Abstract
Neuroimaging biomarkers of depression have potential to aid diagnosis, identify individuals at risk and predict treatment response or course of illness. Nevertheless none have been identified so far, potentially because no single brain parameter captures the complexity of the pathophysiology of depression. Multi-voxel pattern analysis (MVPA) may overcome this issue as it can identify patterns of voxels that are spatially distributed across the brain. Here we present the results of an MVPA to investigate the neuronal patterns underlying passive viewing of positive, negative and neutral pictures in depressed patients. A linear support vector machine (SVM) was trained to discriminate different valence conditions based on the functional magnetic resonance imaging (fMRI) data of nine unipolar depressed patients. A similar dataset obtained in nine healthy individuals was included to conduct a group classification analysis via linear discriminant analysis (LDA). Accuracy scores of 86% or higher were obtained for each valence contrast via patterns that included limbic areas such as the amygdala and frontal areas such as the ventrolateral prefrontal cortex. The LDA identified two areas (the dorsomedial prefrontal cortex and caudate nucleus) that allowed group classification with 72.2% accuracy. Our preliminary findings suggest that MVPA can identify stable valence patterns, with more sensitivity than univariate analysis, in depressed participants and that it may be possible to discriminate between healthy and depressed individuals based on differences in the brain's response to emotional cues.
Keywords: MVPA, LDA, Depression, Valence, Affect, Emotion
Highlights
-
•
Multivariate pattern analysis of the neural correlates of valence in depression.
-
•
Valence classification accuracies around 90% in depressed patients suggest stable patterns of valence.
-
•
Accuracy of 72.2% in discriminating healthy versus depressed individuals based on activation in dorsomedial prefrontal cortex and caudate nucleus.
1. Introduction
Brain imaging studies have traditionally relied on the analysis of the univariate responses of individual voxels in the brain to differing conditions. However, multivariate analyses that incorporate dependencies between multiple voxels (Norman et al., 2006) may be more appropriate for the functional architecture of the human brain, which is characterised by distributed information processing (Haxby et al., 2001; Pinel et al., 2004). Multi-voxel pattern analysis (MVPA) has the ability to detect patterns at a finer resolution, with weaker activations, where they are part of a collective representation of a certain task condition or mental state. Previous studies have applied MVPA for example to detect perceptual (e.g. Haxby et al., 2001; Mourão-Miranda et al., 2005) or cognitive states (e.g. Davatzikos et al., 2005; Haynes and Rees, 2005), predict disease (e.g. Craddock et al., 2009; Mourão-Miranda et al., 2012a, 2012b; Zhang et al., 2005; Zhu et al., 2005) or affective states (Yuen et al., 2012), identify dysfunctional processes in clinical populations (e.g. Yoon et al., 2008) and for clinical response prediction (e.g. Costafreda et al., 2009).
1.1. Studying emotion processing with MVPA
It has been argued that MVPA has superior sensitivity for determining patterns of response compared to univariate methods (De Martino et al., 2008; Hanke et al., 2009; Norman et al., 2006; Yoon et al., 2008). This makes it particularly appealing for emotion research. Emotion processing is assumed to involve a widely distributed network of limbic and prefrontal areas (Damasio, 1998). Its brain correlates have been studied in humans using different models of affect which can be classified as categorical (e.g. Ekman, 1992; Roseman et al., 1990) or dimensional (e.g. Schachter and Singer, 1962). Neuroimaging studies (particularly in combination with MVPA) have the potential to resolve the ongoing debate between both classes of models. Categorical models regard emotions as discrete entities that can be expected to be mediated by distinct brain areas and revealed by univariate analysis. In contrast, dimensional models describe emotions via their placement on two or more dimensions. In terms of brain activation, this would be reflected in changes in the balance of activation between different areas, which can only be picked up by multivariate analyses. In this paper we implemented one of the most influential dimensional models that is based on the emotion circumplex (Russell, 1980) and assumes that emotional states can be described via a combination of arousal (the extent of activation one experiences) and valence (the extent of pleasantness one experiences). In terms of brain imaging, this can be utilised to compute contrasts between different types of affective stimuli and neural correlates of emotions. Previous studies with univariate methodology have shown substantial overlap between the cortical regions that process positive, negative and neutral affect (Johnston et al., 2010; Murphy et al., 2003; Phan et al., 2002), suggesting that univariate/categorical models may not fully capture the complexity of emotion processing in the human brain. Conversely, MVPA studies have suggested that multivariate analysis may be sensitive to differences in neuronal patterns underlying different levels of valence in healthy volunteers (Baucom et al., 2012; Yuen et al., 2012). In the study by Baucom et al. (2012), one classifier predicted whether participants had viewed positive or negative pictures evoking high or low arousal and another discriminated between positive and negative valence. These classifiers reached a maximum within-participant accuracy of 77% and 92% respectively.
1.2. MVPA and pathological emotion processing in depression
Functional imaging has elucidated the brain networks associated with altered emotion processing in affective disorders (Phillips et al., 2003) and has revealed changes in neural activation both in symptomatic and remitted states (Goldapple et al., 2004; Grimm et al., 2008; Siegle et al., 2002). Neuroimaging biomarkers would be of interest to improve diagnosis, for example in the differentiation between unipolar and bipolar depression, or as trait markers of risk for mood disorder in vulnerable individuals (Linden, 2012), and MVPA may be particularly useful for this purpose (Mourão-Miranda et al., 2012b) because no single parameter of brain structure or activation can capture the complexity of the pathophysiology of depression. Such biomarkers would also be potentially useful as predictors of treatment response, for treatment stratification or as surrogate markers in clinical trials (Keedwell and Linden, 2013). Several previous studies have applied MVPA in the context of depression (Fu et al., 2008; Hahn et al., 2011; Marquand et al., 2008). In one study individuals were classified as healthy or depressed (with 86% accuracy) based on the pattern of cortical activity representing the implicit processing of sad facial expression (Fu et al., 2008). Another study applied pattern recognition to the functional magnetic resonance imaging (fMRI) data of healthy and depressed individuals who completed two versions of the monetary incentive delay task and passively viewed facial expression (Hahn et al., 2011). A combination of the conditions involving neutral faces, receiving large rewards and anticipating no loss resulted in the highest group classification accuracy.
