecocq A. Durand JP. Cetre-Sossah C, et al. The nonstructural protein NSs induces a variable antibody response in domestic ruminants naturally infected with Rift Valley fever virus. Clin Vaccine Immunol. 2012;19:5–10. doi: 10.1128/CVI.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filone CM. Heise M. Doms RW. Bertolotti-Ciarlet A. Development and characterization of a Rift Valley fever virus cell-cell fusion assay using alphavirus replicon vectors. Virology. 2006;356:155–164. doi: 10.1016/j.virol.2006.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad AM. Hassan MM. el Said S. Moussa MI, et al. Rift Valley fever virus transmission by different Egyptian mosquito species. Trans R Soc Trop Med Hyg. 1987;81:694–698. doi: 10.1016/0035-9203(87)90460-3. [DOI] [PubMed] [Google Scholar]
- Garcia S. Crance JM. Billecocq A. Peinnequin A, et al. Quantitative real-time PCR detection of Rift Valley fever virus and its application to evaluation of antiviral compounds. J Clin Microbiol. 2001;39:4456–4461. doi: 10.1128/JCM.39.12.4456-4461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargan TP., 2nd Clark GG. Dohm DJ. Turell MJ, et al. Vector potential of selected North American mosquito species for Rift Valley fever virus. Am J Trop Med Hyg. 1988;38:440–446. doi: 10.4269/ajtmh.1988.38.440. [DOI] [PubMed] [Google Scholar]
- Gavrilov BK. Rogers K. Fernandez-Sainz IJ. Holinka LG, et al. Effects of glycosylation on antigenicity and immunogenicity of classical swine fever virus envelope proteins. Virology. 2011;420:135–145. doi: 10.1016/j.virol.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Gerrard SR. Nichol ST. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J Virol. 2002;76:12200–12210. doi: 10.1128/JVI.76.23.12200-12210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard SR. Nichol ST. Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology. 2007;357:124–133. doi: 10.1016/j.virol.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C. In: Molecular biology of phleboviruses. The Bunyaviridae. Eliott RM, editor. Plenum Press; New York: 1996. pp. 105–128. [Google Scholar]
- Grolla A. Mehedi M. Lindsay R. Bosio C, et al. Enhanced detection of Rift Valley fever virus using molecular assays on whole blood samples. J Clin Virol. 2012;54:313–317. doi: 10.1016/j.jcv.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G. Messing J. Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980;10:69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Hubbard KA. Baskerville A. Stephenson JR. Ability of a mutagenized virus variant to protect young lambs from Rift Valley fever. Am J Vet Res. 1991;52:50–55. [PubMed] [Google Scholar]
- Hunter P. Erasmus B. Vorster J. Teratogenicity of a mutagenised Rift Valley fever virus (MVP 12) in sheep. Onderstepoort J Vet Res. 2002;69:95–98. [PubMed] [Google Scholar]
- Ibrahim MS. Turell MJ. Knauert FK. Lofts RS. Detection of Rift Valley fever virus in mosquitoes by RT-PCR. Mol Cell Probes. 1997;11:49–53. doi: 10.1006/mcpr.1996.0075. [DOI] [PubMed] [Google Scholar]
- Ikegami T. Makino S. Rift Valley fever vaccines. Vaccine. 2009;27:69–72. doi: 10.1016/j.vaccine.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen van Vuren P. Potgieter A. Paweska J. van Dijk A. Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J Virol Methods. 2007;140:106–114. doi: 10.1016/j.jviromet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Jansen van Vuren P. Tiemessen CT. Paweska JT. Evaluation of a recombinant Rift Valley fever virus subunit nucleocapsid protein as an immunogen in mice and sheep. The Open Vaccine Journal. 2010;3:114–126. [Google Scholar]
- Jupp PG. Cornel AJ. Vector competence tests with Rift Valley fever virus and five South African species of mosquito. J Am Mosq Control Assoc. 1988;4:4–8. [PubMed] [Google Scholar]
