Abstract
Posttranslational modification of histone proteins in eukaryotes plays an important role in gene transcription and chromatin structure. Dysregulation of the enzymes involved in histone modification has been linked to many cancer forms, making this target class a potential new area for therapeutics. A reliable assay to monitor small-molecule inhibition of various epigenetic enzymes should play a critical role in drug discovery to fight cancer. However, it has been challenging to develop cell-based assays for high-throughput screening (HTS) and compound profiling. Recently, two homogeneous cell-based assay kits using the AlphaLISA® and LanthaScreen® technologies to detect trimethyl histone H3 Lysine 27 have become commercially available, and a heterogeneous cell assay with modified dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA®) format has been reported. To compare their pros and cons, we evaluated, optimized, and validated these three assay formats in three different cell lines and compared their activities with traditional Western blot detection of histone methylation inhibition by using commercial and in-house small-molecule inhibitors. Our data indicate that, although all four formats produced acceptable results, the homogeneous AlphaLISA assay was best suited for HTS and compound profiling due to its wider window and ease of automation. The DELFIA and Western blot assays were useful as validation tools to confirm the cell activities and eliminate potential false-positive compounds.
Introduction
Epigenetics refers to the heritable changes in gene expression or cellular phenotype caused by mechanisms other than alterations to the underlying DNA sequence.1 Epigenetic modifications occur through histone modification and DNA methylation. The nucleosome is composed of an octamer of the four core histones wrapped with 147 bp DNA. The core histone proteins are H2A, H2B, H3, and H4. The N-terminus of the core histone proteins is subject to posttranslational modifications, such as methylation, acetylation, ubiquitination, and phosphorylation.2,3
Acetylation or methylation of lysine residues on histone H3 and H4 affects transcription, that is, it can either increase or decrease gene expression.4,5 Methylation of the N-terminal tail region of histone H3 has been investigated extensively, revealing that several lysine residues (K4, K9, K27, K36, and K79) are subject to modifications that include demethylation and mono-, di-, and trimethylation. Abnormal regulation of these posttranslational modifications has been shown to be linked to various human diseases, including cancer.6–8 For example, hypertrimethylation of K27 in histone H3 [trimethyl histone H3 Lysine 27 (H3K27me3)] was observed in many human cancers.6 Further, a subset of human B-cell lymphoma was linked to point mutation at Y641 in Enhancer of Zeste homolog 2 (EZH2), the catalytic subunit within the five-member Polycomb repressive complex 2 that is responsible for repressing gene transcription by methylating histone H3 at K27.6 Inhibition of histone lysine methyltransferases such as EZH2 can decrease the level of H3K27me3; thus, these methyltransferases constitute a novel class of drug target for cancer therapeutics.9 A recent study supported this concept by reporting that a small-molecule EZH2 inhibitor, GSK126, may provide a treatment option for EZH2 mutant lymphoma.10
Multiple biochemical assays have been reported in the drug discovery effort for small-molecule inhibitors against different epigenetic enzymes.11–15 However, cell-based assays are more biologically relevant and are critical in elucidating the mechanism of action for these inhibitors. Recently, two groups published two cell-based assays detecting H3K27me3 levels; the first was a dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA®)16 and the second was a LanthaScreen® assay.17 Here we report four different assay formats for the measurement of H3K27me3. They are AlphaLISA®, DELFIA, LanthaScreen, and Western blot. The half-maximal inhibitory concentration (IC50) potencies of commercially available and proprietary reference compounds were compared among the four assay formats.
Materials and Methods
Materials
AlphaLISA and LanthaScreen kits for assay development, optimization, and validation were performed in 384-well, tissue culture-treated, white opaque CulturPlates (No. 6007680; PerkinElmer, Waltham, MA). For the DELFIA, the cells were seeded in 384-well, clear-bottom cell culture plates (No. 78109; Greiner Bio-One, Monroe, NC). The cell lysate was transferred to 384-well Nunc high-binding MaxiSorp plates (No. 460518; Rochester, NY). 3-Deazaneplanocin A (DZNep) was purchased from Cayman Chemical (No. 13828; Ann Arbor, MI). RBC081 and RBC124 are internal compounds and part of the Reaction Biology Corporation's compound library collection. Peptide H3K27me3 was purchased from AnaSpec (No. 64378-1; Fremont, CA).
Cell Culture and Cryopreservation
All cell lines in this report were obtained from American Type Culture Collection and maintained in the recommended cell culture media at 37°C in 5% CO2. The cells were transferred to a tube and spun down at 1,000 rpm for 5 min on Beckman (Model # GS-6KR). Cell pellets were resuspended in a freezing medium containing 90% fetal bovine serum (FBS) and 10% dimethylsulfoxide (DMSO), and frozen in a Nalgene® Mr. Frosty device (No. 5100-0001; ThermoFisher Scientific, Rochester, NY) at −80°C for 1–2 h, followed by storage in a liquid N2 tank.