1.3. Current study
To our knowledge no study has investigated whether it is possible to accurately identify specific valence conditions in response to International Affective Picture System (IAPS; Lang et al., 1999) pictures in brain activation data from depressed patients, which thus formed the main aim of the current study. While Mourão-Miranda et al. (2012a, 2012b)) investigated the discriminability of patterns that underlie viewing happy and neutral faces in unipolar and bipolar depressed patients, the current study focused on unipolar depression, was not confined to the processing of facial expressions and included negative valence cues as well. Patients suffering from depression were presented with an emotion localiser composed of positive, negative and neutral images. A support vector machine (SVM) was trained to classify the data as belonging to one of the three picture valence categories. Successfully discriminating brain patterns related to the processing of different valence cues via MVPA is of interest for two reasons. First of all, this would illustrate the ability of MVPA to disentangle closely overlapping neural substrates. This in turn would allow the detection of more fine-grained abnormalities that potentially underlie the dysfunctional emotion processing associated with depression. Furthermore, MVPA offers key ingredients for successful fMRI neurofeedback (Sitaram et al., 2011): detection, decoding and prediction of neural states in a short period of time. Rapidly identifying activation patterns corresponding to specific tasks or states, instead of just focusing on single regions, might lead to more accurate neurofeedback and eventually boost its quality and long-term effects in depression, which is an area of current development (Linden et al., 2012).
To preempt our results, we show a successful application of MVPA in discriminating positive, negative and neutral valence cues in patients with depression. In addition, we demonstrate that less sensitive univariate approaches leave areas undetected that are highly discriminatory in MVPA. Finally, it was possible in our sample to discriminate healthy from depressed individuals based on differences in bivariate response patterns to stimuli of different valence conditions.
2. Materials and methods
2.1. Data
Nine patients (8 male; age range = 21–67 years, mean age = 48.8 years) suffering from unipolar depression as established by the SCID (First et al., 2002) were included in the analysis (see Table S1 for more details). None of the patients had any DSM-IV defined comorbidities. All patients were on anti-depressants, the dose of which remained stable for at least the six weeks preceding the intervention and for the entire duration of the study. Data was acquired on a 3-T Philips Achieva System (Best, The Netherlands) and data acquisition procedures were similar as in Johnston et al. (2011); TR = 2 s, TE = 30 ms, 30 slices, 3-mm slice thickness, inplane resolution 2 × 2 mm). The same localiser as previously described in Johnston et al. (2010, 2011) was adopted, consisting of positive, negative and neutral stimuli adopted from the IAPS. IAPS pictures were employed as these induce the expressive, somatic and autonomic changes that are typically associated with affective expression, in a controlled manner (Lang and Bradley, 2007). Additionally, the IAPS picture set comes along with well-documented ratings of arousal and valence as these two factors have been found to explain most of the variance in evaluative judgments (Lang and Bradley, 2007). During the localiser, 12 trials of each valence type (positive, negative and neutral) were presented in a pseudo-randomised order (see Fig. 1). Per trial either four neutral, negative or positive IAPS pictures were shown for 1.5 s each, alternating with a fixation baseline of 12 s. Two patients participated in three sessions during which they viewed the same localiser and the remaining patients in four sessions. Per patient we thus obtained a total of either 36 of 48 trials per valence condition. The functional data were preprocessed using motion correction and linear detrending to remove signal drift (GLM-Fourier, 2 sines/cosines). The data were then coregistered with the anatomical data and transformed into Talairach Space (Talairach and Tournoux, 1988).
Fig. 1.
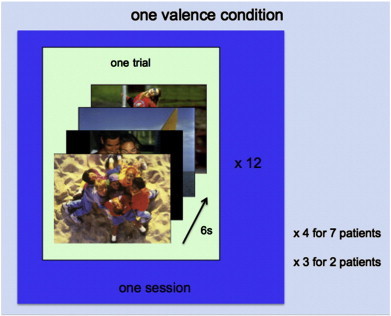
Schematic overview of data compiling one valence condition. Within one trial, four pictures of the same valence type (either positive, negative or neutral) were presented for 1.5 s each. Twelve trials were presented in each localiser session for each valence type amounting to either 36 or 48 trials per valence type patient (depending on whether the patient had participated in three or four localiser sessions). A total of 144 (or 108) trials was obtained per patient collapsed over the three valence conditions and the four (or three) localiser sessions.
2.2. Multi-voxel pattern analysis
2.2.1. Trial estimation/feature extraction
The BrainVoyager QX (Brain Innovation, Maastricht, The Netherlands) software was used to perform supervised multivariate pattern classification via the SVM. A general linear model (GLM) analysis was performed that computed voxel-wise beta estimates for each trial within each valence condition within the following contrasts: positive–negative, positive–neutral and neutral–negative. In total we thus obtained 48 sets of beta estimates per valence condition across all localiser sessions with the exception of two participants who only participated in three localiser sessions (total of 36 beta estimates per valence condition; see Supplementary Information: Materials for more details). No temporal or spatial smoothing was applied to ensure that the selection of informative voxels (and as a consequence the input for the classifier) was not biased as smoothing can hinder the detection of isolated voxels and can instead favour spatially clustered ones. The patterns of estimated beta values (z-normalised) were stored in feature vectors and served as input for the feature selection step.
2.2.2. Feature selection
Feature selection on the dataset is an essential step for the classification of fMRI data (see Mitchell et al., 2004; Norman et al., 2006) for background). The selected parameters are similar to Yuen et al.'s (2012). The visual cortex was masked by excluding all brain tissue posterior to the occipito-parietal sulcus to prevent categorisation driven by differences in visual cortex responses to the dissimilar visual input in each condition. In addition, the ventricles were masked for each patient individually. An initial data reduction step via univariate F-tests followed by multivariate recursive feature elimination (RFE) has been found to result in maximum sensitivity and generalisation performance (De Martino et al., 2008). First a crude selection was made by selecting the top 50% (13 000–27 000) of the voxels that showed the strongest activation, for each of the three contrasts (positive–negative, positive–neutral and neutral–negative).Then a more fine-tuned selection procedure was adopted that selected the top 5% (600–1400 voxels) of the remaining voxels via RFE (see Supplementary Information: Materials for more details). At the end of this stage only the SVM weights of the selected voxels were retained for each trial. Five randomly selected trials out of 48 (or 36) were set apart as testing dataset while the remainder of the trials (either 43 or 31) served as training input.
