Abstract
Recent exome sequencing studies have implicated polymorphic BAF complexes (mammalian SWI/SNF chromatin remodeling complexes) in several human intellectual disabilities and cognitive disorders. However, it is currently unknown how mutations in BAF complexes result in impaired cognitive function. Post mitotic neurons express a neuron specific assembly, nBAF, characterized by the neuron-specific subunit BAF53b. Mice harboring selective genetic manipulations of BAF53b have severe defects in longterm memory and long-lasting forms of hippocampal synaptic plasticity. We rescued memory impairments in BAF53b mutant mice by reintroducing BAF53b in the adult hippocampus, indicating a role for BAF53b beyond neuronal development. The defects in BAF53b mutant mice appear to derive from alterations in gene expression that produce abnormal postsynaptic components, such as spine structure and function, and ultimately lead to deficits in synaptic plasticity. Our studies provide new insight into the role of dominant mutations in subunits of BAF complexes in human intellectual and cognitive disorders.
It is generally accepted that gene expression is required for long-term memory formation; however, it remains unclear how gene expression is orchestrated by chromatin regulation during memory consolidation. Chromatin regulation via histone modification and DNA methylation is critical for controlling gene expression required for normal memory processes1, as demonstrated by animal studies as well as the association of these modifications with human disorders characterized by intellectual disability2. Histone modification and DNA methylation represent two major mechanisms of chromatin regulation, which are known to function intimately with ATP-dependent nucleosome remodeling, another major mechanism of chromatin regulation. Whereas much is known about the former mechanisms in memory formation, the role of nucleosome remodeling in memory processes and intellectual function remains poorly understood.
Nucleosome remodeling complexes have key roles in development, cancer, and stem cell biology3. They utilize ATP hydrolysis to disrupt nucleosome-DNA contacts, move nucleosomes along DNA, and from in vitro studies are thought to remove or exchange nucleosomes. Nucleosome remodeling complexes are comprised of numerous subunits and the exact composition of different subunits is cell-type specific and developmentally regulated4-6. Olave and colleagues (2002) identified the neuron-specific Brg1 Associated Factor (nBAF) complex that contains a neuron-specific subunit BAF53b. BAF53b is an actin-related protein, due to its high homology to β-actin, and both BAF53b and nuclear β-actin are found as monomers in stoichiometric levels bound to Brg/Brm ATPases7 within the nBAF complex. These three proteins form the core of the nBAF complex and are responsible for targeting the Brg/Brm ATPases to specific gene promoters7,8. BAF53b is unique among nucleosome remodeling complex subunits because it is neuron-specific and is not found in any other nucleosome remodeling complex besides the nBAF complex6,8. The unusual dedication of a subunit to a single neuronal complex makes BAF53b an ideal target for elucidating the role of the nBAF complex in learning and memory.
Homozygous deletion of BAF53b in mice is lethal. Neurons cultured from these animals have severe deficits in gene expression, dendritic arborization and synapse formation8. BAF53b begins to be expressed as neural progenitors exit mitosis and terminally differentiate into neurons6,9. A homolog of BAF53b, called BAF53a, is expressed in neural progenitors, and is replaced by BAF53b upon differentiation9. This switch is mediated by the repression of BAF53a via miR-9* and miR-124, which are selectively expressed in post-mitotic neurons10. Failure of miRNA-mediated repression of BAF53a results in decreased BAF53b expression and impaired dendritic outgrowth10. In contrast, expression of miR-9* and miR-124 in human fibroblasts induces their conversion into neurons, indicating that this overall mechanism is instructive in establishing neural cell fate11. Given the highly coordinated regulation of BAF53b during neuronal differentiation and its role in synapse formation and dendritic arborization, it is important to understand the role of BAF53b and the nBAF complex in both the developing and adult brain.
Mutations in various subunits of nucleosome remodeling complexes have recently been implicated in human disorders characterized by intellectual disability. For example, dominant mutations in subunits of mammalian BAF complexes are implicated in Coffin-Siris and Nicolaides-Baraitser syndromes, both of which are associated with intellectual disability and specific digital abnormalities12-14. In addition the nBAF component BAF250b (Arid1b) is mutated frequently in sporadic mental retardation12,13,15,16 and BAF155, BAF170, and REST were found to be mutated in exome sequencing of sporadic autism17. These findings suggest that the BAF nucleosome remodeling complex regulates gene expression required for proper neuronal function, synaptic plasticity, and memory processes. A key open question is whether the BAF complex has a role in memory processes that is distinct from its role in development. In this study, we directly address this question by examining the role of a neuron-specific component of the BAF complex, BAF53b, in synaptic plasticity and memory.
Results
Generation of BAF53b mutant mice
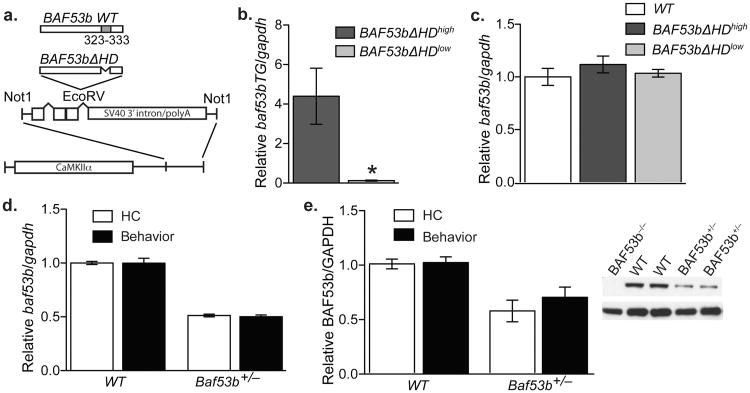
BAF53b is a neuron-specific stoichiometric component of BAF complexes not found in other nucleosome remodeling complexes8. To examine the role of BAF53b in long-term memory processes, we engineered transgenic mice that express a mutant form of BAF53b with a deletion of the hydrophobic domain (BAF53bΔHD; see Fig. 1A). The rationale for this deletion is that the hydrophobic domain is predicted to enable protein-protein interactions with other subunits of nBAF complexes, and deletion of this domain in the non-neuronal homolog BAF53a was shown to generate a dominant negative form of BAF53a18. Given the high degree of homology between BAF53a and BAF53b, we predicted a similar dominant-negative impact of deleting the HD from BAF53b. The BAF53bΔHD transgene (Fig. 1A) is expressed by the CaMKIIα promoter19, which allows for restricted expression to forebrain excitatory neurons and postnatal development20. Thus, we avoided developmental effects due to the known role for BAF53b in post mitotic embryonic development. We developed two independent lines of CaMKIIα-BAF53bΔHD. Both lines have normal brain morphology (Supplemental Fig. S1A), but are characterized by significantly different levels of transgene expression in hippocampal tissue (BAF53bΔHDhigh and BAF53bΔHDlow; Fig. 1B), which is relevant to the hippocampus-dependent experiments in this study. Wildtype BAF53b levels and localization are unaltered in the BAF53bΔHD mice (Fig. 1C & Supplemental Fig. S1B-D).
Figure 1.
Characterization of BAF53bΔHD and Baf53b+/− het mice. (a) Wildtype BAF53b is diagramed with hydrophobic domain (HD) shown in grey. In the BAF53bΔHD construct the amino acids 323-333 within the hydrophobic domain were deleted. The BAF53bΔHD mutant sequence was cloned into a separate vector containing intron and exon sequences with splice sites and the SV40 intron and polyadenylation signal, which was then cloned downstream of the 8.5 kb mouse CaMKIIα promoter. This construct was used to generate BAF53bΔHD transgenic mice. (b) Quantitative RT-PCR was performed with transgene specific primers to measure transgene expression in dorsal hippocampus of two independently derived lines of BAF53bΔHD mice. We identified two significantly different lines (Mann-Whitney U (18)=0.00, p<0.0001): a low expressing line (n=10) and a high expressing line (n=10) (2 out of 12 founder lines). (c) Wildtype BAF53b expression in the dorsal hippocampus of BAF53bΔHDhigh (n=10) and BAF53bΔHDlow (n=11) is not significantly different (Kruskal-Wallis H2,33=4.03, p=0.13) between mutant mice and wildtype littermates (n=15). (d) Quantitative RT-PCR shows that wildtype Baf53b expression in the dorsal hippocampus of Baf53b+/− het mice (n=13) is significantly (2-way ANOVA main effect of genotype F1,18=17.87, p<0.001) reduced compared to wildtype littermates (n=8). There was no effect of behavior (F1,18=0.81, p=0.38) or interaction (F1,18=0.40, p=0.53) Mean (± SEM). (e) Western blot analysis shows that BAF53b protein in dorsal hippocampus of Baf53b+/− het mice (n=13) is significantly (2-way ANOVA main effect of genotype F1,17=345.0, p<0.0001) reduced compared to wildtype littermates (n=9). There was no effect of behavior (F1,17=0.05, p=0.82) or interaction (F1,17=0.03, p=0.85).
In addition to the BAF53bΔHD mice, we also examined BAF53b heterozygous null animals (Baf53b+/−)8. Baf53b−/− homozygous knockout mice have a complete loss of BAF53b mRNA and protein, and die within the first two postnatal days8. Consistent with the exclusive expression of BAF53b in post mitotic neurons, they have no detectable defects outside of the nervous system8. In contrast, Baf53b+/− het mice develop normally (Supplemental Fig. S1) and express approximately 50% of the wildtype levels of Baf53b mRNA (Fig. 1D) and protein (Fig. 1E). Baf53b mRNA and protein expression in Baf53b+/− het mice was half of that in wildtype littermates both from homecage samples and after training (sacrificed 30min post OLM training; see Fig 2A for schematic diagram) (Fig 1D & 1E). Importantly, there are no neuron-specific homologs of BAF53b that could compensate for its decreased expression. Thus, these results demonstrate that there is no compensation in expression in the Baf53b+/− het mice following behavior. In addition, both anxiety and motor function were found to be normal in BAF53bΔHD and Baf53b+/− het mice as compared to wildtype controls (Supplemental Fig. S2), allowing us to examine memory processes in these mice.