AlphaLISA Assay of H3K27me3
The AlphaLISA cell assay was performed utilizing the H3K27me3 Cellular Detection Kit from PerkinElmer (No. AL722C/F). Cryopreserved cells from the SU-DHL-6 human lymphoma cell line were thawed at 37°C for 2 min, followed by centrifugation at 1,500 rpm for 5 min on Beckman (Model # GS-6KR) to remove freezing media. The cell pellet was resuspended at 2.5×105 cells/mL in alpha minimum essential medium assay media (No. SH30265.02; ThermoFisher Scientific) with 15% FBS. The cell suspension (10 μL) was dispensed into assay plates at 2,500 cells per well. DZNep and the internal compound RBC124 were serially diluted (10 concentrations with 1:3 dilution factor) at a 3× final assay concentration in 384-well plates. Five microliters of a compound or DMSO was transferred to the assay plate, with a final DMSO concentration of 1%. The assay plate was incubated at 37°C in 5% CO2 for 3 days, before adding 5 μL of lysis buffer to each well. The plate was then incubated at room temperature for 15 min. Next, 10 μL of Cell-Histone Extraction buffer was added to each well of the plates, followed by incubation at room temperature for 10 min. Then, a 10-μL mix of Acceptor beads (100 μg/mL) coated with the anti-H3K27me3 antibody and biotinylated anti-H3 antibody (15 nM) diluted in 1× Cell-Histone Detection buffer was dispensed to each well followed by incubation at room temperature for 1 h. The detection buffer/AlphaLISA streptavidin (SA) donor beads (10 μL at 100 μg/mL) were then added to each well in the dark, followed by incubation with shaking at room temperature for 2 h. Finally, the assay plate was read on an EnVision™ 2101 multilabel plate reader (PerkinElmer) using an AlphaScreen® mirror and laser as a light source at 680 nm excitation and 570 nm emission (Table 1). IC50 values were determined using the four-parameter logistic equation in GraphPad Prism 5 (La Jolla, CA).
Table 1.
AlphaLISA Assay Protocol Table
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 10 μL | 2,500 Su-DHL-6 cells |
| 2 | Control | 5 μL | DMSO |
| 3 | Library compounds | 5 μL | DZNep, RBC124 |
| 4 | Incubation time | 3 days | 37°C, 5% CO2 |
| 5 | Lysis buffer | 5 μL | |
| 6 | Incubation time | 15 min | Ambient temperature |
| 7 | Extraction buffer | 10 μL | |
| 8 | Incubation time | 10 min | Ambient temperature |
| 9 | Acceptor beads+antibody | 10 μL | 100 μg/mL Acceptor beads; 15 nM antibody |
| 10 | Incubation time | 1 h | Ambient temperature |
| 11 | Donor beads | 10 μL | 100 μg/mL Donor beads |
| 12 | Incubation time | 2 h | Ambient temperature |
| 13 | Assay readout | 680 and 570 nm | EnVision plate reader, AlphaScreen mode |
Step Notes
1. 384-well white opaque CulturPlates.
3. 100 μM to 15 nM dilution series.
4. Plates were sealed with sterile breathable tape.
5,7. From the H3K27me3 Cellular Detection Kit.
6,8,10. Plates lidded.
9,11. In 1× Cell-Histone Detection buffer from the H3K27me3 Cellular Detection Kit.
12. Plates lidded until read.
13. Excitation with laser (680 nm), emission filter (570/100 nm).
DMSO, dimethylsulfoxide; DZNep, 3-Deazaneplanocin A.
LanthaScreen Assay of H3K27me3
The LanthaScreen cell assay was performed utilizing the BacMam Histone H3K27me3 Cellular Assay Kit from Life Technologies Corporation (No. A14159; Carlsbad, CA). Cryopreserved cells from the HeLa human cervical cancer cell line were thawed at 37°C for 2 min, followed by centrifugation at 1,500 rpm for 5 min on Beckman (Model # GS-6KR) to remove freezing media. The cell pellet was resuspended at 3.7×105 cells/mL in assay media (Dulbecco's modified Eagle's medium with 10% dialyzed FBS, 0.1 mM nonessential amino acid, 100 U/mL penicillin, 100 μg/mL streptomycin) containing a 0.5× BacMam enhancer solution. The cell suspension (14 μL) was dispensed into 384-well assay plates at 5,000 cells per well. The cells were incubated at 37°C in 5% CO2 overnight. The next day, 6 μL BacMam Histone H3 reagent was dispensed to each well. DZNep and the internal compound RBC124 were serially diluted (10 concentrations with 1:3 dilution factor) at a 5× final assay concentration in 384-well plates. Five microliters of a compound or DMSO was transferred to the assay plate, with a final DMSO concentration of 1%. The assay plate was incubated at 37°C in 5% CO2 for 3 days. Cells were lysed with 5 μL 6× complete lysis buffer containing a 3× protease inhibitor and 12 nM Terbium-anti-Histone H3K27me3 antibody, followed by incubation in the dark at room temperature for 2–3 h. The assay was read on an EnVision plate reader with excitation wavelength at 340 nm and emission wavelengths at 520 and 492 nm (Table 2).
Table 2.