2.2.3. Classifier training
In the third step a linear SVM, known for their good generalisation performance even in studies with relatively small datasets, was trained (see Belousov et al., 2002; Misaki et al., 2010 for background). A cross-validation procedure testing a series of different SVMs using different values for the regularisation parameter C was run for each of the three contrasts separately (see Supplementary Information: Materials for more details). The C-value, which establishes the trade-off between classification accuracy and generalisability, that resulted in the maximum cross-validation accuracy determined the optimal hyperplane that separated the training trials of the valence conditions. This hyperplane, or decision boundary, can be described by the linear discriminant function f(x) = wx + b, where w is the vector containing the SVM weights, x the training patterns containing the beta estimates of the valence conditions in the contrast (of the selected voxels) and b the bias term. Depending on the side of the decision boundary at which the training pattern of a trial appeared in feature space, a trial was assigned to one of the two valence conditions in the contrast. The class assignment was then checked with the experimental protocol to determine the correctness of the classification. In the last phase of this third step an overall accuracy score indicating the proportion of correctly allocated training trials was calculated for each contrast separately.
2.2.4. Classifier testing
In the final step of the MVPA the remaining data served as input for the trained SVM to test the performance on a set of input new to the classifier. This SVM was used to predict the categories of the test trials for each contrast and individual separately. Overall prediction accuracy scores for each contrast were computed based on the prediction accuracy of all trials together. For all contrasts five trials (out of 48/36 trials) of each condition were randomly selected to be left out of the training stage (decoding and classifying) which later served as testing (predicting) data.
2.2.5. Group-level activation probability maps
Group-level probability maps were generated to determine the areas that were driving the classifications. For this purpose, individual discriminative maps based on the SVM weights were created first for each of the three contrasts. These served as the basis for masks that contained all discriminative voxels. Group-level probability maps were then calculated via these masks and were smoothed with a 4 mm FWHM Gaussian kernel to adjust for individual differences in neural anatomy. These maps were thresholded at 60%, entailing that a voxel only appeared on the probability map if it was discriminative in more than five individuals.
2.2.6. Permutation tests
Finally permutation tests were performed to compare the performance of the classifier to a null-distribution. The trials were randomly categorised after which the classifier was retrained with these new and possibly wrong categorisations. This classifier was repeatedly tested (200 permutations) with a ‘leave one out’ cross-validation method and provided a null distribution that showed the probability of gaining a correct classification result while the conditions were randomly allocated. Classification accuracies of above the 95th percentile of the null distribution indicated that a significant classifier accuracy result was obtained.
2.3. Univariate analysis
The sensitivity of the multivariate method was compared with a univariate analysis. In accordance with standard univariate analysis procedures the data were corrected for head motion, linearly detrended and temporally (3 s) and spatially smoothed (4 mm FWHM Gaussian Kernel) to increase the signal-to-noise ratio for the group analysis. The same mask as in the multivariate method was applied for each patient. A conventional single-subject GLM (p < .05) was performed for the three contrasts. This served as the basis for individual masks of the activated voxels in each valence condition in each contrast. Group-level activation probability maps (thresholded at 60%) were generated based on these masks to allow for comparison with the multivariate probability maps.
2.4. Group classification
In follow up of the MVPA results we investigated whether it was possible to discriminate activation patterns in response to different valence conditions in different groups of people. Due to software limitations this was not examined via MVPA but a bivariate differentiation analysis similar to Ihssen et al.'s (2011). Since the healthy controls only participated in 12 trials it was not possible to train a separate classifier on the data collected in healthy controls either. Instead, the localiser data of one session of nine healthy controls (7 male, age range = 30–56 years; mean age = 38 years) were added to the localiser dataset of the first session of the depressed patients. The data had been collected during a previous study (Johnston et al., 2010) that applied the same localiser protocol as the depression study. There was no significant difference of age between the two groups (t(16) = 2.023, p > .05) and both groups were matched for gender. Activity maps were created via a two-way ANOVA to identify areas that showed a significant interaction between group and valence contrast. As this was an exploratory analysis an arbitrary threshold of p < .002 was chosen that would ensure that only the most discriminative areas would be maintained in the analysis. Subsequently two stepwise linear discriminant analyses (LDAs) were conducted in SPSS 18.0 (SPSS, Chicago, IL, USA) that searched for the brain areas with the highest discriminative power and thereby investigated how well the different areas discriminate between the healthy and depressed group. One LDA was conducted with all areas identified on the activity maps and another with all areas that survived multiple comparison correction via cluster thresholding (p < .05, cluster size threshold of 108 mm3 for all three valence contrasts). After this exploratory analysis another stepwise LDA was conducted that is not affected by a potential bias from circularity. For each of the three valence pairs the five clusters that showed the most significant main effect of valence were identified. From these 15 areas the clusters surviving cluster threshold correction (p < .05, cluster size threshold of 108 mm3 for the contrast [positive–negative], 81 mm3 for the contrast [positive–neutral] and 135 mm3 for the contrast [neutral–negative]) were selected as input for the LDA. Areas that also showed a significant interaction between group and valence contrast were excluded from the LDA. The generalisation of the classifier was tested via a leave-one-out cross-validation procedure.
3. Results
3.1. Multi-voxel pattern analysis
For all valence discriminations the SVM achieved accuracy levels between 80 and 100% (positive–negative: 92%; negative–neutral: 86%; neutral–positive 89%). The permutation tests demonstrated the statistical significance of the SVM accuracy results as the obtained discrimination accuracy for all valence labels and all depressed subjects was significantly higher than at the chance level of .05 (see Fig. S1).
The emotional valence information for neutral, negative and positive stimuli in depressed patients was reflected in a highly distributed activity pattern across the brain and covered areas that have previously been linked to emotional processing (see Table S2 for complete overview). Several areas were identified that are part of the fronto-limbic system such as the VLPFC, insula, striatum, cingulate cortex, amygdala and hippocampus (see Fig. 2). None of these areas showed selectivity for only one valence condition (see Table 1). Instead they contained voxels weighted for several valence conditions in several contrasts, albeit at varying locations within that area.
Fig. 2.