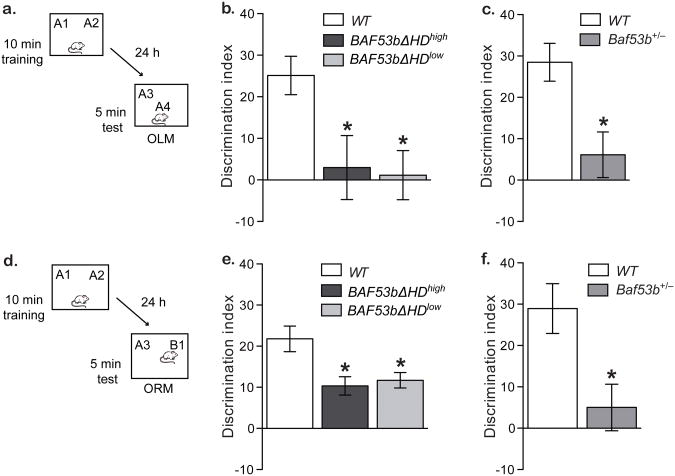
Figure 2.
BAF53bΔHD and Baf53b+/− het mice have impaired long-term memory. (a) Mice received 10 min training in an environment with 2 identical objects and received a retention test 24 hrs later in which one object is moved to a new location (OLM). (b) BAF53bΔHDhigh mutant mice (n=9) and BAF53bΔHDlow (n=13) exhibit a significant 24 hr long-term OLM deficit (ANOVA F2,34=5.79, p<0.01) in a hippocampus-dependent task as compared to wildtype littermates (n=15) and were not significantly different from zero (BAF53bΔHDhigh t-test t(8)=0.39, p=0.71 and BAF53bΔHDlow t-test t(12)=0.19, p=0.85). There were no significant differences in total exploration time between the groups during testing (ANOVA F2,34=0.45, p=0.64) or training (ANOVA F2,34=1.13, p=0.33) (c) Baf53b+/− het mice (n=16) exhibit impaired long-term OLM compared to wildtype mice (n=6; t-test t(20)=2.35, p<0.05). There was no difference in overall exploration between the groups at training t-test t(20)=0.40, p=0.70 or testing t-test t(20)=0.95, p=0.35. (d) Mice received 10 min training in an environment with 2 identical objects and received a retention test 24 hrs later in which one object is replaced with a novel one. (e) In a hippocampus-independent object recognition task, BAF53bΔHDhigh mutant mice (n=12) and BAF53bΔHDlow (n=11) exhibit significant ORM deficits as compared to wildtype mice (n=18; Kruskal-Wallis H2,38=10.72, p<0.01; t(11)=13.06, p<0.05 and t(10)=11.43, p<0.05, respectively, Dunn's post hoc tests). There were no differences in total exploration time between the groups at training Kruskal-Wallis H2,38=0.61, p=0.98 or testing ANOVA F(2,30)=2.53, p=0.09. (f) Baf53b+/− het mice (n=9) exhibit impaired long-term OLM compared to wildtype mice (n=10; t-test t(17)=2.88, p<0.05). There was no difference in the overall exploration time between the groups at training t-test t(17)=0.60, p=0.56 or testing t-test t(17)=0.36, p=0.72. Mean (± SEM).
Baf53b is required for long-term memory
To examine the role of BAF53b in memory, BAF53bΔHD and Baf53b+/− het mice were tested for both long- and short-term memory for object location (OLM; Fig. 2A) and object recognition (ORM; Fig. 2D). Wildtype mice exhibited long-term OLM (Fig. 2B & 2C; as measured by preference for the novel location over the familiar location and calculated as the discrimination index). However, both the high and low expressing lines of BAF53bΔHD mice exhibited significantly reduced discrimination indices compared to wildtype littermates (Fig. 2B). Similar to the BAF53bΔHD lines, Baf53b+/− het mice also exhibited significant deficits in long-term OLM (Fig. 2C). In contrast to the long-term memory deficits, short-term OLM (tested at 90 min) was normal in BAF53bΔHD mice (Supplemental Fig. S3B) and Baf53b+/− het mice (Supplemental Fig. S3C) as compared to respective wildtype littermates. Deficits in long-term memory formation were also found in the object recognition paradigm (Fig. 2D). Wildtype mice exhibited normal long-term ORM (as measured by preference for the novel object over the familiar object and calculated as the discrimination index). Both the BAF53bΔHD high and low expressing lines had significantly reduced discrimination indices compared to the wildtype littermates (Fig. 2E). Baf53b+/− het mice also showed significant long-term ORM deficits (Fig. 2F). In contrast, short-term ORM (tested at 90 min) was normal in BAF53bΔHD mice (Supplemental Fig. S3E) and Baf53b+/− het mice as compared to respective wildtype littermates (Supplemental Fig. S3F).
Together, these results indicate that BAF53b has a critical role in long-term memory formation. Importantly, all BAF53b mutant mice (BAF53bΔHD and Baf53b+/− hets) have normal short-term memory, indicating that they can perform these tasks and that their deficits at 24hrs are due to a failure of learning and memory and not performance. These results also support the hypothesis that BAF53b is involved in regulating gene expression required for long-term memory, as short-term memory in these tasks is transcription-independent.
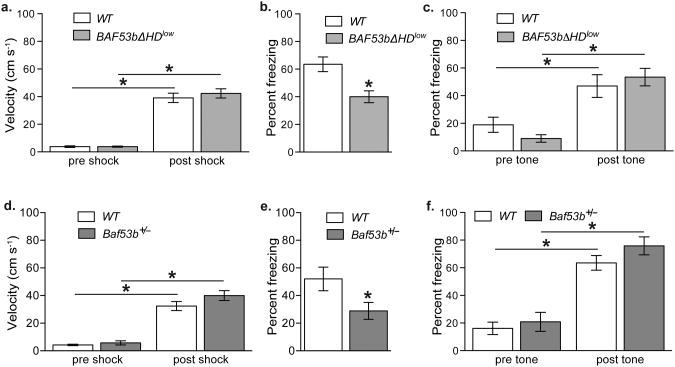
In addition to ORM and OLM tasks, we examined long-term memory for contextual and cued fear conditioning in BAF53bΔHDlow and Baf53b+/− het mice. Both lines exhibited normal response to shock, as measured by pre- and post-shock velocity (Fig. 3A & 3D). BAF53bΔHDlow mice exhibited a significant decrease in freezing in a 24 hr retention test, indicating impaired long-term memory for contextual fear (Fig. 3B). In contrast, long-term memory for cued fear was normal in BAF53bΔHDlow mice as compared to wildtype littermates in a 24 hr retention test (Fig. 3C). Similar results were observed in Baf53b+/− het mice. Baf53b+/− het mice exhibited a significant decrease in freezing in the conditioned context in a 24 hr retention test, indicating impaired long-term memory for contextual fear (Fig. 3E). In contrast, long-term memory for cued fear was normal in Baf53b+/− het mice as compared to wildtype littermates in a 24 hr retention test (Fig. 3F). Together, these results suggest that BAF53b has a role in long-term memory for contextual fear, but perhaps not cued fear memory. Because contextual fear conditioning is hippocampus-dependent, whereas cued fear conditioning is hippocampus-independent, the results also suggest that the hippocampus may be more sensitive to alterations in BAF53b.
Figure 3.
BAF53bΔHDlow and Baf53b+/− het mice have impairments in long-term memory for contextual fear, but normal cued fear memory. (a) During contextual fear training velocity (cm s−1) did not differ between BAF53bΔHDlow (n=10) and wildtype (n=9) mice for the five second prior to shock (Pre-Shock) nor during the shock (Shock) (Repeated Measures ANOVA F1,17=234.2, p<0.0001, bonferroni post hoc t-test pre-shock vs. post shock for wildtypes t-test t(8)=10.09, p<0.001 and BAF53bΔHDlow t-test t(9)=11.60, p<0.001, and no effect of genotype ANOVA F1,17=0.44, p=0.51 or interaction F1,17=0.44, p=0.51). (b) Animals were tested in the conditioned context 24 hours after conditioning. BAF53bΔHDlow mutant mice froze significantly less then wildtypes (t-test t(17)=3.46, p<0.05). At test there was a significant main effect of sex (ANOVA F1,15=12.39, p=0.003) but no interaction with genotype (F1,15=0.03, p=0.86) with males freezing more then females for BAF53bΔHDlow (bonferroni post hoc t-test t(9)=2.58, p<0.05) and a similar (but not significant) trend in wildtypes (bonferroni post hoc t-test t(8)=2.40, p>0.05). (c) 24hr memory test for cued fear conditioning (test in novel context). Both groups exhibited similar levels of freezing prior to tone onset (Pre-Tone) and after Tone onset (Tone) (Repeated Measures ANOVA F1,16=0.98, p=0.77), with a significant increase in freezing following tone onset in both groups (F1,16=38.21, p<0.0001, bonferroni post hoc t-test Pre-Tone vs. Tone for wildtype (n=8) t(7)=3.21, p<0.05 and BAF53bΔHDlow (n=10) t(9)=5.68, p<0.001). (D) Baf53b+/− het mice (n=9) have a normal response to the shock during contextual fear training compared to wildtype littermates (n=8) with a significant increase in velocity following shock for both groups (Repeated Measures ANOVA F1,15=183.3, p<0.0001), bonferroni post hoc t-test pre-shock vs. post shock for wildtype t(7)=8.38, p<0.001 and Baf53b+/− het mice t(8)=10.85, p<0.001, and no effect of genotype (F1,15=2.57, p=0.13) or interaction F1,15=1.80, p=0.20). (E) Baf53b+/− het mice froze significantly less then wildtypes at the 24hr long-term contextual fear memory test (t-test t(15)=2.25, p<0.05) (F) 24hr memory test for cued fear conditioning (test in novel context). Both groups exhibited similar levels of freezing prior to tone onset (Pre-Tone) and after Tone onset (Tone) (Repeated Measures ANOVA F1,22=0.53, p=0.48) with a significant increase in freezing following tone onset in both groups (ANOVA F1,22=63.29, p<0.0001, bonferroni post hoc t-test Pre-Tone vs. Tone for wildtype (n=12) t(11)=6.56, p<0.001 and Baf53b+/− het mice (n=12) t(11)=4.69, p<0.001). Mean (± SEM).