LanthaScreen Assay Protocol Table
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 14 μL | 5,000 HeLa cells |
| 2 | Incubation time | Overnight | 37°C, 5% CO2 |
| 3 | Virus | 6 μL | BacMam Histone H3 Reagent |
| 4 | Control | 5 μL | DMSO |
| 5 | Library compounds | 5 μL | DZNep, RBC124 |
| 6 | Incubation time | 3 days | 37°C, 5% CO2 |
| 7 | Lysis buffer+antibody | 5 μL | 6× complete lysis buffer with 3× protease inhibitor, 12 nM antibody |
| 8 | Incubation time | 2–3 h | Ambient temperature |
| 9 | Assay readout | 340 and 520, 492 nm | EnVision plate reader, TR-FRET mode |
Step Notes
1. 384-well white opaque CulturPlates.
2,6. Plates were sealed with sterile breathable tape.
3. From the BacMam Histone H3K27me3 Cellular Assay Kit.
5. 33 μM to 5 nM dilution series.
7. 6× stock concentration lysis buffer, 100× stock concentration protease inhibitor, 3,400 nM stock concentration Terbium-anti-Histone H3K27me3 antibody.
8. Plates lidded until read, in the dark.
9. Excitation with laser (340 nm), emission filter (520/25, 492/8 nm).
TR-FRET, time-resolved fluorescence resonance energy transfer.
DELFIA of H3K27me3
The experimental procedure for the DELFIA cell assay was modified based on the publication by Xie et al.16 Cryopreserved cells from the PC3 human prostate carcinoma cell line were thawed at 37°C for 2 min, followed by centrifugation at 1,500 rpm for 5 min on Beckman (Model # GS-6KR) to remove freezing media. The cell pellet was resuspended at 1.1×105 cells/mL in assay media (RPMI-1640 medium supplemented with 10% FBS). The cell suspension (45 μL) was dispensed into 384-well clear-bottom Greiner Bio-One assay plates (No. 78109) at 5,000 cells per well. The cells were incubated at 37°C in 5% CO2 overnight. DZNep and the internal compound RBC124 were serially diluted (10 concentrations with 1:3 dilution factor) at a 10× final assay concentration in 384-well plates. Five microliters of a compound or DMSO was transferred to the assay plate, with a final DMSO concentration of 1%. The assay plate was incubated at 37°C in 5% CO2 for 3 days. The medium was aspirated and the plates were frozen at −80°C for at least 1 h. Before assay, the frozen plates were equilibrated at room temperature for 30 min. The cells were lysed with 50 μL of 0.2 normality (N) hydrochloric acid (HCl) at 4°C overnight. After lysis, the cell pellets were pipetted up and down for 10 cycles. Twenty microliters of cell lysate was transferred to two 384-well high-binding MaxiSorp plates. One copy was used for detection of H3K27me3; the other was used for detection of total histone H3. After cell lysate transfer, 5 μL/well of 5× neutralization buffer was added. The plates were incubated for 1 h at room temperature, followed by one wash with Tris-buffered saline Tween-20 (TBS-T) at 50 μL/well and two washes with TBS-T at 100 μL/well. For blocking, 100 μL/well of SuperBlock blocking buffer was added and incubated for at least 1 h at room temperature. The plates were washed three times with 100 μL/well TBS-T washing buffer. The detection antibody, rabbit anti-H3K27me3 (No. ABE44; Millipore, Billerica, MA) or rabbit anti-histone H3 antibody (No. 9715S; Cell Signaling, Danvers, MA), was then added at 25 μL/well to the whole plate. The rabbit anti-H3K27me3 antibody was diluted in SuperBlock-T20 to 1 μg/mL. The rabbit anti-total histone H3 antibody was diluted in SuperBlock-T20 to 0.29 μg/mL. After 1 h incubation at room temperature, the plates were washed three times at 100 μL/well of TBS-T. Then, 25 μL/well europium-N1-labeled anti-rabbit IgG antibody (No. AD0105; PerkinElmer), which was diluted 3,000-fold in SuperBlock-T20, was added. After 1 h incubation, the plates were washed three times with 100 μL/well of TBS-T and two times with 120 μL/well of TBS-T. Next, 25 μL/well of DELFIA Enhancement solution was added. After a 30-min incubation at room temperature, the plates were read with an EnVision plate reader with excitation wavelength at 320 nm, emission wavelength at 615 nm, and a 505 nm dichroic mirror (Table 3).
Table 3.
Dissociation-Enhanced Lanthanide Fluorescence Immunoassay Protocol Table
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Plate cells | 45 μL | 5,000 PC3 cells |
| 2 | Incubation time | Overnight | 37°C, 5% CO2 |
| 3 | Control | 5 μL | DMSO |
| 4 | Library compounds | 5 μL | DZNep, RBC124 |
| 5 | Incubation time | 3 days | 37°C, 5% CO2 |
| 6 | Remove media | ||
| 7 | Freeze plates | >1 h | −80°C |
| 8 | Lysis buffer | 50 μL | 0.2 N HCl |
| 9 | Incubation time | Overnight | 4°C |
| 10 | Transfer lysate | 20 μL | |
| 11 | Neutralization buffer | 5 μL | |
| 12 | Incubation time | 1 h | Ambient temperature |
| 13 | Wash plates | 50 μL once, 100 μL twice | TBS-T |
| 14 | Blocking | 100 μL | SuperBlock blocking buffer |
| 15 | Incubation time | 1 h | Ambient temperature |
| 16 | Wash plates | 100 μL three times | TBS-T |
| 17 | Primary antibody | 25 μL | 1 μg/mL rabbit anti-H3K27me3 or 0.29 μg/mL rabbit anti-histone H3 |
| 18 | Wash plates | 100 μL three times | TBS-T |
| 19 | Secondary antibody | 25 μL | europium-N1-labeled anti-rabbit IgG antibody |
| 20 | Wash plates | 100 μL three times, 120 μL two times | TBS-T |
| 21 | Enhancement solution | 25 μL | |
| 22 | Incubation time | 30 min | Ambient temperature |
| 23 | Assay readout | 320 and 615 nm | EnVision plate reader, TRF mode |
Step Notes
1. 384-well clear-bottom Greiner Bio-One assay plate.