- Lagerqvist N. Naslund J. Lundkvist A. Bouloy M, et al. Characterisation of immune responses and protective efficacy in mice after immunisation with Rift Valley Fever virus cDNA constructs. Virol J. 2009;6:6. doi: 10.1186/1743-422X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N. Dubaele S. De Santis LP. Billecocq A, et al. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116:541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- Le May N. Mansuroglu Z. Leger P. Josse T, et al. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008;4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Celma C. Roy P. Rift Valley fever virus structural proteins: expression, characterization and assembly of recombinant proteins. Virol J. 2008;5:82. doi: 10.1186/1743-422X-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo G. Martin-Folgar R. Rodriguez F. Brun A. Priming with DNA plasmids encoding the nucleocapsid protein and glycoprotein precursors from Rift Valley fever virus accelerates the immune responses induced by an attenuated vaccine in sheep. Vaccine. 2008;26:5255–5262. doi: 10.1016/j.vaccine.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Lorenzo G. Martin-Folgar R. Hevia E. Boshra H, et al. Protection against lethal Rift Valley fever virus (RVFV) infection in transgenic IFNAR(−/−) mice induced by different DNA vaccination regimens. Vaccine. 2010;28:2937–2944. doi: 10.1016/j.vaccine.2010.02.018. [DOI] [PubMed] [Google Scholar]
- Magurano F. Nicoletti L. Humoral response in Toscana virus acute neurologic disease investigated by viral-protein-specific immunoassays. Clin Diagn Lab Immunol. 1999;6:55–60. doi: 10.1128/cdli.6.1.55-60.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BM. Jupp PG. Epidemiological aspects of Rift Valley fever in South Africa with reference to vectors. Contrib Epidemiol Biostat. 1981;3:92–99. [Google Scholar]
- Meegan JM. Yedloutschnig RJ. Peleg BA. Shy J, et al. Enzyme-linked immunosorbent assay for detection of antibodies to Rift Valley fever virus in ovine and bovine sera. Am J Vet Res. 1987;48:1138–1141. [PubMed] [Google Scholar]
- Morrill JC. Jennings GB. Caplen H. Turell MJ, et al. Pathogenicity and immunogenicity of a mutagen-attenuated Rift Valley fever virus immunogen in pregnant ewes. Am J Vet Res. 1987;48:1042–1047. [PubMed] [Google Scholar]
- Morrill JC. Jennings GB. Johnson AJ. Cosgriff TM, et al. Pathogenesis of Rift Valley fever in rhesus monkeys: role of interferon response. Arch Virol. 1990;110:195–212. doi: 10.1007/BF01311288. [DOI] [PubMed] [Google Scholar]
- Morrill J. Mebus C. Peters C. Safety and efficacy of a mutagen-attenuated Rift Valley fever virus vaccine in cattle. Am J Vet Res. 1997;58:1104–1109. [PubMed] [Google Scholar]
- Nfon CK. Marszal P. Zhang S. Weingartl HM. Innate immune response to Rift Valley fever virus in goats. PLoS Negl Trop Dis. 2012;6:e1623. doi: 10.1371/journal.pntd.0001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklasson B. Grandien M. Peters CJ. Gargan TP., 2nd Detection of Rift Valley fever virus antigen by enzyme-linked immunosorbent assay. J Clin Microbiol. 1983;17:1026–1031. doi: 10.1128/jcm.17.6.1026-1031.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklasson B. Peters CJ. Grandien M. Wood O. Detection of human immunoglobulins G and M antibodies to Rift Valley fever virus by enzyme-linked immunosorbent assay. J Clin Microbiol. 1984;19:225–229. doi: 10.1128/jcm.19.2.225-229.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklasson B. Peters C. Bengtsson E. Norrby E. Rift Valley fever virus vaccine trial: study of neutralizing antibody response in humans. Vaccine. 1985;3:123–127. doi: 10.1016/0264-410x(85)90061-1. [DOI] [PubMed] [Google Scholar]
- Papin JF. Verardi PH. Jones LA. Monge-Navarro F, et al. Recombinant Rift Valley fever vaccines induce protective levels of antibody in baboons and resistance to lethal challenge in mice. Proc Natl Acad Sci. 2011;108:14926–14931. doi: 10.1073/pnas.1112149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweska JT. Barnard BJ. Williams R. The use of sucrose-acetone-extracted Rift Valley fever virus antigen derived from cell culture in an indirect enzyme-linked immunosorbent assay and haemagglutination-inhibition test. Onderstepoort J Vet Res. 1995;62:227–233. [PubMed] [Google Scholar]
- Paweska J. Smith S. Wright I. Williams R, et al. Indirect enzyme-linked immunosorbent assay for the detection of antibody against Rift Valley fever virus in domestic and wild ruminant sera. Onderstepoort J Vet Res. 2003;70:49–64. [PubMed] [Google Scholar]