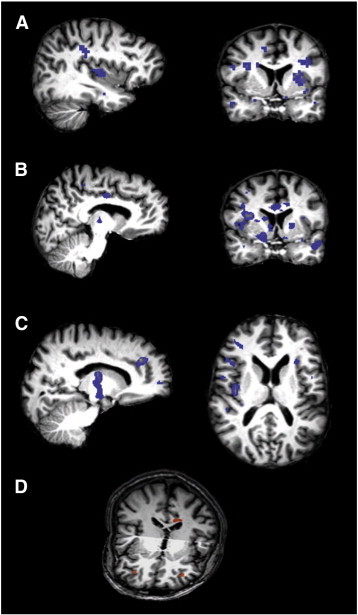
Areas underlying the valence patterns for the three different contrasts as identified via MVPA.
a) Contrast positive versus negative. Areas carrying information about positive valence included the insula, parahippocampal gyrus and postcentral gyrus (x = − 37). For negative valence the areas included the putamen, inferior frontal gyrus (IFG), middle frontal gyrus (MFG), middle temporal gyrus (MTG), parahippocampal gyrus, ventral striatum and cingulate gyrus (y = 4, radiological convention). The areas that form part of the valence patterns are coloured in blue.
b) Contrast positive versus neutral. The cingulate gyrus and hypothalamus were amongst other regions part of the pattern underlying positive valence (x = − 9). The cingulate gyrus, insula, putamen, superior temporal gyrus (STG) and occipitotemporal gyrus (OTG) formed part of the pattern underlying neutral valence (y = 4, radiological convention).
c) Contrast neutral versus negative. For neutral valence the anterior cingulate gyrus (ACG), thalamus and midbrain carried information related to neutral valence (x = 15). For negative valence the areas included the orbitofrontal cortex (OFC), IFG, temporoparietal junction (TPJ) and insula (z = 12, radiological convention).
d) Differential activation patterns in the depressed and healthy group, in the contrast positive versus negative, were found in the bilateral ventrolateral cortex (VLPFC) and dorsal cingulate gyrus (all represented in orange) and allowed group classification with an accuracy of 100%.
Table 1.
Fronto-limbic areas aiding classification per valence condition per contrast.
| Positive vs Negative |
Positive vs Neutral |
Neutral vs Negative |
||||
|---|---|---|---|---|---|---|
| Pos | Neg | Pos | Neu | Neu | Neg | |
| Insula | x | x | x | x | x | x |
| IFG | x | x | x | x | x | x |
| Amygdala | x | x | x | |||
| Caudate nucleus | x | x | x | |||
| Putamen | x | x | x | x | x | |
| Hippocampus | x | x | x | x | x | |
| ACG | x | x | x | |||
IFG = inferior frontal gyrus, ACG = anterior cingulate gyrus.
3.2. Univariate analysis
The group-level activation probability maps across all patients created by univariate analysis were thresholded at the same level (60%) as in the multivariate approach. Only activation related to negative valence survived this threshold. For positive valence, the first activation appeared at 50% (contrasted to the neutral condition) and at 40% (contrasted with the negative condition).
3.3. Group classification
An LDA was performed to investigate the differentiability between affect processing areas in depressed and healthy participants. Exploratory activation maps were constructed for the interaction ‘group’ × ‘valence contrast’ to identify areas that would serve as input for the LDA (see Table 2A). Based on the activation levels in four areas, the stepwise LDA was able to correctly classify all participants. These areas were the right VLPFC and dorsal cingulate gyrus in the [neutral–negative] contrast, the left VLPFC in the contrast [positive–neutral] and the dorsal cingulate gyrus in the contrast [positive–negative]. For both areas located on the cingulate gyrus the depressed group had higher activation levels in the negative than neutral or positive conditions, whereas the healthy controls had lower activation levels in the negative than neutral or positive conditions (see Fig. 3A). When the LDA was conducted with the areas that had survived cluster threshold correction (see Table 2B), the cluster in the right VLPFC in the contrast [neutral–negative] was retained and was able to classify all individuals with 100% accuracy in conjunction with a cluster in the MFG and IPL in the contrast [positive–neutral] (see Fig. 3B).
Fig. 3.
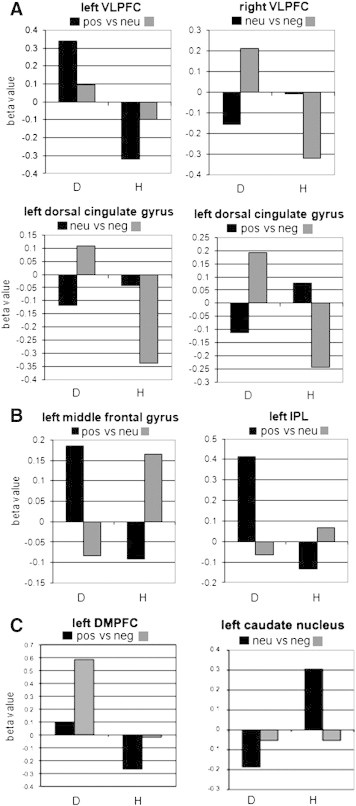
Activation patterns of the areas underlying successful group classification.
a) Areas identified via a ‘group’ × ‘valence contrast’ interaction, without cluster threshold correction. Based on the bivariate response patterns in four areas (in different contrasts) all participants were classified in the correct group. Elevated activation levels were found for negative valence in the depressed group in comparison to the healthy control group.
b) Areas identified via a ‘group’ × ‘valence contrast’ interaction, surviving cluster threshold correction. The bivariate response patterns of three areas that survived cluster threshold correction allowed successful group classification. Apart from a cluster in the right VLPFC that was identified without cluster threshold correction, the left MFG and left IPL showed a marked difference in the activation pattern in both groups.
c). Areas identified via a main effect of valence, surviving cluster threshold correction. The group classification was based on the bivariate response patterns in the left DMPFC and left caudate nucleus.
D = depressed, H = healthy.
Table 2.
Areas selected as LDA input.