2,5. Plates were sealed with sterile breathable tape.
4. 100 μM to 15 nM dilution series.
6. Use aspiration.
9,12,15. Plates lidded.
10. Transfer cell lysates to two 384-well high-binding MaxiSorp plates.
17,19. Diluted in SuperBlock-T20.
22. Plates lidded until read.
23. Excitation with laser (320 nm), emission filter (615 nm).
HCl, hydrochloric acid; TBS-T, Tris-Buffered Saline Tween-20; TRF, time-resolved fluorescence technology.
Western Blot of H3K27me3 and Unmodified Histone H3
SU-DHL-6 cells were seeded in 12-well plates at 1×106 cells/well with 2 mL of the RPMI-1640 medium containing 10% FBS, followed by incubation at 37°C in 5% CO2 overnight. DZNep and the internal compound RBC124 were serially diluted (six concentrations with 1:5 dilution factor). Twenty microliters of a compound or DMSO were added to cells for 3 days, with a final DMSO concentration of 1%. The cells were harvested and lysed with 0.2 N HCl at 4°C overnight and sonicated briefly. Two sets of samples were run side-by-side for probing with anti-H3K27me3 and anti-histone H3 antibodies. Fifteen microliters of cell lysates were electrophoresed on a 10%–20% Tris-Glycine mini gel (No. EC61385BOX) and transferred onto nitrocellulose membranes (No. ib801001) by the iBlot dry blotting system (No. ib1001), all from Life Technologies. All primary antibodies were from the same source as for the DELFIA. The membranes were incubated with primary antibodies against tri-methyl histone H3 (host: rabbit, 1:1,000 dilution) or total histone H3 (host: rabbit, 1:500 dilution). The Alexa Fluor 633 goat anti-rabbit IgG secondary antibody from Life Technologies (No. A-21070) was used to detect tri-methyl histone H3 and total histone H3 proteins. The membranes were scanned with a Typhoon 9410 (GE HealthCare, Piscataway, NJ). The specific signals of bands of interest were calculated by utilizing the software Gel-Pro Analyzer (Media Cybernetics, Inc, Rockville, MD).
Results and Discussion
AlphaLISA Assay of H3K27me3
The AlphaLISA cell assay has a homogeneous format, and is amenable to high-throughput screening (HTS). It detects epigenetic markers in cellular extracts. Following a homogeneous histone extraction protocol, the marker of interest is detected by the addition of a biotinylated anti-Histone H3 (C-terminus) antibody and AlphaLISA Acceptor beads conjugated to an antibody (Ab) specific to the marker. The biotinylated antibody is then captured by SA conjugated Donor beads, bringing the two beads into proximity. Upon laser irradiation of the Donor beads at 680 nm, short-lived singlet oxygen molecules produced by the Donor beads can reach the Acceptor beads in proximity to generate an amplified chemiluminescent signal at 615 nm.