Reintroduction of BAF53b into dorsal hippocampus rescues OLM but not ORM
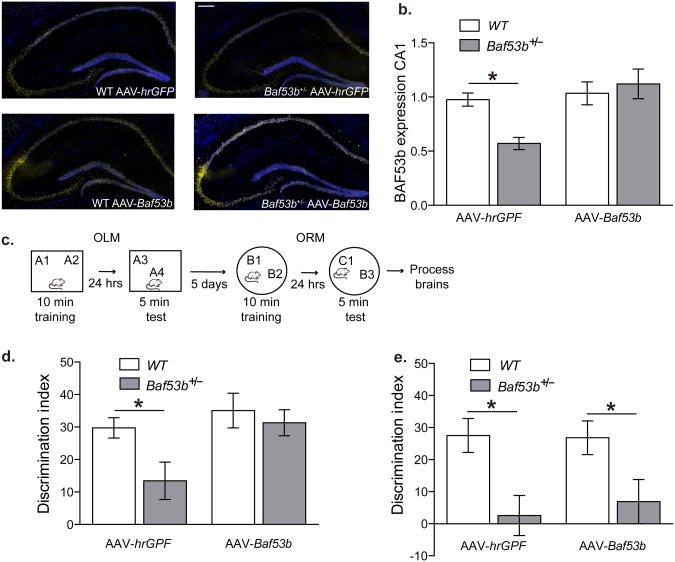
To assess whether BAF53b is sufficient for long-term memory formation in the hippocampus we reintroduced wildtype BAF53b into the dorsal hippocampus of adult Baf53b+/− het mice and wildtype littermates using adeno-associated virus (AAV). AAV expressing BAF53b (AAV-Baf53b) or the control virus (AAV-hrGFP) were surgically delivered into dorsal hippocampus 14 days prior to behavior. As shown in Figure 4, AAV-Baf53b was robustly expressed in the dorsal hippocampus and BAF53b protein levels in the Baf53b+/− het mice with AAV-Baf53b were increased to wildtype levels in CA1 (Fig 4A and 4B). Importantly all four groups of animals (WT AAV-hrGFP, Baf53b+/− AAV-hrGFP, WT AAV-Baf53b, Baf53b+/− AAV-Baf53b) showed normal motor function and anxiety (Supplemental Fig S2). All four groups of animals were tested for long-term OLM (hippocampal dependent) followed by long-term ORM (in a novel context with different objects) (hippocampal independent) (Fig4C). As shown in Figure 4D, WT AAV-hrGFP animals showed a robust preference for the displaced object indicating long-term OLM. The Baf53b+/− het mice with the control virus showed significantly impaired OLM, replicating our previous findings (Fig 2C). Wildtype animals with AAV-Baf53b also show a robust preference for the displaced object similar to wildtype animals with control virus. Critically, Baf53b+/− het mice with AAV-Baf53b show a robust preference for the displaced object that is indistinguishable from wildtype animals, demonstrating a complete rescue of long-term OLM formation (Fig 4D). These findings indicate that the memory deficits observed in the Baf53b+/− het mice (Fig 4D & 2C) are due to a role for BAF53b in regulating gene expression in the adult brain and not a consequence of BAF53b's role in development.
Figure 4.
Hippocampal AAV-Baf53b rescues OLM but not ORM deficits in Baf53b+/− het mice. (a) Representative images of immunofluorescence of BAF53b (yellow) expression in wildtype and Baf53b+/− het mice with control (AAV-hrGFP) or AAV-Baf53b. Nuclei (blue) were counterstained with DAPI. Scale bar 200μm. (b) Mean intensity of BAF53b immunofluorescence from CA1 cell layer normalized to background (corpus collosum) and wildtype AAV-hrGFP. There is a complete return of BAF53b expression in CA1 of Baf53b+/− het mice to wildtype levels (ANOVA no main effect genotype F1,38=2.23, p=0.14, a main effect of virus F1,38=8.14, p<0.01, and a significant interaction F1,38=5.34, p<0.05). Bonferroni post hoc t-test WT vs. Baf53b+/− het mice for AAV-hrGFP t-test t(16)=2.5, p<0.05 and AAV-Baf53b t-test t(22)=0.62, p>0.05. WT AAV-hrGFP (n=10), WT AAV-Baf53b (n=12), Baf53b+/− het mice AAV-hrGFP (n=9), Baf53b+/− het mice AAV-Baf53b (n=12). (C) Schematic of behavioral testing. OLM was conducted as described in the methods. Following a five day rest period, animals then were habituated to a novel context and then underwent ORM training and testing. (D) Long-term Object Location Memory (OLM) (24hrs). There is a complete rescue of OLM in Baf53b+/− het mice with AAV-Baf53b (2-way ANOVA main effect genotype F1,40=4.49, p<0.05, virus F1,40=6.04, p<0.05, and no interaction F1,40=1.76 p=0.19). Bonferroni post hoc t-test WT vs. Baf53b+/− het mice for AAV-hrGFP t-test t(18)=2.33, p<0.05 and AAV-Baf53b t-test t(22)=0.59, p>0.05. There was no difference between any of the groups for total exploration at training (2-way ANOVA no effect genotype F1,40=3.56, p=0.07, virus F1,40=0.02, p=0.90, and no interaction F1,40=0.63 p=0.43) or testing (2-way ANOVA no effect genotype F1,40=0.29, p=0.59, virus F1,40=0.53, p=0.47, and no interaction F1,40=4.02 p=0.05). WT AAV-hrGFP (n=10), WT AAV-Baf53b (n=12), Baf53b+/− het mice AAV-hrGFP (n=10), Baf53b+/− het mice AAV-Baf53b (n=12). (E) Long-term Object Recognition Memory (ORM) (24hrs). There is a no rescue of ORM in Baf53b+/− het mice with AAV-Baf53b expression in dorsal hippocampus (2-way ANOVA main effect genotype F1,33=12.79, p<0.01, no main effect of virus F1,33=0.08 p=0.77, and no interaction F1, 33=0.16, p=0.69). Bonferroni post hoc t-test WT vs. Baf53b+/− het mice for AAV-hrGFP t-test t(14)=2.63, p<0.05 and AAV-Baf53b t-test t(19)=2.42, p<0.05). There was no difference between any of the groups for total exploration at training (2-way ANOVA no effect genotype F1,33=1.08, p=0.31, virus F1,33=2.85, p=0.10, and no interaction F1,33=0.04 p=0.84) or testing (2-way ANOVA no effect genotype F1,33=4.13, p=0.05, virus F1,33=1.78, p=0.19, and no interaction F1,33=0.51 p=0.48). WT AAV-hrGFP (n=7), WT AAV-Baf53b (n=10), Baf53b+/− het mice AAV-hrGFP (n=9), Baf53b+/− het mice AAV-Baf53b (n=11). Mean (± SEM).
To examine the specificity of the long-term memory rescue, the same animals were then tested on the hippocampal independent long-term ORM task (Fig 4E). Similar to our previous findings with the Baf53b+/− mice (Fig 2F), WT AAV-hrGFP animals showed a robust preference for the displaced object indicating long-term OLM. The Baf53b+/− het mice with the control virus showed significantly impaired OLM, replicating our previous findings (Fig 2F). Wildtype animals with AAV-Baf53b also show long-term ORM; however in the ORM task the Baf53b+/− het mice with AAV-Baf53b fail to show a preference for the novel object (Fig 4E). Given the spatially restricted viral expression in dorsal hippocampus of the Baf53b+/− AAV-Baf53b mice, the failure to rescue a hippocampal independent memory task (ORM) demonstrates specificity for the OLM rescue and not a global change in brain state or processing. Overall, these rescue experiments provide strong evidence that BAF53b plays a critical role in long-term memory formation in the adult animal.
LTP fails to stabilize in slices from BAF53b mutant mice
To assess the potential contributions of BAF53b in synaptic plasticity, a cellular mechanism thought to underlie memory processes, we examined long-term potentiation in acute hippocampal slices from BAF53bΔHD and Baf53b+/− het mice. Wildtype slices were tested concurrently with a subset of cases for each of the three experimental groups. Results for all wildtype controls (n=24) are included in the summary graphs of the latter groups. Theta burst stimulation (TBS) was delivered to a single collection of Schaffer-commissural fibers in all slices for many of which a second input received only low frequency (3pulses/min) stimulation (LFS). We detected no changes over time in the EPSP slopes elicited by LFS in any of the slice groups (Supplemental FigS4A), and these data are also combined in the data summaries.
A single train of ten theta bursts, which past studies have shown is well above the threshold (number of bursts) needed to induce LTP21,22 produced a robust and immediate potentiation in wildtype slices that steadily decayed over 10 min to a plateau level that was approximately 60% above the pre-TBS baseline; the mean (± SEM) % LTP for minutes 50-60 post-TBS was 59 ± 4%. Hippocampal slices prepared from Baf53b+/− het mice exhibited a very different pattern: short-term potentiation (STP) was not measurably different from wildtype values but the enhanced responses failed to stabilize and instead gradually fell back to baseline (Fig 5A). Percent facilitation for the last ten minutes of the one hour post-TBS session was 14 ± 6% (p<0.0001 vs. WT).
Figure 5.