| Region | Side | TAL coordinates | No. of voxels |
|---|---|---|---|
| A. Group × valence contrast interaction (p < .002) | |||
| A. Group × (positive–negative) interaction | |||
| Dorsal cingulate gyrus | L | − 13/−21/29 | 326 |
| Amygdala | R | 23/4/− 18 | 4 |
| VLPFC | R | 27/41/0 | 31 |
| L | − 31/43/1 | 99 | |
| Precentral gyrus | R | 24/−7/48 | 348 |
| B. Group × (positive–neutral) interaction | |||
| MFG | L | − 41/4/50 | 139 |
| VLPFC | L | − 55/6/20 | 60 |
| Postcentral gyrus | R | 42/−28/44 | 229 |
| L | − 30/− 20/42 | 56 | |
| Subgyral region | L | − 37/− 32/0 | 12 |
| SFG | L | − 19/36/45 | 34 |
| IPL | L | − 39/−39/37 | 151 |
| L | − 47/−33/39 | 342 | |
| DMPFC | L | − 12/34/42 | 20 |
| Insula | L | − 40/− 8/− 9 | 4 |
| C. Group × (neutral–negative) interaction | |||
| VLPFC | R | 33/41/0 | 225 |
| L | − 34/38/− 2 | 108 | |
| DMPFC | R | 10/44/37 | 81 |
| Caudate nucleus | L | − 15/− 3/20 | 58 |
| Precuneus | R | 15/− 54/38 | 66 |
| L | − 17/− 62/30 | 57 | |
| Dorsal cingulate gyrus | L | − 13/13/27 | 6 |
| Precentral gyrus | R | 51/− 16/47 | 24 |
| IPL | R | 28/− 53/27 | 13 |
| Posterior cingulate gyrus | R | 17/− 42/9 | 12 |
| B. Main effect valence per contrast (p < .001) | |||
| A. Contrast (positive–negative) | |||
| MFG | R | 42/15/30 | 352 |
| DMPFC | L | − 4/43/46 | 424 |
| MTG | L | − 57/−20/−6 | 253 |
| VLPFC | R | 35/23/19 | 141 |
| Insula | L | − 33/3/10 | 60 |
| B. Contrast (positive–neutral) | |||
| DMPFC | R | 3/24/45 | 11 |
| MFG | R | 41/19/31 | 227 |
| Putamen | R | 32/− 10/13 | 56 |
| Midbrain | R | 19/− 17/− 9 | 22 |
| Insula | L | − 33/− 23/15 | 19 |
| C. Contrast (neutral–negative) | |||
| Caudate nucleus | L | − 11/5/14 | 224 |
| STS | L | − 38/− 52/9 | 87 |
| VLPFC | L | − 30/27/4 | 254 |
| MTG | L | − 42/−70/−5 | 772 |
| ITS | R | 39/−61/−11 | 515 |
The areas surviving cluster threshold correction are printed in bold.
VLPFC = ventrolateral prefrontal cortex, MFG = middle frontal gyrus.
SFG = superior frontal gyrus, IPL = intraparietal lobule.
DMPFC = dorsal medial prefrontal cortex, MTG = middle temporal gyrus.
STS = superior temporal sulcus, ITS = inferior temporal sulcus.
The stepwise LDA based on only areas showing a main effect of valence (see Table 3) correctly classified all participants based on activation levels in the caudate nucleus and dorsomedial prefrontal cortex (DMPFC; see Fig. 3C). In the cross-validation procedure that was run to test the generalisation of the classifier, 72.2% (chance level: 50%) of all cases were correctly classified as either belonging to the depressed or healthy group.
Table 3.
Stepwise LDA results.
| Wilks λ | χ2 | Significance level | Classification accuracy | |
|---|---|---|---|---|
| Input based on areas showing a ‘group’ × ‘valence contrast’ interaction | ||||
All contrasts before cluster threshold correction
|
0.067 | 37.067 | p < 0.001 | 100% |
All contrasts after cluster threshold correction
|
0.159 | 26.768 | p < 0.001 | 100% |
| Input based on areas showing a main effect of valence | ||||
All contrasts after cluster threshold correction
|
0.477 | 11.094 | p = 0.004 | 100% |
VLPFC = ventrolateral prefrontal cortex, MFG = middle frontal gyrus, IPL = intraparietal lobule, DMPFC = dorsomedial prefrontal cortex.
To summarise, the SVM classified the different valence conditions in patients with depression with high accuracy. Several of the neural correlates underlying these conditions form part of a fronto-limbic system. Conversely, a standard univariate analysis did not pick up any activation differences in any of the contrasts. Based on the activity patterns in the DMPFC and caudate nucleus the LDA was able to perfectly separate healthy from depressed individuals.
4. Discussion
4.1. MVPA findings
This study identified activation patterns of specific valence conditions elicited by IAPS pictures in patients with unipolar depression. The group-level probability maps obtained via MVPA showed that a distributed pattern of brain regions contributed to the representation of each valence condition, with overlap in the brain areas associated with the different valence conditions. The overlap could indicate that an area fulfills a more general role in affective processing such as emotion reappraisal (Ochsner and Gross, 2005) or may indicate emotion specific involvement in both valence conditions, for example arousal responses that are similar across valence directions. Our findings in patients are thus generally in line with previous studies in healthy individuals (Baucom et al., 2012; Johnston et al., 2010; Murphy et al., 2003; Yoon et al., 2008; Yuen et al., 2012) and confirm the view that neural correlates of affective states are dispersed across the brain. Anderson and Oates (2010) have criticised the identification of neural correlates via MVPA arguing that MVPA can yield unstable results. However, Li et al. (2012) demonstrate the effectiveness of recursive feature elimination in combination with permutation tests to improve the overlap between informative features obtained in different folds and to limit the chance of including irrelevant features. If the MVPA results had indeed been unstable like Anderson and Oates argue then the likelihood of measuring substantial overlap of discriminative voxels across participants, as shown on the group-level probability maps, would have been slim. We thus argue that our RFE approach allowed us to obtain discriminative voxels that were indeed informative in different individuals. This approach alone does not refute the other main critique brought forward by Anderson and Oates, that successful classifier performance does not imply that the brain solves a task in the same way. Although we did not address this issue by testing performance of different classifiers, we are confident that the identified brain areas contributed to the perceptual-affective response to emotional valence because several of the discriminatory areas identified in the current study form part of a fronto-limbic system of areas involved in emotion processing and/or dysfunctional in patients with depression (Damasio, 1998; Drevets, 2001; Phan et al., 2002; Phillips et al., 2003), including the insula, amygdala, striatum, thalamus, hippocampus, SFG, (anterior) cingulate cortex and VLPFC.
Our study explored the neural underpinnings of the processing of IAPS pictures in a depressed sample via multivariate analysis. Several other studies have previously investigated the discriminability of different types of valence via a multivariate approach in healthy individuals (e.g. Baucom et al., 2012; Sitaram et al., 2011; Yuen et al., 2012) or patients with mood disorders (e.g. Mourão-Miranda et al., 2012a). The classifier accuracies that we obtained are comparable with similar studies. Baucom et al. (2012) presented IAPS pictures of different levels of arousal and valence to healthy participants and achieved accuracy scores around 80%. One explanation for the high classification accuracies in their study may be that the physical properties of the visual stimuli were more similar within than across valence conditions. Although they matched the stimuli set for hue, saturation and intensity values, identical stimuli could be repeated in the training and test trials, and the same was true for our study. To prevent the low level visual properties of the stimulus set from interfering with the classification of the emotional content we therefore excluded the posterior cortex from the analysis. Mourão-Miranda et al. (2012a) trained a classifier to discriminate between happy and neutral faces presented to groups composed of healthy controls, unipolar depressed patients and bipolar patients. Prediction rates of 81%, 70% and 61% respectively were obtained. In our study we show that high classification accuracies can be obtained even when comparing positive or negative emotions with a neutral condition in patients with unipolar depression.