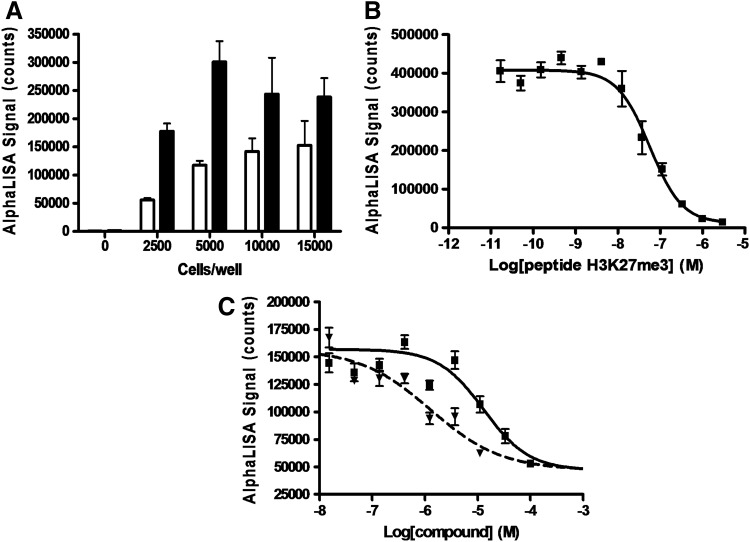
To compare different assay formats, determination of optimal assay conditions was necessary. Su-DHL-6 is a lymphoma cell line that carries the EZH2 Y641N mutation. This cell line was shown to be better than another cell line, OCI-LY-19, and thus used by the manufacturer in the assay validation. We used Su-DHL-6 cells in our study to be consistent and for ease of data comparison. First, a cell titration experiment was performed by incubating SU-DHL-6 cells with DMSO and an internal control compound RBC081 for 24 h at 37°C in 5% CO2. RBC081 and RBC124 are part of the Reaction Biology Corporation's compound library collection. Their structures are not revealed in this article due to their proprietary nature. They were discovered to have inhibitory activities against EZH2 with enzymatic potency of about 9.9 and 19.9 μM for RBC081 and RBC124, respectively (data not shown). A graph of the AlphaLISA signals for the positive (RBC081, 100% inhibition) and negative (DMSO, 0% inhibition) controls at various cell densities is shown in Figure 1A. The assay window was calculated as the ratio of negative versus positive controls. At 2,500 cells per well, an assay window of 3.2 was obtained. Second, peptide H3K27me3 [sequence: KAAR-K(Me3)-SAPATGG] was used to compete for the interaction between the Acceptor beads, which are conjugated to the anti-H3K27me3 antibody, and the modified histone proteins, to verify that the kit was working properly and the antibody recognized the correct marker. The data showed that peptide H3K27me3 competed with high affinity (Fig. 1B). The mean±standard error of the mean (SEM) IC50 value was 56.1±13.0 nM, consistent with the data from the manufacturer (47.8 nM). Last, a commercial compound (DZNep) and an internal compound (RBC124) were used to treat the cells for 3 days to evaluate the inhibitory activity against the H3K27me3 marker. The concentration–response curves are shown in Figure 1C. The mean±SEM IC50 values for RBC124 and DZNep were 13.0±1.3 and 1.4±0.4 μM, respectively. All these IC50 values were the average of at least three determinations. Comparison of 1-day versus 3-day treatments of internal compound IC50's was carried out and the values were within twofold of one another (data not shown), suggesting that the conditions for 1 day or 3 days are similar.
Fig. 1.
Measurement of H3K27me3 level in SU-DHL-6 cells using AlphaLISA format. (A) Titration of SU-DHL-6 cells in H3K27me3 AlphaLISA assay. The bar graph illustrates the responses of the negative (uninhibited) controls (▪) versus positive (100% inhibited) controls (□) at various cell densities (0–15,000 cells per well). (B) Response of AlphaLISA assay at various peptide concentrations. Five thousand SU-DHL-6 cells were seeded to assay plates. Serially diluted peptides from 30 pM to 3 μM were added to the assay plate just before the addition of the AlphaLISA detection reagents. (C) The concentration response of reference inhibitors in H3K27me3 AlphaLISA cell-based assay. Cells were incubated with a compound for 3 days at 37°C in 5% CO2. Evaluation of the cellular inhibitory activity of the reference inhibitor generated average IC50 values of 13.0±1.3 and 1.4±0.4 μM for RBC124 (▪) and DZNep (▾), respectively. The average±SEM IC50 value from three independent determinations was calculated. Error bars denote SEM. H3K27me3, trimethyl histone H3 Lysine 27; DZNep, 3-deazaneplanocin A; IC50, half-maximal inhibitory concentration; SEM, standard error of the mean.
LanthaScreen Assay of H3K27me3
The LanthaScreen assay is another homogeneous cell assay and is easy to automate for HTS purposes. The BacMam histone H3K27me3 cellular assay kit is a terbium-based time-resolved fluorescence resonance energy transfer (TR-FRET) technology in conjunction with BacMam gene delivery system. The histone H3 H3K27me3 protein is expressed as a fusion with the green fluorescent protein (GFP), and the anti-H3K27me3 antibody is labeled with terbium. The advantage of this assay format is that the histone can be introduced to different cell backgrounds via transient transfection. The posttranslational modification was detected upon cell lysis in the presence of a terbium anti-Histone H3K27me3 antibody. In this system, GFP acts as a FRET acceptor, and terbium as a FRET donor. Data have been published utilizing this technology for several histone H3 site-specific modifications.17
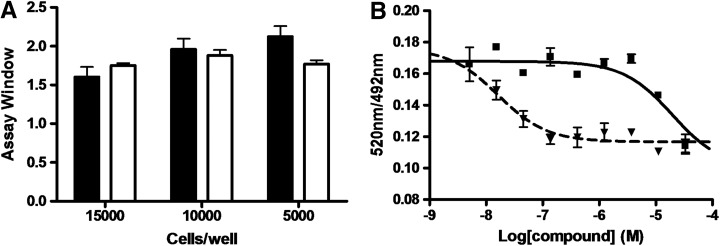
To compare this assay with the AlphaLISA and DELFIA cell assays, we optimized the LanthaScreen assay in terms of BacMam GFP-Histone H3 virus concentrations and cell density. HeLa cells were shown to be the optimal cell line for the LanthaScreen assay format for the detection of H3K27me3.17 HeLa cells were purchased from the American Type Culture Collection and used in this assay as published.17 The assay development was conducted by comparing assay windows of 10% and 30% virus transduction at 5,000, 10,000, and 15,000 cells per well when treated with RBC124 (100% inhibition, positive control) and DMSO (0% inhibition, negative control). The assay window was calculated as the emission ratio of 520 and 490 nm of a negative control/emission ratio of 520 and 490 nm of a positive control. Immediately after virus transduction, compounds were dispensed and the cells were treated for 3 days, consistent with other formats. The results are shown in Figure 2A. It appears that the assay window for 30% virus was higher than 10% virus at most cell densities (1.6, 2.0, and 2.1 for 15,000, 10,000, and 5,000 cells per well, respectively, for 30% virus, vs. 1.8, 1.9, and 1.8 for 15,000, 10,000, and 5,000 cells per well, respectively, for 10% virus). In addition, with 30% virus, 5,000 cells per well provided the highest assay window among all the cell densities tested (2.1-fold). Therefore, the optimal condition was 30% virus with 5,000 cells per well. With the optimized assay conditions, the internal compound RBC124 and a commercial compound DZNep were tested for the inhibitory activity for H3K27me3 levels in cells. The results showed that they both inhibited in a dose-proportioned fashion, with mean±SEM IC50 values of 20.3±3.5 μM and 21.0±6.9 nM for RBC124 and DZNep, respectively (Fig. 2B). All these IC50 values were the average of at least three determinations.