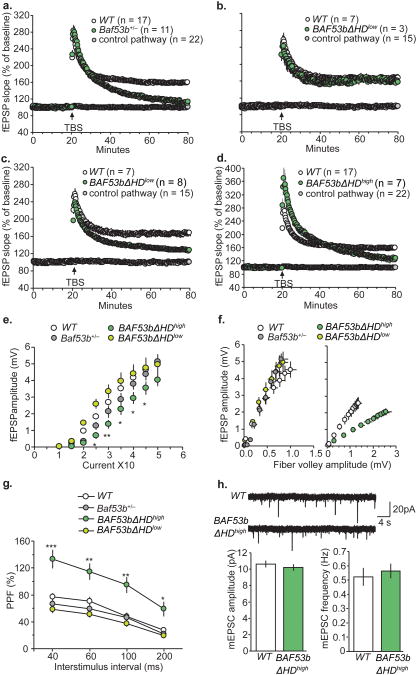
BAF53bΔHD and Baf53b+/− het mice have disrupted LTP in hippocampal slices. (A, B) Simultaneous recordings of fEPSP slope from slices receiving baseline stimulation (control, grey circle) and 10 theta bursts (TBS) for wildtype (WT) (open circle) and Baf53b+/− het slices (green circle). Baf53b+/− het slices fail to maintain stable LTP (average for last 5min) t-test t(26)=6.48, p<0.0001. BAF53bΔHDlow slices produced stable potentiation similar to wildtype slices t(8)=0.30, p=0.79. (C) LTP induced with 5 theta bursts delivered to one of the two stimulation electrodes (control, grey circles) produced stable potentiation in wildtype slices (open circles) but not in BAF53bΔHDlow slices (green circles) t(13)=3.77, p<0.01. (D) Simultaneous recordings of fEPSP slope in slices receiving low-frequency stimulation (grey circles) and 10 theta bursts (open and green circles) in wildtype and BAF53bΔHDhigh slices t(22)=3.49, p<0.01. (E) Input/Output curves measuring the magnitude of the fEPSP response across a range of stimulation currents (10-50 μA) was comparable between Baf53b+/− het, BAF53bΔHDlow and wildtype slices, but not BAF53bΔHDhigh mice. (2-way Repeated ANOVA main effect genotype F3,35=4.47, p<0.01, time F8,35=246.7, p<0.0001, and significant interaction F24,35=2.01 p<0.005). For each stimulation current bonferroni post hoc t-test for wildtype (n=20) vs. Baf53b+/− het slices (n=6), wildtype (n=20) vs. BAF53bΔHDhigh slices (n=7), and wildtype (n=20) vs. BAF53bΔHDlow slices (n=6) are given in Supplemental FigS4. (**p<0.01; *p<0.05). (F) Input/Output curves compare amplitudes of the presynaptic fiber volley to the fEPSP amplitude across a range of stimulation currents. Left, Input/output curves were not different between Baf53b+/− het mice (n=6), BAF53bΔHDlow (n=6), and wildtype (n=6) (ANOVA F2,15=1.04, p=0.38). Right, The slope was significantly reduced in the Input/Output curve produced from BAF53bΔHDhigh slices (n=12) relative to wildtype (n=12) bathed in aCSF containing 3mM Mg+ and 1mM Ca2+ (Mann-Whitney U(22)=3.00, p<0.0001). (G) Paired pulse facilitation of the initial slope of the synaptic response was comparable (40, 60, 100, and 200 ms inter-pulse intervals) in slices from Baf53b+/− het (n=9), BAF53bΔHDlow (n=7), and wildtype mice (n=14) but not in BAF53bΔHDhigh slices (n=6) (2-way Repeated ANOVA main effect genotype F3,32=11.60, p<0.0001, interval F3,32=192.6, p<0.0001, and significant interaction F9,32=3.31 p<0.005). Bonnferoni post hoc comparisons in Supplemental FigS4 (***p<0.001; ** p<0.01; *p<0.05). (H) Top; sample traces of mini excitatory postsynaptic currents (mEPSCs) recorded from BAF53bΔHDhigh (n=14) and wildtype (n=15) neurons. Bottom; There were no differences in mEPSCs from BAF53bΔHDhigh and wildtype for the amplitude (left, t(27)=0.75, p=0.46) and frequency (right, t(27)=0.52, p=0.61) of the events in slices.
Initial studies with slices from BAF53bΔHDlow did not detect any abnormalities in the LTP elicited by ten theta bursts from that obtained in wildtype cases (Fig 5B). We therefore repeated the experiment using trains of five theta bursts, a protocol that is closer to the threshold for eliciting stable LTP in mice. A pronounced defect was found under these conditions. Five bursts delivered to wildtype slices produced STP and LTP effects comparable to those induced by ten bursts; slices from the BAF53bΔHDlow exhibited normal STP but potentiation failed to stabilize and over the next hour decayed to levels that were markedly lower than those recorded for the controls (p<0.001 at 50-60 min post TBS; Fig 5C). We conclude from these results that low expression of the BAF53bΔHD mutant is sufficient to markedly impair the machinery required to consolidate LTP and that this impairment can be overcome by supra-threshold stimulation.
We next tested if the higher expression of mutant BAF53bΔHD in BAF53ΔHDhigh mice would reproduce the total loss of LTP found in the Baf53b+/− het mice. Slices prepared from BAF53ΔHDhigh mice had unexpected responses to ten theta bursts. Initial potentiation was greatly amplified (% facilitation at two minute post-TBS was 239±31% compared to 156±10% for widltypes; p=0.003) but then showed a failure to stabilize similar to the BAF53bΔHDlow and Baf53b+/− het mice (Fig 5D). Collectively, the above findings suggest that BAF53b is required for consolidation of lasting changes in synaptic strength and that somewhat lower levels are needed to maintain STP within normal boundaries.
We then investigated which aspects of the complex series of events leading to induction, expression, and consolidation of LTP are negatively affected by manipulations of BAF53b by examining conventional measures of transmission and the responses to theta bursts. Input / output curves (number of axons stimulated / magnitude of post-synaptic response) were comparable for wildtypes vs. Baf53b+/− het mice or BAF53bΔHDlow but there was a marked depression of the curve in the BAF53bΔHDhigh mice (Fig 5E). The most straightforward interpretation of these results is that high expression of the BAF53ΔHD mutation affects axon excitability. In accord with this idea, the slope of the relationship between stimulation current and the amplitude of the fiber volley, a measure of the number of responsive axons, was reduced for the BAF53ΔHDhigh slices, but not for the other two genotypes, relative to controls (Fig 5F). We then tested the genotypes for differences in transmitter release kinetics using paired pulse facilitation, a measure of the extent to which release triggered by a single stimulation pulse increases release by a slightly delayed second pulse. The Baf53b+/− het slices and BAF53bΔHDlow slices did not differ from wildtypes, but the BAF53ΔHDhigh group was enhanced (p<0.0001) (Fig 5G). However, whole cell recording indicated that the frequency and magnitude of miniature excitatory postsynaptic potentials in the BAF53ΔHDhigh slices were comparable to wildtype values (Fig 5H). In all, BAF53ΔHDhigh slices are characterized by depressed axon excitability and exaggerated transmitter mobilization while spontaneous release and associated post-synaptic receptor responses appear to be normal. The Baf53b+/− het slices and BAF53bΔHDlow cases were comparable to controls on all tests of baseline physiology.
The above measures suggest that the failure of LTP consolidation in the Baf53b+/− het mice and BAF53ΔHDlow groups cannot be attributed to presynaptic variables. We therefore examined the magnitude and within train facilitation of the post-synaptic responses to theta bursts in these two groups; this analysis uncovered no differences between the two genotypes and wildtype slices (Supplemental FigS4B&C). It thus appears that the loss of LTP in the mutants involves factors that operate after induction and initial expression of potentiation, a conclusion that accords with the above observation that STP is unaffected in these animals (see Fig. 5A, B, C).
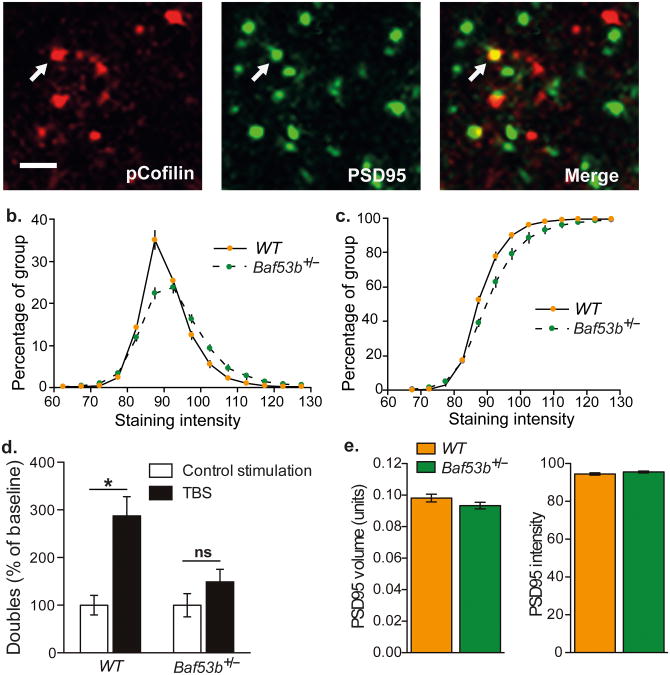
The Baf53b+/− het mice show robust failure in the expression of LTP. Thus, we wanted to determine if the mice showed deficits in activity induced actin signaling. TBS stimulation activates actin regulatory pathways and alters synapse morphology within the same dendritic spines as shown by a TBS induced increase in spines containing phosphorylated p21-activated kinase (PAK) or its downstream target Cofilin23. Using wide field deconvolution microscopy and immunofluorescent labeling we found that phosphorylated (p) Cofilin has punctate labeling within stratum radiatum of CA1 and colocalizes with PSD95, a marker for excitatory synapses (Fig 6A). Quantification revealed that the intensity profiles of pCofilin puncta colocalized with PSD95 are right shifted in the Baf53b+/− het mice, indicating an increase in more intensely labeled puncta (Fig 6B & 6C). pCofilin has been shown to peak seven min post-TBS24. Therefore, we examined pCofilin at this timepoint in Baf53b+/− het mice and wildtype littermates. Wildtype mice showed a clear, TBS induced increase in the number of densely labeled pCofilin puncta co-localized to PSD95 seven minutes following TBS, while the Baf53b+/− het mice did not show a TBS induced increase in the double labeled puncta (Fig 6D). This difference was not due to any changes detected in PSD95 volumes or intensities (Fig 6E), suggesting that the post synaptic terminal between the two groups is structurally similar.