Our study also shows that high classification accuracies can be obtained across sessions, which attests to the good reliability of the procedure. The stable representation of valence across the brain and across time is relevant for further clinical applications such as fMRI neurofeedback. Future neurofeedback studies will have the option to provide feedback of brain patterns (LaConte, 2011; Shibata et al., 2011; Sitaram et al., 2011) instead of restricted ROIs which might lead to more pronounced behavioural effects. As an example, the design of this study could be translated into a neurofeedback paradigm in which depressed patients would receive neurofeedback on their emotion regulation in response to the presentation of the positive, negative or neutral IAPS pictures. As depression has been associated with physiological abnormalities that are dispersed across the brain, it might be crucial to attempt to regulate patterns of brain activity across the whole brain with the aid of MVPA. Given the time constrains that apply to neurofeedback, the feasibility of real-time feedback from a pattern classifier was tested by running the SVM procedure only once. This resulted in very high prediction accuracies suggesting that the future for neurofeedback experiments applying real-time classification looks promising.
4.2. Univariate versus multivariate analysis
Although the sensitivity threshold of the univariate and multivariate analysis seems to differ only marginally, the group-level probability maps resulting from the univariate analyses were based on a substantially higher number of voxels due to the feature elimination steps conducted in the multivariate analyses. While the masks used to create the univariate-based group maps contained on average 109,701 voxels, the number of voxels that survived RFE ranged from 600 to 1400. Hence the likelihood of spatial overlap between discriminative voxels was much higher in the univariate compared to the multivariate analysis. Yet, we found the opposite: the group-level probability maps based on the univariate analysis showed less spatial overlap than the multivariate-based group maps. Because of the ability of MVPA to detect fine-grained activation patterns, MVPA thus seems more sensitive to detect stable representations. One reason for the lower stability of the univariate analysis might be that the activity levels in the areas for different conditions cancel each other out because of the high overlap between the conditions (Murphy et al., 2003). Another explanation could be that relatively weak activations that discriminated between conditions are too subtle to be picked up by univariate approaches. Several studies that compared the neural correlates of different valence conditions via a passive IAPS picture viewing paradigm in healthy subjects did find suprathreshold clusters. A direct comparison of negative over positive valence for instance resulted in clusters in the bilateral VLPFC and along the left middle and bilateral superior temporal gyrus in one study (Kensinger and Schacter, 2006), yet did not result in any significant clusters in another (Gerdes et al., 2010). It must be noted however that different studies selected different pictures from the IAPS database and that these studies were conducted in healthy individuals.
4.3. Group classification findings
Although we cannot make any claims about the predictive performance of the LDA analysis as it was based on activity maps created from the same data, our results suggest that significant differences exist between healthy and depressed individuals with respect to valence processing. The exploratory LDAs demonstrated that based on clusters in the bilateral VLPFC and dorsal cingulate cortex a perfect separation between the data of depressed patients and healthy controls was obtained. One previous classification study investigated which mixture of task conditions resulted in the maximum discrimination between healthy and depressed individuals (Hahn et al., 2011). While the single level classifiers performed above chance when taking neutral, happy or sad faces into account, it was only the responsiveness to neutral faces that served as a vital discriminatory criterion. It thus seemed that the differences between patients and controls in response to viewing happy and sad faces were highly similar to those in response to neutral faces. In contrast, the results from the current study suggest that the responsiveness to all valence levels was distinct in healthy and depressed participants. A potential explanation for the discrepancy in results is that our study used broader emotion categories because of which there were more facets along which healthy and depressed individuals could have differed, thereby being more suitable for group classification purposes. An alternative reason might be that in order to identify the most discriminative classifier, Hahn et al. (2011) predicted the accuracy for each of the 15 single condition classifiers as well as a decision tree algorithm that combined the descriptive probabilities of all single classifiers first. It has been demonstrated that the testing and selection of (most relevant) dependent variables from a subset inflates false-positive results (Simmons et al., 2011) yet the study did not take any measure to correct for potential type I errors. Its findings should thus be interpreted with caution. In contrast to Hahn et al. we obtained high group classification accuracies while solely depending on the neural patterns underlying emotion processing. This is intriguing since symptom constellations can vary considerably across patients with depression. Nevertheless our results suggest some common ground in emotion processing across patients with depression. We have also provided preliminary evidence that this neural basis differs from that observed in healthy controls. The replication of successful classifiers with independent samples could contribute to the development of biomarkers of mood state that might be used in the diagnosis and longitudinal monitoring of mood disorders.
4.4. Limitations and future studies
The major limitation of this study is that the datasets were not optimally designed for classification purposes. Consequently the test data in the MVPA was not composed of an independent sample. In addition, any medication-related differences cannot be ruled out due to the nature of both groups. Another limitation is that even though the visual cortex was excluded in the classification and prediction processes it cannot be ruled out that neural responses to the identical physical features of the stimuli in each session may have aided classification. The limited number of trials acquired in our healthy sample unfortunately did not allow a comparison of MVPA results obtained in healthy and depressed participants. Even though this was not the aim of our paper, which was to investigate the feasibility of pattern classification of valence in a depressed sample, future studies should attempt to contrast healthy and depressed individuals via MVPA. This may reveal potential differences previously unidentified by univariate methods. It would be beneficial for future studies to adopt larger datasets since the small number of participants in the current study limits the generalisability of our findings. Finally, future studies are required to confirm the advantage of multivariate over univariate analysis in other forms of emotion processing and other domains.
4.5. Conclusion
In conclusion, this study illustrated the capacity of multivariate analysis of brain activation data to successfully differentiate between highly overlapping neural activations that carry information about emotional valence in patients with depression with a limited numbers of trials and its superior sensitivity compared to the univariate analysis conducted in this study. Moreover, it appears that in our sample depressed patients could be separated from healthy controls with the use of regional activity patterns and the appropriate valence contrast. However, we did not test the discriminatory power of the same patterns in an independent patient group, which would be necessary to infer valid classification in the general population. The long-term goal of this research programme is the development of reliable diagnostic markers that allow the discrimination between healthy and depressed individuals, the identification of current mood state and predictions of which individuals are most likely to benefit from certain type of treatments ((Mourão-Miranda, Oliveira, et al., 2012).