Fig. 2.
Measurement of H3K27me3 level in HeLa cells using LanthaScreen technology. (A) Optimization of BacMam histone H3 transduction versus HeLa cell plating density in H3K27me3 LanthaScreen cell-based assay. Cells were plated for 5,000, 10,000, and 15,000 cells per well. The bar graph illustrates the assay windows of 10% (□) and 30% (▪) BacMam histone H3 virus transduction, followed by incubation with an internal control compound (100% inhibition, positive control) and DMSO (0% inhibition, negative control) for 3 days at 37°C in 5% CO2. The assay window was calculated as the emission ratio of 520 and 490 nm of the negative control/emission ratio of 520 and 490 nm of positive control. (B) The concentration response of reference inhibitors in H3K27me3 LanthaScreen cell-based assay. Cells were transduced with 30% BacMam histone H3 virus, followed by treatment of compounds RBC124 (▪) and DZNep (▾) for 3 days. Evaluation of the cellular inhibitory activity of the reference inhibitors generated average IC50 values of 20.3±3.5 μM and 21.0±6.9 nM for RBC124 (▪) and DZNep (▾), respectively. The IC50 value (average±SEM) from three independent determinations was calculated. Error bars denote SEM. DMSO, dimethylsulfoxide.
DELFIA of H3K27me3
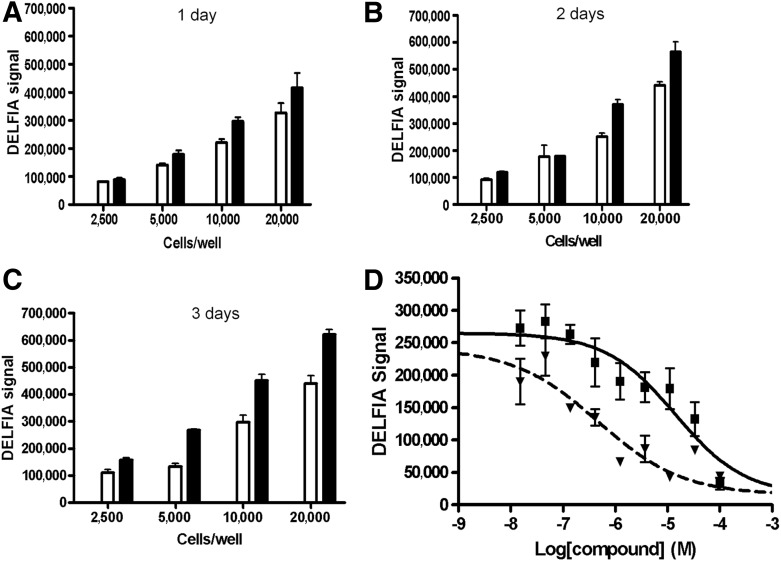
A modified DELFIA detecting cellular H3K27me3 level was published by Xie et al.16 DELFIA is a time-resolved fluorescence technology (TRF), and a heterogeneous assay that can measure the modified histone and total histone simultaneously. As with other enzyme-based immunosorbent assays, the disadvantage of this assay is low throughput and difficulty of adaptation to HTS. To compare this assay with the AlphaLISA and LanthaScreen assays, the assay was optimized in terms of cell density and compound treatment time. PC3 cells are prostate cancer cells. They were used in the assay validation by Xie et al.16 To compare and confirm the results, we used the same PC3 cells for our study. PC3 cells were plated at 2,500, 5,000, 10,000, and 20,000 cells per well. Positive (DZNep, 100% inhibition) and negative (DMSO, 0% inhibition) controls were dispensed to cell wells for treatment time of 1, 2, and 3 days. The signal-to-background ratio (S/B) was calculated as a ratio of the DELFIA signal with negative and positive controls. Based on the results, the S/B ratio for 1-day treatment was 1.1, 1.3, 1.3, and 1.3 for 2,500, 5,000, 10,000, and 20,000 cells/well, respectively. The S/B ratio for 2-day treatment was 1.3, 1.0, 1.5, and 1.3 for 2,500, 5,000, 10,000, and 20,000 cells/well, respectively. The S/B ratio for 3-day treatment was 1.4, 2.0, 1.5, and 1.4 for 2,500, 5,000, 10,000, and 20,000 cells/well, respectively. In addition, for the 3-day treatment, 5,000 cells per well displayed a higher S/B ratio than 2,500, 10,000, and 20,000 cells per well (Fig. 3A–C). The commercial compound DZNep and internal compound RBC124 were tested in PC3 cells for their potency in lowering the level of H3K27me3. The mean±SEM values were 11.7±1.6 and 0.32±0.07 μM for RBC124 and DZNep, respectively (Fig. 3D). All these IC50 values were the average of at least three determinations. DELFIA signal data for the total histone was also collected. The normalized IC50 values of both compounds were similar before normalization (data not shown), indicating that the inhibitors only affected the modified histone protein, but not the total histone. Thus, it eliminates the possibility that the compounds are false-positives.16
Fig. 3.