Figure 6.
TBS induced phosphorylation (p) of Cofilin is altered in Baf53b+/− het mice. Adult mouse hippocampal slices are stimulated electrophysiologically and immunolabeled. (A) Immunocytochemical labeling of pCofilin (left) and PSD95-immunoreactive puncta (green) display some co-localization (Scale 2.5um). (B) Distribution of double labeled pCofilin intensities, show that Baf53b+/− het mice have a different baseline distribution, with an increase in the more intensely labeled puncta. (C) Cumulative probability distributions show that the Baf53b+/− het mice have curves that are shifted to the right relative to their wildtype counterparts, thus favoring the more intense puncta. (D) Bar graph shows values of double labeled puncta 7 minutes after control stimulation or TBS, with values normalized to respective control stimulation group. ANOVA main effect of stimulation F1,25=14.92, p<0.005, genotype F1,25=5.13, p<0.05, and a significant interaction F1,25=5.11, p<0.05. Bonnferoni corrected post hoc t-test wildtype control vs. TBS t(12)=4.24, p<0.001; Baf53b+/− het control vs. TBS t(13)=1.16, p>0.05 wildtype control (n=8), wildtype TBS (n=8), Baf53b+/− het control (n=7), Baf53b+/− het TBS (n=8). Mean +/- SEM. (E) Left Quantification of the mean volumes of PSD95-immuno reactive puncta that were colocalized with pCofilin for Baf53b+/− het mice (n=15) and wildtype littermates (n=16) t-test t(29)=1.39, p=0.19. Right Mean intensities of PSD95 labeled elements also show no difference between the Baf53b+/− het mice (n=15) and wildtype littermates (n=16) t-test t(29)=1.47, p=0.15. “n” refers to the number of images analyzed with ∼40,000 PSD95 immunoreactive puncta per image.
In view of the localization of the profound functional synaptic deficits to the post-synaptic compartment in BAF53ΔHDhigh mice (Fig 5), we assessed more closely the number and structure of dendritic spines, the sites of post-synaptic elements. Crossing the BAF53ΔHDhigh mice with a line expressing YFP under the Thy1 promoter in pyramidal cells, we confirmed the overall normal anatomy of the hippocampus (Supplemental Fig S5A). We then analyzed the spectrum of spine types (Supplemental Fig S5B) encompassing subpopulations of thin, stubby and mushroom-like spines25-27. The overall density of dendritic spines was modestly lower in BAF53bΔHDhigh mice compared with controls (Supplemental Fig S5F). However, a significant decrease in thin spines was apparent in the assessed CA1 pyramidal cells, including both main (stem) and oblique apical branches, resulting in an abnormally low ratio of thin / mushroom spines (Supplemental Fig S5C-E). The abnormal distribution of dendritic spine subpopulations already in juvenile BAF53bΔHDhigh mice is consistent with the problems in synaptic function and plasticity found in these mice. Together with data from Figure 6, these results indicate that BAF53b may have roles in both spine morphology/structure as well as synaptic events depending on the type of Baf53b mutation.
BAF53b-dependent gene expression disrupted during memory consolidation
Together, the above data indicate that reduced BAF53b function interferes significantly with the functional and structural foundations of long-term memory, i.e., memory consolidation. Therefore we set out to identify which genes BAF53b regulates during memory consolidation. We performed an RNA Sequencing experiment (Fig. 7) from dorsal hippocampal tissue from four groups of animals: Baf53b+/− homecage (Baf53b+/− HC); Baf53b+/− behavior (Baf53b+/− Beh); wildtype homecage (WT HC); and wildtype behavior (WT Beh). All Baf53b+/− and wildtype animals were handled and habituated as described for OLM (Fig. 2A, supplemental Fig S2). On the training day, half the animals from each genotype were sacrificed 30min after OLM training (Beh) or directly from their home cage without training (HC). We have previously observed significant gene expression changes in the dorsal hippocampus 30 min following OLM training28. Mean PHRED quality scores indicate high quality sequencing data for each replicate (Supplemental FigS6A). After mapping and considering the haploid genome29,30, RNA Sequencing successfully covered transcriptome 298.50 times (Supplemental Fig S6B) and significant differences in expression profiles were examined between all pairs of samples for p<0.0531-33.
Figure 7.
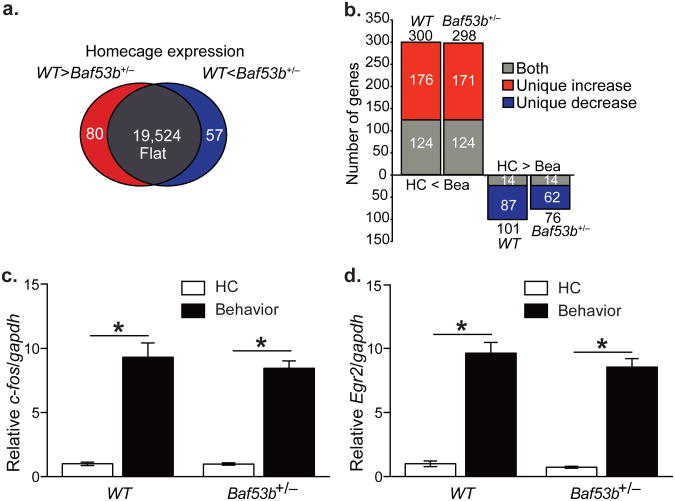
Differential gene expression in Baf53b+/− het mice by RNA Sequencing. (A) Gene expression diagram for wildtype compared to Baf53b+/− het mice mutant mice sacrificed directly from the homecage. (B) Gene expression for genes that increased or decreased expression following behavior (sacrificed 30min post training) compared to homecage. Genes with differential expression at homecage were removed prior to analysis. “Both” indicates genes regulated similarly in wildtype and Baf53b+/− het mice. “Unique Increase” comprises genes that increase in only the indicated genotype. “Unique Decrease” comprises genes that decrease in only the indicated genotype. Groups: Baf53b+/− het mice homecage (Baf53b+/− HC) (n=6); Baf53b+/− het mice Behavior (Baf53b+/− Beh) (n=6); wildtype homecage (WT HC) (n=6); and wildtype behavior (WT Beh) (n=6). Total gene counts for each genotype given above or below each column. (C) qRTP-PCR validation of the IEG c-fos. ANOVA main effect of behavior F1,20=157.6, p<0.0001, no effect of genotype F1,20=0.49, p=0.49, and no interaction F1,20=0.45, p=0.51. Expression relative to gapdh and normalized to wildtype homecage. (D) qRTP-PCR validation of the IEG Egr2. ANOVA main effect of behavior F1,20=224.2, p<0.0001, no effect of genotype F1,20=1.53, p=0.23, and no interaction F1,20=0.55, p=0.47. Expression relative to gapdh and normalized to wildtype homecage.
We first compared the expression profiles of the wildtype and Baf53b+/− het mice homecage groups and found that the majority of genes (19,524) were equivalently expressed at baseline in the two groups (Fig. 7A). There were also groups of genes that showed increased expression (80) in the wildtype compared to Baf53b+/− het mice and the reverse (57) at homecage (Supplemental Tables S1 & S2). We next examined differences in gene expression following training in the wildtype mice. In agreement with numerous studies demonstrating similar findings34, wildtype mice showed robust changes in gene expression including many immediate early genes (IEGs) following OLM training (compared to homecage) (Fig7B, Supplemental Table S3 & 4) indicating that the training period was sufficient to induce activity-dependent gene expression during memory consolidation. In addition, many of the activity-regulated genes (124) increased in the wildtype were also significantly induced in Baf53b+/− het mice following training (Supplemental Table S5). These genes were enriched for Gene Ontology (GO)35-38 terms for regulation of transcription, RNA processing, and intracellular signaling and included the majority of IEGs (Supplemental Table S6, Fig 7C & D). This suggests that BAF53b and nucleosome remodeling do not affect IEG expression during memory consolidation and that the long-term memory impairments observed in Baf53b+/− het mice are caused by different mechanisms. Of the 300 genes that were increased in the wildtype following OLM training, 176 failed to significantly increase in the Baf53b+/− het mice (Fig 7B, Supplemental Table S7). These genes were enriched for GO terms involving transcription regulation and neurogenesis, as well as chromosome organization and chromatin modification, indicating a potential role for BAF53b in organizing higher order chromatin structure. In the Baf53b+/− het mice there were also a group of genes (171) that were induced following behavior that are not normally increased in the wildtype (Fig 7B, Supplemental Table S8). These genes were enriched for GO terms involving regulation of cell death, glutamate release, behavioral response to drugs of abuse, synaptic transmission, and regulation of neurogenesis.
In addition to increases in gene expression, there are 101 genes that decrease expression in the wildtypes following OLM training compared to homecage (Fig7B) and 76 genes that decrease in the Baf53b+/− het mice. Of the 101 genes that decrease in the wildtype, 14 also decrease in the Baf53b+/− het mice (Supplemental Table S9) and 87 do not (Supplemental Table S10). The 87 genes that fail to show an activity-dependent decrease in expression in the Baf53b+/− het mice are enriched for GO terms involving cell homeostasis, postsynaptic cell membrane, and cytoskeleton. In addition to the impaired decrease in gene expression, the Baf53b+/− het mice also have 62 genes that decrease in expression following behavior that do not decrease in the wildtype (Supplemental Table S11). These aberrantly decreased genes are enriched for GO terms involving mitochondria function.