Acknowledgements
This work was supported by a PhD studentship to I.H. from the National Institute for Social Care and Health Research (NISCHR) HS/10/25 and MRC grant G 1100629. We would like to thank Niklas Ihssen for the helpful suggestions on LDA analysis.
The authors report no conflicts of interest.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary data
Supplementary materials
References
- Anderson M.L., Oates T. Proceedings of the 32nd Annual Meeting of the Cognitive Science Society. 2010. A critique of multi-voxel pattern analysis. [Google Scholar]
- Baucom L.B., Wedell D.H., Wang J., Blitzer D.N., Shinkareva S.V. Decoding the neural representation of affective states. NeuroImage. 2012;59:718–727. doi: 10.1016/j.neuroimage.2011.07.037. [DOI] [PubMed] [Google Scholar]
- Belousov A.I., Verzakov S.A., Von Frese J. A flexible classification approach with optimal generalisation performance: support vector machines. Chemometrics and Intelligent Laboratory Systems. 2002;64:15–25. [Google Scholar]
- Craddock R., Holtzheimer P., III, Hu X., Mayberg H. Disease state prediction from resting state functional connectivity 1645. Magnetic Resonance in Medicine. 2009;62:1619–1628. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damasio A. Emotion in the perspective of an integrated nervous system. Brain Research Reviews. 1998;26:83–86. doi: 10.1016/s0165-0173(97)00064-7. [DOI] [PubMed] [Google Scholar]
- Davatzikos C., Ruparel K., Fan Y., Shen D.G., Acharyya M., Loughead J.W., Gur R.C., Langleben D.D. Classifying spatial patterns of brain activity with machine learning methods: application to lie detection. NeuroImage. 2005;28:663–668. doi: 10.1016/j.neuroimage.2005.08.009. [DOI] [PubMed] [Google Scholar]
- De Martino F., Valente G., Staeren N., Ashburner J., Goebel R., Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fMRI spatial patterns. NeuroImage. 2008;43:44–58. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- Drevets W.C. Neuroimaging and neuropathological studies of depression: implications for the cognitive–emotional features of mood disorders. Current Opinion in Neurobiology. 2001;11:240–249. doi: 10.1016/s0959-4388(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Ekman P. An argument for basic emotions. Cognition Emotion. 1992;6:169–200. [Google Scholar]
- First M., Spitzer R., Gibbon M., Williams J. Biometrics Research, New York State Psychiatric Institute; New York: 2002. Structured Clinical Interview for DSM-IV-TR Axis I Disorders Research Version. [Google Scholar]
- Fu C.H.Y., Mourao-Miranda J., Costafreda S.G., Khanna A., Marquand A.F., Williams S.C.R., Brammer M.J. Pattern classification of sad facial processing: toward the development of neurobiological markers in depression. Biological Psychiatry. 2008;63:656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Gerdes A.B.M., Wieser M.J., Mühlberger A., Weyers P., Alpers G.W., Plichta M.M., Breuer F., Pauli P. Brain activations to emotional pictures are differentially associated with valence and arousal ratings. Frontiers in Human Neuroscience. 2010;4:175. doi: 10.3389/fnhum.2010.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldapple K., Segal Z., Garson C., Lau M., Bieling P., Kennedy S. Modulation of cortical–limbic pathways in major depression. Archives of General Psychiatry. 2004;61:34–41. doi: 10.1001/archpsyc.61.1.34. [DOI] [PubMed] [Google Scholar]
- Grimm S., Beck J., Schuepbach D., Hell D., Boesiger P., Bermpohl F., Niehaus L., Boeker H., Northoff G. Imbalance between left and right dorsolateral prefrontal cortex in major depression is linked to negative emotional judgment: an fMRI study in severe major depressive disorder. Biological Psychiatry. 2008;63:369–376. doi: 10.1016/j.biopsych.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Hahn T., Marquand A.F., Ehlis A.-C., Dresler T., Kittel-Schneider S., Jarczok T.A., Lesch K.-P., Jakob P.M., Mourao-Miranda J., Brammer M.J., Fallgatter A.J. Integrating neurobiological markers of depression. Archives of General Psychiatry. 2011;68:361–368. doi: 10.1001/archgenpsychiatry.2010.178. [DOI] [PubMed] [Google Scholar]
- Hanke M., Halchenko Y.O., Sederberg P.B., José S., Haxby J.V., Pollmann S. PyMVPA: a python toolbox for multivariate pattern analysis of fMRI data. Neuoinfomatics. 2009;7:37–53. doi: 10.1007/s12021-008-9041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby J.V., Gobbini M.I., Furey M.L., Ishai A., Schouten J.L., Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- Haynes J.-D., Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nature Neuroscience. 2005;8:686–691. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- Ihssen N., Cox W.M., Wiggett A., Fadardi J.S., Linden D.E.J. Differentiating heavy from light drinkers by neural responses to visual alcohol cues and other motivational stimuli. Cerebral Cortex. 2011;21:1408–1415. doi: 10.1093/cercor/bhq220. [DOI] [PubMed] [Google Scholar]
- Johnston S., Linden D.E.J., Healy D., Goebel R., Habes I., Boehm S.G. Upregulation of emotion areas through neurofeedback with a focus on positive mood. Cognitive, Affective, & Behavioral Neuroscience. 2011;11:44–51. doi: 10.3758/s13415-010-0010-1. [DOI] [PubMed] [Google Scholar]
- Johnston S.J., Boehm S.G., Healy D., Goebel R., Linden D.E.J. Neurofeedback: a promising tool for the self-regulation of emotion networks. NeuroImage. 2010;49:1066–1072. doi: 10.1016/j.neuroimage.2009.07.056. [DOI] [PubMed] [Google Scholar]
- Keedwell P.A., Linden D.E.J. Integrative neuroimaging in mood disorders. Current Opinion in Psychiatry. 2013;26:27–32. doi: 10.1097/YCO.0b013e32835a0b63. [DOI] [PubMed] [Google Scholar]
- Kensinger E., Schacter D.L. Processing emotional pictures and words: effects of valence and arousal. Cognitive, Affective, & Behavioral Neuroscience. 2006;6:110–126. doi: 10.3758/cabn.6.2.110. [DOI] [PubMed] [Google Scholar]
- LaConte S.M. Decoding fMRI brain states in real-time. NeuroImage. 2011;56:440–454. doi: 10.1016/j.neuroimage.2010.06.052. [DOI] [PubMed] [Google Scholar]