Measurement of H3K27me3 level in PC3 cells using the DELFIA format. (A–C) Titration of PC3 cells in H3K27me3 DELFIA cell-based assay. The bar graph illustrates the responses of the negative (uninhibited) controls (▪) versus positive (100% inhibited) controls (□) at various cell densities (2,500–20,000 cells per well) for 1-, 2-, and 3-day treatment. (D) The concentration response of reference inhibitors in H3K27me3 DELFIA cell-based assay. Cells were incubated with a compound for 3 days at 37°C in 5% CO2. Evaluation of the cellular inhibitory activity of the reference inhibitors generated average IC50 values of 11.7±1.6 and 0.32±0.07 μM for RBC124 (▪) and DZNep (▾), respectively. The IC50 value (average±SEM) from three independent determinations was calculated. Error bars denote SEM. DELFIA, dissociation-enhanced lanthanide fluorescence immunoassay.
Western Blot of H3K27me3 and Unmodified Histone H3
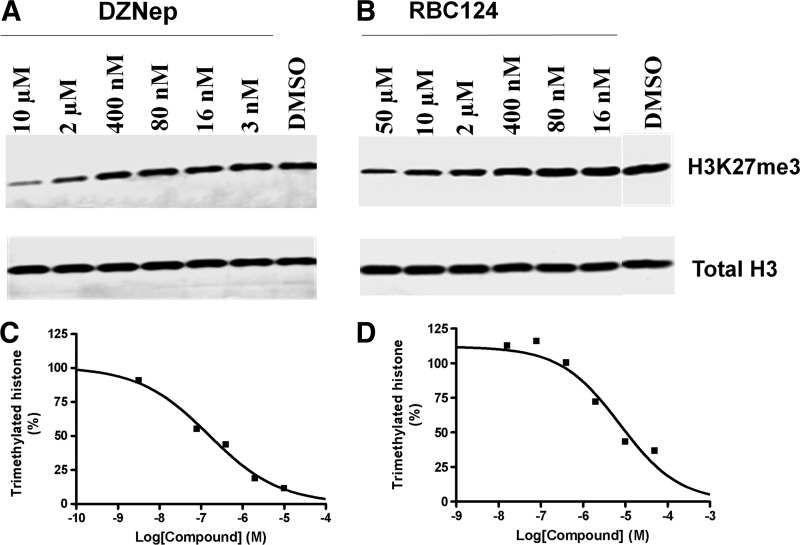
A Western blot analysis for H3K27me3 against SU-DHL-6 cells was performed to compare and confirm the data from other formats. DZNep and RBC124 were tested starting at 10 and 50 μM, respectively, along with DMSO control. The data showed that there was a dose response with each compound treatment for H3K27me3, while the level of total histone H3 remained the same (Fig. 4), indicating that the inhibitors only affected the modified histone protein, but not the total histone. This is consistent with the results from the DELFIA. Data for the modified and total histone were quantitated using the Gel-Pro Analyzer; the concentration–response curves are shown in Figure 4. The IC50 values for RBC124 and DZNep were 7.4 and 0.16 μM, respectively, consistent with results for the other three formats (except for DZNep in AlphaLISA assay).
Fig. 4.
Measurement of H3K27me3 level in SU-DHL-6 cells using Western blot following treatment for 3 days at 37°C in 5% CO2. The concentration response of reference inhibitors DZNep (A) and RBC124 (B) in H3K27me3 Western blot cell-based assay. Total histone H3 is shown as a loading control. Data for the modified and total histone were quantitated using the Gel-Pro Analyzer. Evaluation of the cellular inhibitory activity of the reference inhibitors generated an IC50 value of 7.4 and 0.16 μM for RBC124 (C) and DZNep (D), respectively.
Comparison of the Four Cell-Based Assay Formats
All four assays were able to detect the H3K27me3 marker with reasonable results. They are all antibody-based technologies, and all can utilize cryopreserved cells. Internal (RBC124) and commercial (DZNep) compounds were used to treat all three cell lines for the same length (3 days). AlphaLISA and LanthaScreen are homogeneous assays, while DELFIA and Western blot are heterogeneous formats; therefore, AlphaLISA and LanthaScreen are more HTS friendly than DELFIA and Western blot assays. AlphaLISA and DELFIA had more robust assay window than LanthaScreen (>3 for AlphaLISA and DELFIA, and ≤2 for LanthaScreen assay), although LanthaScreen is a ratiometric assay, so a lower assay window is acceptable.