To further explore the link between the impairments in maintenance of long-term potentiation and cofilin phosphorylation we examined gene expression for GO terms involved in actin cytoskeleton and the post synaptic density. Most genes examined showed similar expression between the Baf53b+/− het mice and the wildtype littermates. However, there were several key genes that showed misregulation either at baseline (homecage) or following OLM training that are involved in regulating the Rac-PAK and RhoA-LIMK pathways that both culminate in phosphorylation of cofilin and actin cytoskeleton reorganization23 (Supplemental Table S12). For example, mir132 has previously been shown to regulate spine plasticity39,40 and long-term OLM41 by regulating Rac1 activity through translational repression of p250GAP39. Additional regulators of this pathway were also disrupted in the Baf53b+/− het mice including Citron (Rho interacting kinase) and Fhl2 (a member of the four-and-a-half LIM only protein family implicated in linking signaling pathways to transcriptional regulation). Components upstream of the Rac-PAK and RhoA-LIMK were also altered in the Baf53b+/− het mice including the NMDA receptor subunits Grinb2 and Grin2a and Ephrin type-A receptor.
Discussion
Epigenetic mechanisms of gene regulation are emerging as a critical mechanism underlying long-term memory processes. The majority of the evidence supporting this idea comes from examination of chromatin modification (e.g. histone acetylation) and DNA methylation. However, nucleosome remodeling, which represents a third major epigenetic mechanism involved in regulating gene expression that has been shown to work together with chromatin modification and DNA methylation in yeast and cancer research, has yet to be investigated with regard to cognitive function. It has become increasingly important to understand the role of nucleosome remodeling considering the recent studies showing that mutations in the BAF complex are associated with intellectual disability disorders in humans12-14. In this study, we show that both a dominant negative and heterozygous knockout of BAF53b produces severe long-term memory and LTP consolidation impairments, providing the first evidence that nucleosome remodeling plays a critical role in synaptic plasticity and memory. The reintroduction of BAF53b into dorsal hippocampus using adeno-associated virus rescued long-term memory deficits in the Baf53b+/− het mice, providing clear evidence for the role of BAF53b in memory formation in the adult, independent of its role in neuronal development. The rescue effect was specific to the hippocampus-dependent OLM task, further supporting a specific role for BAF53b in adult memory formation.
The transgenic approach allowed us to avoid potential effects of expressing mutant BAF53blt, independent of its role in neuronal development. The rescue effect was specific to the hippocampus-dependents in long-term memory for hippocampus-dependent tasks (object location and contextual fear conditioning). In contrast, long-term memory for cued fear was normal in both lines. This suggests that the hippocampus may be more sensitive to alterations in BAF53b function, as cued fear memory is considered hippocampus-independent. BAF53b may also have a key role in cortex-dependent long-term memory as well considering that mutant BAF53b mice also exhibited impaired long-term memory for object recognition. When performed in this way the task mainly depends on the perirhinal and insular cortices indicating a potential role for BAF53b mediated gene expression in these cortical regions.
The observed long-term memory impairments were associated with specific hippocampal long-term potentiation deficits and altered dendritic spine morphology. In general, BAF53b appears to be necessary for LTP consolidation as induction and initial expression of potentiation are normal. However, we did observe significantly increased initial potentiation in the high expressing transgenic line, which then showed a failure to stabilize similar to the low expressing line and the heterozygous knockout line. Interestingly, the more severe LTP phenotype in the BAF53bΔHDhigh mice compared to the BAF53ΔHDlow and Baf53b+/− het mice suggests that higher transgene expression alters the degree to which BAF53b effects induction, initial expression, and consolidation of LTP. Given recent findings that mutations in subunits of the mammalian BAF complex are linked with sporadic autism, sporadic mental retardation and syndromic intellectual disability12-16, the spectrum of synaptic plasticity and memory impairments found with our different manipulations of BAF53b could serve as a novel model for disorders such as Coffin-Siris and Nicolaides-Baraitser syndrome12-14, and intellectual disability in general.
Importantly, the impaired long-term memory and LTP consolidation deficits were associated with significantly altered spine dynamics in the adult Baf53b+/− het mice following TBS. It is generally considered that pCofilin marks spines undergoing activity-dependent actin reorganization and also show an expansion in spine size thought to underlie the maintenance of LTP24,42. Similarly, BAF53bΔHDhigh mice had reduced availability of thin spines, consistent with the post-synaptic impairments in long-term synaptic plasticity. In juvenile and mature hippocampus, it is generally considered that thin spines are those where afferent patterned stimulation induces structural and functional plasticity, leading to incorporation of AMPA-type glutamate receptor in the synapse and a conversion from a thin to a mushroom type spine8,24,25,43. The specific loss of thin spines may indicate a loss in the ability to respond to afferent patterned stimulation by functional and structural plasticity, namely, incorporation of AMPA-type glutamate receptor in the synapse, synapse growth and the conversion of a thin to a mushroom type spine24-27,43. Given that these spine changes occurred relatively early in life (by 3 weeks of age) may contribute to the more severe LTP phenotype observed in this line of mutant mice, more closely mimicking the deficits observed in individuals with intellectual disability.
The RNA Seq data indicates that spine abnormalities may be due to altered BAF53b-mediated gene expression. In the Baf53b+/− het mice several key postsynaptic density genes involved in spine plasticity showed altered expression, including multiple regulators of the Rac-PAK and RhoA-LIMK pathways. For example, mir132 expression was increased following OLM training in wildtype mice, but not in Baf53b+/− het mice. Mir132 has previously been shown to regulate spine plasticity39,40 and long-term OLM41 by regulating Rac1 activity through translational repression of p250GAP39. Mir132 also blocks the translation of methyl CpG-binding protein 2 (MeCP2)44, a transcriptional repressor that interacts with methylated DNA. MeCP2 mutant mice have impairments in a synaptic plasticity and a variety of behaviors including long-term memory formation (for review see 45) indicating a critical role for MeCP2 in normal brain function. In humans mutations in MeCP2 have been linked to Rett syndrome, a developmental disorder characterized by cognitive deficits similar to autism46. In addition to mir132, Baf53b+/− het mice have alterations in gene expression of other key regulators of the Rac-PAK and RhoA-LIMK pathways, both at the level of the PSD (Grinb2 and Grin2a and Ephrin type-A receptor) and at the level of pathway regulation (Citron and Fhl2). Understanding how exactly BAF53b and the nBAF complex is involved in the regulation of these and other genes will be critical, especially considering that spine abnormalities appear to be a common feature in intellectual disabilities including nonsyndromic intellectual disability, Down, Fragile X, and Rett syndromes47.
In the transgenic animals, we targeted a hydrophobic domain of BAF53b. Further analysis will be necessary to fully understand how deletion of the hydrophobic domain impairs nBAF-mediated nucleosome remodeling. Nucleosome remodeling experiments are almost exclusively carried out in vitro and in cell culture, however nucleosome positioning experiments using high-density sequencing are emerging, which may make this more feasible in heterogeneous cell populations of various brain regions associated with learning and memory. In addition, currently available antibodies are insufficient to perform a chromatin immunoprecipitation making the localization of BAF53b to specific DNA binding sites unfeasible. It is also unclear how neuronal activity induced by a learning event activates nBAF-mediated nucleosome remodeling. In cultured neurons nBAF interacts with the calcium-regulated transcriptional activator Calcium-Response Transactivator (CREST)8. Calcium influx activates CREST-mediated transcription that is required for normal activity-dependent dendritic development48,49. In vitro, BAF53b is not required for the CREST-nBAF interaction, but loss of BAF53b does disrupt targeting of both nBAF and CREST to target gene promoters8. CREST expression in the hippocampus continues into adulthood48 and may serve as potential mechanism to link neuronal activity to nBAF mediated nucleosome remodeling.
In summary, recent studies have shown that mutations in various subunits of the BAF complex cause intellectual disability disorders in humans12-17 and elegant studies in culture have demonstrated that BAF53b in particular is necessary for dendritic outgrowth8. However, there remained no clear evidence linking BAF-mediated nucleosome remodeling to cognition. In this study, we demonstrate for the first time that the neuron-specific BAF53b subunit of the neuronal BAF complex is necessary for synaptic plasticity and long-term memory processes, likely via the regulation of gene expression required for spine structure and function. These findings support the overall idea that impaired nucleosome remodeling may be a key underlying mechanism to intellectual disability disorders.
Database
The RNA Sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus50 and are accessible through GEO Series accession number GSE44229 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44229).
Online Methods
Animals
All animals were between 8-15 weeks old at the time of behavioral testing. Mice had free access to food and water and lights were maintained on a 12:12 h light/dark cycle, with all behavioral testing performed during the light portion of the cycle. All experiments were conducted according to National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. All animals were group housed and backcrossed at least five generations to C57BL/6 (Jackson Labs). Official Mouse Genome Database strain name for BAF53b Δ HD mutant mice: B6J.CB6-Tg(CamK2α-Actl6b*)Mwood.
Object Location and Novel Object Recognition Paradigms
Object location and object recognition were performed as previously described28,51. Prior to training, mice are handled 1-2 min for 5 days and then habituated to the experimental apparatus for 5 min a day for 4-6 days in the absence of objects. During the training period, mice were placed into the experimental apparatus with two identical objects (100 ml beakers or light bulbs or vases) and allowed to explore for 10 min51. During the retention test, (24 hrs for long-term memory or 90 minutes for short-term memory), mice were allowed to explore the experimental apparatus for 5 min. Exploration was scored when a mouse's head was oriented towards the object within a distance of 1 cm or when the nose was touching the object. The relative exploration time (t) is recorded and expressed as a discrimination index (D.I. = (tnovel − tfamiliar) / (tnovel + tfamiliar) × 100%). Mean exploration times were then calculated and the discrimination indexes between treatment groups compared. Animals that explored less then 3 sec total for both objects during either training or testing were removed from further analysis. Animals that demonstrated an object preference during training (D.I. > +/− 20) were also removed.
Elevated plus maze
The plus-maze consisted of two open arms, 30 × 5 cm, and two enclosed arms, 30 (1) × 5 (w) × 15 (h) cm. The arms extended from a central platform 5 × 5 cm, and the apparatus was raised to a height of 40 cm above the floor. The light level was adjusted to 15 Lux. Testing (5min) consisted of placing an animal onto the central platform of the maze facing an open arm. Between subjects the maze was cleaned with 70% ethanol. The percentage of time spent in the closed and open arms was scored using EthoVision 3.1 (Noldus Information Technology).