- Paweska J. Jansen van Vuren P. Swanepoel R. Validation of an indirect ELISA based on a recombinant nucleocapsid protein of Rift Valley fever virus for the detection of IgG antibody in humans. J Virol Methods. 2007;146:119–124. doi: 10.1016/j.jviromet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Paweska J. van Vuren P. Kemp A. Buss P, et al. Recombinant nucleocapsid-based ELISA for detection of IgG antibody to Rift Valley fever virus in African buffalo. Vet Microbiol. 2008;127:21–28. doi: 10.1016/j.vetmic.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Pepin M. Bouloy M. Bird BH. Kemp A, et al. Rift Valley fever virus(Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res. 2010;41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper ME. Sorenson DR. Gerrard SR. Efficient cellular release of Rift Valley fever virus requires genomic RNA. PLoS One. 2011;6:e18070. doi: 10.1371/journal.pone.0018070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn CS. Bunyaviridae: The viruses and their replication. In: Fields BN, editor; Knippe DM, editor; Howley DM, editor. Field Virology. 3rd. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 1447–1471. [Google Scholar]
- Schwarz TF. Gilch S. Pauli C. Jager G. Immunoblot detection of antibodies to Toscana virus. J Med Virol. 1996;49:83–86. doi: 10.1002/(SICI)1096-9071(199606)49:2<83::AID-JMV2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Shoemaker T. Boulianne C. Vincent MJ. Pezzanite L, et al. Genetic analysis of viruses associated with emergence of Rift Valley fever in Saudi Arabia and Yemen, 2000–01. Emerg Infect Dis. 2002;8:1415–1420. doi: 10.3201/eid0812.020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shope RE. Sather GE. Arboviruses. In: Lenette EH, editor; Schmidt NJ, editor. Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infection. 5th. American Health Association; Washington, DC: 1979. pp. 767–814. [Google Scholar]
- Smith DR. Bird BH. Lewis B. Johnston SC, et al. Development of a novel nonhuman primate model for Rift Valley fever. J Virol. 2012;86:2109–2120. doi: 10.1128/JVI.06190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithburn KC. Rift Valley fever; the neurotropic adaptation of the virus and the experimental use of this modified virus as a vaccine. Br J Exp Pathol. 1949;30:1–16. [PMC free article] [PubMed] [Google Scholar]
- Suzich JA. Kakach LT. Collett MS. Expression strategy of a phlebovirus: Biogenesis of proteins from the Rift Valley fever virus M segment. J Virol. 1990;64:1549–1555. doi: 10.1128/jvi.64.4.1549-1555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R. Struthers JK. Erasmus MJ. Shepherd SP, et al. Comparison of techniques for demonstrating antibodies to Rift Valley fever virus. J Hyg (Lond) 1986a;97:317–329. doi: 10.1017/s0022172400065414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R. Struthers JK. Erasmus MJ. Shepherd SP, et al. Comparative pathogenicity and antigenic cross-reactivity of Rift Valley fever and other African phleboviruses in sheep. J Hyg (Lond) 1986b;97:331–346. doi: 10.1017/s0022172400065426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara K. Morikawa S. Bishop DH. Characterization of baculovirus-expressed Rift Valley fever virus glycoproteins synthesized in insect cells. Virus Res. 1990;17:173–90. doi: 10.1016/0168-1702(90)90063-h. [DOI] [PubMed] [Google Scholar]
- Turell MJ. Faran ME. Cornet M. Bailey CL. Vector competence of Senegalese Aedes fowleri (Diptera: Culicidae) for Rift Valley fever virus. J Med Entomol. 1988;25:262–266. doi: 10.1093/jmedent/25.4.262. [DOI] [PubMed] [Google Scholar]
- Turell MJ. Wilson WC. Bennett KE. Potential for North American mosquitoes (Diptera: Culicidae) to transmit rift valley fever virus. J Med Entomol. 2010;47:884–889. doi: 10.1603/me10007. [DOI] [PubMed] [Google Scholar]
- Vialat P. Billecocq A. Kohl A. Bouloy M. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol. 2000;74:1538–1543. doi: 10.1128/jvi.74.3.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DB. Ellis CE. Espach A. Smith SJ, et al. Protective immune responses induced by different recombinant vaccine regimes to Rift Valley fever. Vaccine. 2006;24:7181–7189. doi: 10.1016/j.vaccine.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Won S. Ikegami T. Peters CJ. Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol. 2007;81:13335–13345. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]