DZNep has published data for cell-based assay against H3K27me3 using DELFIA.16 Our result showed that the IC50 value for DZNep was 0.32±0.07 μM with this format, which agrees with that reported by Xie et al., both with pIC50 of 7.16 The data for RBC124 were consistent among AlphaLISA, LanthaScreen, DELFIA, and Western blot (13.0, 20.3, 11.7, and 7.4 μM, respectively), validating that all four assays are suitable for compound profiling. The IC50 values for DZNep were more varied among AlphaLISA, DELFIA, Western blot, and LanthaScreen (1.4, 0.32, 0.16, and 0.021 μM, respectively), than those for RBC124. Each format utilizes different cell lines, and the differences in the metabolism of DZNep or in its cellular uptake could contribute to the discrepancy in IC50 values. It has been reported that DZNep is a global histone methylation inhibitor.18 However, RBC124 might have a different inhibitory mechanism than DZNep. The combination of mechanism of action difference and the assay system could contribute to the different IC50's observed for DZNep.
Overall, AlphaLISA is a homogeneous assay, and it has a better assay window than LanthaScreen assay for measuring histone H3K27me3. Besides H3K27me3, more assay kits for different epigenetic markers are available from PerkinElmer using AlphaLISA assay technology. We successfully developed and validated five more kits (H3K4me2, H3K36me2, H3K79me2, H3K9ac, and H3K27ac) by first optimizing the assays, and testing with internal and commercial compounds. All assays worked as expected. In summary, AlphaLISA is our preferred assay format for HTS and compound profiling. DELFIA and Western blot formats are useful tools to confirm hits and eliminate false-positives.
Abbreviations
- Ab
antibody
- DELFIA
dissociation-enhanced lanthanide fluorescence immunoassay
- DMSO
dimethylsulfoxide
- EZH2
Enhancer of Zeste homolog 2
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HTS
high-throughput screening
- IC50
half-maximal inhibitory concentration
- SA
streptavidin
- S/B
signal-to-background ratio
- SEM
standard error of the mean
- TBS-T
Tris-buffered saline Tween-20
- TRF
time-resolved fluorescence technology
- TR-FRET
time-resolved fluorescence resonance energy transfer
Acknowledgment
This work was funded, in part, by U.S. National Institutes of Health Small Business Innovation Research grant R44CA139621 to H.M.
Disclosure Statement
No competing financial interests exist.
References
- 1.Wolffe AP. Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou VW. Goren A. Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 4.Roth SY. Allis CD. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T. Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Sneeringer CJ. Scott MP. Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bissinger EM. Heinke R. Sippl W. Jung M. Histone methyltransferase inhibitors. Med Chem Commun. 2010;1:114–124. [Google Scholar]
- 8.Chang MJ. Wu H. Achille MJ, et al. Histone H3 lysine 79 methyltransferase dot1 is required for immortalization by mll oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Q. Patel MP. Semus SF, et al. Perspectives on the discovery of small-molecule modulators for epigenetic processes. J Biomol Screen. 2012;17:555–571. doi: 10.1177/1087057112437763. [DOI] [PubMed] [Google Scholar]
- 10.McCabe MT. Ott HM. Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 11.Wang C. Caron M. Burdick D, et al. A sensitive, homogeneous, and high-throughput assay for lysine-specific histone demethylases at the H3K4 site. Assay Drug Dev Technol. 2012;10:179–186. doi: 10.1089/adt.2011.0395. [DOI] [PubMed] [Google Scholar]
- 12.Diaz E. Machutta CA. Chen S, et al. Development and validation of reagents and assays for EZH2 peptide and nucleosome high-throughput screens. J Biomol Screen. 2012;17:1279–1292. doi: 10.1177/1087057112453765. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi KY. Eason MM. Ferry JJ, et al. Assay development for histone methyltransferases. Assay Drug Dev Technol. 2013;11:227–236. doi: 10.1089/adt.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn AM. Allali-Hassani A. Vedadi M. Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors. Mol Biosyst. 2010;6:782–788. doi: 10.1039/b921912a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn AM. Bedford MT. Espejo A, et al. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res. 2010;38:e11. doi: 10.1093/nar/gkp899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie W. Ames RS. Li H. A cell-based high-throughput screening assay to measure cellular histone H3 lys27 trimethylation with a modified dissociation-enhanced lanthanide fluorescent immunoassay. J Biomol Screen. 2012;17:99–107. doi: 10.1177/1087057111422378. [DOI] [PubMed] [Google Scholar]
- 17.Machleidt T. Robers MB. Hermanson SB. Dudek JM. Bi K. TR-FRET cellular assays for interrogating posttranslational modifications of histone H3. J Biomol Screen. 2011;16:1236–1246. doi: 10.1177/1087057111422943. [DOI] [PubMed] [Google Scholar]
- 18.Miranda TB. Cortez CC. Yoo CB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]