Fear Conditioning
Contextual and cued fear conditioning were performed as previously described28,51,52. For contextual fear conditioning, a single 2sec, 0.70mA scrambled footshock was given in at 2:28 of a 3:00min training period followed by a 5min testing period 24 hrs later. For cued fear conditioning, animals were in the chamber for two minutes followed by a 30sec tone that co-terminated with a 2sec, 0.70mA scrambled footshock. Animals were allowed to remain in the chamber for an additional 30sec. Twenty-four hours after conditioning, animals were tested in a novel context (2min) followed by a 3min tone. Freezing behavior was measured using EthoVision 3.1 (Noldus Information Technology).51
Phosphorylated cofilin Analysis
Hippocampal slices were prepared as described above. Following a baseline recording, slices were either given control stimulation (3 pulses/min) or TBS. All the slices were collected 7 minutes after stimulation and placed into cold 4% paraformaldehyde. As previously described53, hippocampal slices were subsection of a freezing microtome and slide mounted. Immunocytochemical labeling was done by washing slices with 0.1M phosphate buffer, then placed in a cocktail of primary antibodies pCofilin (ABCam, AB12866; 1:250) and PSD95 (ThermoScientific, MA1-045, 1:800) in diluent which included 0.1M PB, 0.3% Triton, and 1.8% bovine serum albumin. Sections were exposed to primary antibody for two nights at 4°C. Tissue was then rinsed 3 times with 0.1M PB and incubated in secondary antibodies for donkey anti-mouse Alexafluor 488 (Life Technologies; A-21202) and donkey anti-rabbit Alexafluor 594 (Life Technologies; A-2107).
Imaging and quantification were completed as described in Babayan et al 2012. Briefly, images were acquired on a Leica DM6000B using a 63× Plan Apo objective (1.4 numerical aperture). Image acquisition and deconvolution (99% confidence) were done using Volocity 6.0 (Perkin-Elmer) at which time deconvolved images were exported and quantified using previously described in-house analysis software.
Spine Analysis
Immunohistochemistry was carried out on 20-μm free-floating sections from perfused, fixed brains as described previously28,51. Mouse anti-GFP (1:8,000, Sigma) and anti-mouse IgG conjugated to Alexa Fluor 488 (1:200, Molecular Probes) antibodies were used. Sections from all experimental groups were run concurrently in the same conditions, and analyzed without knowledge of treatment group. The analysis was performed as previously described54.
Immunohistochemistry
Immunohistochemistry was carried out as previously described28,51. Antibodies included: mouse anti-NeuN (1:1000; MAB377, Millipore Corporation), rabbit anti-BAF53b (1:500; generous gift from G.R. Crabtree), goat anti-mouse IgG–FITC secondary antibody (1:1000; Millipore Bioscience Research Reagents), and donkey anti-rabbit IgG-CY3 (1:1000; Jackson ImmunoResearch).
Crestyl Violet and Luxol Blue staining
Animals were transcardially perfused with saline (2min) followed by 4% paraformaldehyde (10min). Brains were post-fixed overnight (4% PFA) followed by 30% sucrose (two days) and then flash frozen. Luxol Blue and Crestyl Violet staining were performed as previously described55. Briefly, free floating sections (50um) were washed in 0.1mPBS (3×10min), 70% ethanol (2×5min) and incubated in 70% ethanol overnight (room temperature). Sections were transferred to prewarmed Luxol Fast Blue solution (0.1% Luxol Fast Blue Solvent 38 (Sigma) dissolved in 96% ethanol and 10% acetic acid) and incubated overnight at 56C. Sections were washed sequentially (3min) in water, 0.1M PBS, and lithium carbonate solution (0.05% lithium carbonate in water). Sections were differentiated in 70% ethanol (3min), washed twice (5min) in 0.1M PBS, mounted and dried overnight. The next day sections were crestyl violet stained: defatting (30min 50% chloroform/50% ethanol), dehydrations (100%, 95%, 70% and 50% ethanol sequentially), rinsed (water), Cresyl Violet stained (6 dips), rehydrated (50%, 70%, 95%, 100% ethanol), cleared (Hemo-De) and coversliped.
Western Blot
Dorsal hippocampus was homogenized in RIPA buffer (Boston BioProducts, BP-115) with protease and phosphatase inhibitors (ThermoScientific, 1861281) using a dounce homogenizer. BAF53b homozygous knockout brain tissue was collected from P2 pups to verify antibody specificity. Protein lysates were quantified using a modified Bradford assay (BioRad, 500-0006). 10ug of total protein lysate were loaded per lane on a 4-12% Bis Tris gel (Novex) and run for 50min at 200V. Blots were then transferred overnight at 15V at 4C on to 0.45um nitrocellulose membranes (Novex). Total protein transfer was verified using PonseauS staining. Membranes were blocked in StartingBlock T20 (ThermoScientific, 37543) for 30min and then probed with rabbit anti-BAF53b (1:1,000; generous gift from G.R. Crabtree). The membranes were washed (3×10min in 0.1MPBS with .05%Tween20) and probed with donkey anti-rabbit-HRP (1:10,000; Jackson ImmunoResearch, 711-036-152). The membranes were washed and developed using Pierce SuperSignal West Pico Chemiluminescent Substrate (Pierce, 34077). Multiple film exposures were used to verify linearity. Blots were washed and then reprobed with rabbit anti-GAPDH (1:1,000; Santa Cruz Biotechnology, SC25778). Relative densities were taken from scanned film using ImageJ (NIH). All values were normalized to GAPDH expression levels.
Slice preparation and recording
Hippocampal slices were prepared as previously described28 from Baf53b+/− het mice,BAF53ΔHDlow, BAF53ΔHDhigh, and wildtype mice (approximately 2 months of age). Transverse hippocampal slices (300 μm) through the mid-third of the septotemporal axis of the hippocampus were placed in an interface recording chamber containing preheated artificial cerebrospinal fluid (ACSF; in mM): 124 NaCl, 3 KCl, 1.25 KH2PO4, 1.5 MgSO4, 2.5 CaCl2, 26 NaHCO3, and 10 D-glucose and maintained at 31 ± 1°C). Slices were continuously perfused with at a rate of 1.75-2 ml/min while the surface was exposed to warm, humidified 95% O2 / 5% CO2. Recordings began following at least 2 hr of incubation.
Field excitatory postsynaptic potentials (fEPSPs) were recorded from CA1b stratum radiatum using a single glass pipette (2-3 MΩ). Bipolar stainless steel stimulation electrodes (25 μm diameter, FHC) were positioned at two sites (CA1a and CA1c) in the apical Schaffer collateral-commissural projections to provide activation of separate converging pathways of CA1b pyramidal cells. Pulses were administered in an alternating fashion to the two electrodes at 0.03 Hz using a current that elicited a 50% maximal response. After establishing a stable baseline, long-term potentiation (LTP) was induced by delivering 5 or 10 ‘theta’ bursts (each burst was four pulses at 100 Hz and bursts were separated by 200 msec). Data were collected and digitized by NAC 2.0 Neurodata Acquisition System (Theta Burst Corp.).
Slices used for whole-cell recordings were prepared as previously described23,56. Briefly, slices were placed in a submerged recording chamber and continuously perfused at 2–3 ml/min with oxygenated (95% O2/5% CO2) ACSF at 32°C. Whole-cell recordings were made with 3–5 MΩ recording pipettes filled with solution of the following composition (in mM): 130 K-gluconate, 0.1 EGTA, 0.5 MgCl2, 10 HEPES, 2 ATP (pH 7.25, 285 mosM) using an Axopatch 200A amplifier (Molecular Devices). Miniature excitatory postsynaptic currents (mEPSCs) were recorded at a holding potential of −70 mV in the presence of tetrodotoxin (1 μM) and bicuculline (50 μM). Data were filtered at 2 kHz, digitized at 1–5 kHz, stored on a computer, and analyzed off-line using Mini Analysis Program (Synaptosoft), Origin (OriginLab), and pCLAMP 7 (Molecular Devices) software.
AAV Production and Infusion
The wildtype BAF53b was V5 tagged and cloned into the pAAV-IRES-hrGFP Vector (Agilent Technologies) under the CMV promoter. Adeno-associated virus (AAV) serotype 2/1 for AAV-Baf53b was purchased from Penn Vector Core; University of Pennsylvania. AAV-hrGFP serotype 2/1 was generated and viral titer was determined by qPCR with primers specific to hrGFP57. A titer of 1010-1013 genome copies/ul was used for all injections.
As previously described two weeks prior to the experiments, 1.0ul of virus was infused into the dorsal hippocampus (AP 2.0 mm; ML ±1.5 mm; 1.5mm DV from bregma) at a rate of 6 ml/h28,51. All infusions were confirmed by immunohistochemistry.
RNA-Sequencing
RNA was isolated from bilateral, dorsal hippocampus using the using RNeasy minikit (Qiagen, 74104). RNA quality was assessed by Bioanalyzer, and only samples with a RIN greater than 9 were included for analysis. cDNA libraries for each group were prepared as described in the TruSeq RNA Sample Preparation v2 Guide (Illumina). Briefly, 3ug of total RNA from each animal was used as starting material. The mRNA was purified with poly-T oligo-attached magnetic beads and heat fragmented. The first and second strand cDNA were then synthesized and purified. The ends were blunted and the 3′ end was adenylated to prevent concatenation of the template during adapter ligation. For each group a unique adapter set was added to the ends of the cDNA and the libraries amplified by PCR. The quality of the library was assessed by Bioanalyzer and quantified using qRT-PCR with a standard curve prepared from a purchased sequencing library (Illumina). Samples were multiplexed so that each behavioral group was represented in each flow cell of the sequencer. 10nM of each library was pooled together in four multiplex libraries and sequenced on an IlluminaHiSeq 2000 instrument during a single-end 50 cycles sequencing run by the Genomics High-Throughput Facility (GHTF) at the University of California, Irvine. The resulting sequencing data for each library were post-processed to produce FastQ files, then demultiplexed and filtered using both Illumina software CASAVA 1.8.2 and in-house software. Control reads successfully aligned to the PhiX control genome or poor quality reads failing Illumina's standard quality tests were removed from the following analyses. The quality of the remaining sequences was further assessed using the PHRED quality scores produced in real-time during the base-calling step of the sequencing run (Supplemental FigS6A).
Alignment to the Reference Genome and Transcriptome
The reads from each replicate experiment were separately aligned to the reference genome mm929 and corresponding transcriptome using the short-read aligners ELAND v2e (Illumina) and Bowtie30. Reads uniquely aligned by both tools to known exons or splice junctions with no more than two mismatches were included in the transcriptome. Reads uniquely aligned but with more than two mismatches, or reads matching several locations in the reference genome, were removed from the analysis. The percentage of reads assigned to the reference genome and transcriptome using this protocol is reported for each group of replicates (Supplemental Fig S6B) with the corresponding percentage of covered genes (Supplemental Fig S6B).
Gene Expression and Differential Analysis
Gene expression levels were directly computed from the read alignment results for each replicate in the different groups. Standard RPKM values31 (reads per kilobase of exon model per million mapped reads) were extracted for each gene covered by the sequencing data and each replicate used in this study.
Differential transcriptional analyses were performed using CyberT32,33 across each pair of groups: WT HC, WT Beh, Baf53b+/− HC, and Baf53b+/− Beh. Additional control analyses were performed using CASAVA. For different threshold values the corresponding number of genes predicted to be differentially regulated, up-regulated, or down-regulated was examined. Enrichment for specific tissue expression, GO terms35, and KEGG pathways36,37 was assessed using DAVID38.
qRT-PCR
Quantitative RT-PCR (qRT-PCR) was performed using methods previously described 28,51. Tissue was collected from dissections of dorsal hippocampal hippocampus and stored in RNALater (Invitrogen, AM7020) at −80C until processed. RNA was isolated using RNeasy minikit (Qiagen, 74104). cDNA was made from 500-1000ng total RNA using the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Sciences, 04379012001). Primers and probes were derived from the Roche Universal ProbeLibrary (Supplemental Table S13) and used for multiplexing in the Roche Light- Cycle 480 II machine (Roche Applied Sciences). All values were normalized to gapdh expression levels. Analysis and statistics were performed using the Roche proprietary algorithms and REST 2009 software based on the Pfaffl method58,59.
Statistical Analysis
Statistical analysis was conducted as indicated in the text and figure legends using Prism (GraphPad). Parametric tests (t-test or ANOVA) were used where assumptions of normality (Shapiro-Wilk) and equal variance (F test or Bartlett's test) were met and were replaced by nonparametric equivalents where appropriate (Mann Whitney U or Kruskal-Wallis). Main effects and interactions for all ANOVAs or Kruskal-Wallis tests are described in the text. The appropriate post hoc tests (Bonferroni or Dunn's) are listed for each condition examined. All t-tests were two-tailed using a p value of 0.05.
Supplementary Material
Acknowledgments
This work was supported by a grant from NIMH (MH081004) to M.A.W. and training grant support from NIMH (T32-AG00096-29) to A.V.C. T.Z.B. was supported by NIH NS 28912 and MH73136. The work of M.Z., C.M., and P.B. was supported by grants NSF (IIS-0513376), NIH (LM010235), and NIH NLM (T15 LM07443) to PB. We wish also to acknowledge the UCI Institute for Genomics and Bioinformatics, the UCI Genomics High-Throughput Facility, and Jordan Hayes for additional computing support. G.L., E.K. and Y.J. were supported by NIH (P01 NS045260) and ONR (#N00014-10-1-0072). We would especially like to thank Dr. John Guzowski and Dr. Teiko Miyashita for their technical expertise and the use of Dr. Guzowski's Olympus BX61 microscope and XC10 camera.
Footnotes
Author Contributions: A.V.C, D.P.M., R.M.B. and M.A.W. designed the experiments. A.V.C, D.P.M., and R.M.B. conducted experiments. A.V.C. and M.A.W. wrote the manuscript. E.K. and Y.J. conducted electrophysiological experiments. A.B. conducted pCofilin experiments. Y.C. and T.Z.B. designed and conducted the spine analysis. C.N.M., M.Z., and P.B. performed the RNA Sequencing analysis. S.A., A.S., J.H., A.T., R.D., and R.J.P. performed behavioral experiments. M.C. made the AAV-hrGFP virus. J.I.W. and G.R.C. provided technical assistance and assisted in manuscript preparation. P.B., T.Z.B. and G.L. assisted in experimental design, data analysis, and manuscript preparation.
The authors have no competing financial interests.
References
- 1.Day JJ, Sweatt JD. Cognitive neuroepigenetics: a role for epigenetic mechanisms in learning and memory. Neurobiol Learn Mem. 2011;96:2–12. doi: 10.1016/j.nlm.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahir FR, Brown CJ. Epigenetic impacts on neurodevelopment: pathophysiological mechanisms and genetic modes of action. Pediatr Res. 2011;69:92R–100R. doi: 10.1203/PDR.0b013e318213565e. [DOI] [PubMed] [Google Scholar]
- 3.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Nature Publishing Group. 2011:1–25. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu JI, Lessard J, Crabtree GR. Understanding the Words of Chromatin Regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. The EMBO Journal. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 6.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & Development. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 8.Wu JI, et al. Regulation of Dendritic Development by Neuron-Specific Chromatin Remodeling Complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 13.Santen GWE, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 14.Van Houdt JKJ, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–9. S1. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- 15.Halgren C, et al. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clinical genetics. 2011;82:248–255. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer J, Ekici AB, Endele S, Popp B, Zweier C. Haploinsufficiency of ARID1B, a Member of the SWI/SNF-A Chromatin-Remodeling Complex, Is a Frequent Cause of Intellectual Disability. The American Journal of Human Genetics. 2012;90:565–572. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Molecular and Cellular Biology. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayford M, et al. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 20.Kojima N, et al. Rescuing impairment of long-term potentiation in fyn-deficient mice by introducing Fyn transgene. Proc Natl Acad Sci USA. 1997;94:4761–4765. doi: 10.1073/pnas.94.9.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson J, Wong D, Lynch G. Patterned stimulation at the theta frequency is optimal for the induction of hippocampal long-term potentiation. Brain Research. 1986;368:347–350. doi: 10.1016/0006-8993(86)90579-2. [DOI] [PubMed] [Google Scholar]
- 22.Lauterborn JC, et al. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. Journal of Neuroscience. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rex CS, et al. Different Rho GTPase–dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J Cell Biol. 2009;186:85–97. doi: 10.1083/jcb.200901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in Synaptic Morphology Accompany Actin Signaling during LTP. Journal of Neuroscience. 2007;27:5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- 27.Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett RM, et al. Hippocampal Focal Knockout of CBP Affects Specific Histone Modifications, Long-Term Potentiation, and Long-Term Memory. Neuropsychopharmacology. 2011;36:1545–1556. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 32.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012;40:W553–9. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 34.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dennis G, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 39.Wayman GA, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Impey S, et al. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedulov V, et al. Evidence That Long-Term Potentiation Occurs within Individual Hippocampal Synapses during Learning. Journal of Neuroscience. 2007;27:8031–8039. doi: 10.1523/JNEUROSCI.2003-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukazawa Y, et al. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- 44.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 45.Nelson ED, Monteggia LM. Epigenetics in the mature mammalian brain: effects on behavior and synaptic transmission. Neurobiol Learn Mem. 2011;96:53–60. doi: 10.1016/j.nlm.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amir RE, Van den Veyver IB, Wan M, Tran CQ. Rett syndrome is caused by mutations in X-linked MECP2, encoding methylCpG-binding protein 2. Nature. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 47.Levenga J, Willemsen R. Perturbation of dendritic protrusions in intellectual disability. Prog Brain Res. 2012;197:153–168. doi: 10.1016/B978-0-444-54299-1.00008-X. [DOI] [PubMed] [Google Scholar]
- 48.Aizawa H. Dendrite Development Regulated by CREST, a Calcium-Regulated Transcriptional Activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- 49.Qiu Z, Ghosh A. A Calcium-Dependent Switch in a CREST-BRG1 Complex Regulates Activity-Dependent Gene Expression. Neuron. 2008;60:775–787. doi: 10.1016/j.neuron.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. Journal of Neuroscience. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vecsey CG, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. Journal of Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babayan AH, et al. Integrin dynamics produce a delayed stage of long-term potentiation and memory consolidation. Journal of Neuroscience. 2012;32:12854–12861. doi: 10.1523/JNEUROSCI.2024-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, et al. Impairment of synaptic plasticity by the stress mediator CRH involves selective destruction of thin dendritic spines via RhoA signaling. Mol Psychiatry. 2012:1–12. doi: 10.1038/mp.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geisler S, Heilmann H, Veh RW. An optimized method for simultaneous demonstration of neurons and myelinated fiber tracts for delineation of individual trunco-and palliothalamic nuclei in the mammalian brain. Histochemistry and cell biology. 2002;117:69–79. doi: 10.1007/s00418-001-0357-z. [DOI] [PubMed] [Google Scholar]
- 56.Colgin LL, Jia Y, Sabatier JM, Lynch G. Blockade of NMDA receptors enhances spontaneous sharp waves in rat hippocampal slices. Neuroscience Letters. 2005;385:46–51. doi: 10.1016/j.neulet.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Lawlor PA, Bland RJ, Das P, Price RW. Novel rat Alzheimer's disease models based on AAV-mediated gene transfer to selectively increase hippocampal Abeta levels. Molecular Neurodegeneration. 2007;2 doi: 10.1186/1750-1326-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfaffl MW, et al. Real-time RT-PCR quantification of insulin-like growth factor (IGF)-1, IGF-1 receptor, IGF-2, IGF-2 receptor, insulin receptor, growth hormone receptor, IGF-binding proteins 1, 2 and 3 in the bovine species. Domest Anim Endocrinol. 2002;22:91–102. doi: 10.1016/s0739-7240(01)00128-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.