- Lang P., Bradley M. The International Affective Picture System (IAPS) in the study of emotion and attention. In: Coan J., Allen J., editors. Handbook of Emotion Elicitation and Assessment. Oxford University Press; 2007. pp. 29–46. [Google Scholar]
- Lang P.J., Bradley M.M., Cuthbert B. Center for the Study of Emotion and Attention; Gainesville, FL: 1999. International Affective Picture System (IAPS): Technical Manual and Affective Ratings. [Google Scholar]
- Li Y., Long J., He L., Lu H., Gu Z., Sun P. A sparse representation-based algorithm for pattern localization in brain imaging data analysis. PLoS One. 2012;7:e50332. doi: 10.1371/journal.pone.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden D.E.J. The challenges and promise of neuroimaging in psychiatry. Neuron. 2012;73:8–22. doi: 10.1016/j.neuron.2011.12.014. [DOI] [PubMed] [Google Scholar]
- Linden D.E.J., Habes I., Johnston S.J., Linden S., Tatineni R., Subramanian L., Sorger B., Healy D., Goebel R. Real-time self-regulation of emotion networks in patients with depression. PloS One. 2012;7:e38115. doi: 10.1371/journal.pone.0038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquand A.F., Mourão-Miranda J., Brammer M.J., Cleare A.J., Fu C.H.Y. Neuroanatomy of verbal working memory as a diagnostic biomarker for depression. Neuroreport. 2008;19:1507–1511. doi: 10.1097/WNR.0b013e328310425e. [DOI] [PubMed] [Google Scholar]
- Misaki M., Kim Y., Bandettini P.A., Kriegeskorte N. Comparison of multivariate classifiers and response normalization for pattern-information fMRI. NeuroImage. 2010;53:103–118. doi: 10.1016/j.neuroimage.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T.M., Hutchinson R., Niculescu R.S., Pereira F., Wang X., Just M., Newman S. Learning to decode cognitive states from brain images. Machine Learning. 2004;57:145–175. [Google Scholar]
- Mourão-Miranda J., Almeida J.R.C., Hassel S., De Oliveira L., Versace A., Marquand A.F., Sato J.R., Brammer M., Phillips M.L. Pattern recognition analyses of brain activation elicited by happy and neutral faces in unipolar and bipolar depression. Bipolar Disorders. 2012;14:451–460. doi: 10.1111/j.1399-5618.2012.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão-Miranda J., Bokde A.L.W., Born C., Hampel H., Stetter M. Classifying brain states and determining the discriminating activation patterns: support vector machine on functional MRI data. NeuroImage. 2005;28:980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J., Oliveira L., Ladouceur C.D., Marquand A., Brammer M., Birmaher B., Axelson D., Phillips M.L. Pattern recognition and functional neuroimaging help to discriminate healthy adolescents at risk for mood disorders from low risk adolescents. PLoS One. 2012;7:e29482. doi: 10.1371/journal.pone.0029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F.C., Nimmo-Smith I., Lawrence A.D. Functional neuroanatomy of emotions: a meta-analysis. Cognitive, Affective, & Behavioral Neuroscience. 2003;3:207–233. doi: 10.3758/cabn.3.3.207. [DOI] [PubMed] [Google Scholar]
- Norman K.A., Polyn S.M., Detre G.J., Haxby J.V. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends in Cognitive Science. 2006;10:424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Ochsner K.N., Gross J.J. The cognitive control of emotion. Trends in Cognitive Science. 2005;9:242–249. doi: 10.1016/j.tics.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Phan K.L., Wager T., Taylor S.F., Liberzon I. Functional neuroanatomy of emotion: a meta-analysis of emotion activation studies in PET and fMRI. NeuroImage. 2002;16:331–348. doi: 10.1006/nimg.2002.1087. [DOI] [PubMed] [Google Scholar]
- Phillips M.L., Drevets W.C., Rauch S.L., Lane R. Neurobiology of emotion perception II: implications for major psychiatric disorders. Biological Psychiatry. 2003;54:515–528. doi: 10.1016/s0006-3223(03)00171-9. [DOI] [PubMed] [Google Scholar]
- Pinel P., Piazza M., Le Bihan D., Dehaene S. Distributed and overlapping cerebral representations of number, size, and luminance during comparative judgments. Neuron. 2004;41:983–993. doi: 10.1016/s0896-6273(04)00107-2. [DOI] [PubMed] [Google Scholar]
- Roseman I.J., Spindel M.S., Jose P.E. Appraisals of emotion-eliciting events: testing a theory of discrete emotions. Journal of Personality and Social Psychology. 1990;59:899–915. [Google Scholar]
- Russell A circumplex model of affect. Journal of Personality and Social Psychology. 1980;39:1066–1072. [Google Scholar]
- Schachter S., Singer J.E. Cognitive, social and physiological determinants of emotional state. Psychology Review. 1962;69:379–399. doi: 10.1037/h0046234. [DOI] [PubMed] [Google Scholar]
- Shibata K., Watanabe T., Sasaki Y., Kawato M. Perceptual learning incepted by decoded fMRI neurofeedback without stimulus presentation. Science. 2011;334:1413–1415. doi: 10.1126/science.1212003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegle G.J., Steinhauer S.R., Thase M.E., Stenger V.A., Carter C.S. Can't shake that feeling: event-related fMRI assessment of sustained amygdala activity in response to emotional information in depressed individuals. Biological Psychiatry. 2002;51:693–707. doi: 10.1016/s0006-3223(02)01314-8. [DOI] [PubMed] [Google Scholar]
- Simmons J.P., Nelson L.D., Simonsohn U. False-positive psychology: undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychological Science. 2011;22:1359–1366. doi: 10.1177/0956797611417632. [DOI] [PubMed] [Google Scholar]
- Sitaram R., Lee S., Ruiz S., Rana M., Veit R., Birbaumer N. Real-time support vector classification and feedback of multiple emotional brain states. NeuroImage. 2011;56:753–765. doi: 10.1016/j.neuroimage.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Talairach J., Tournoux P. Thieme Medical Publishers; Stuttgart: 1988. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: an Approach to Cerebral Imaging. [Google Scholar]
- Yoon J.H., Tamir D., Minzenberg M.J., Ragland J.D., Ursu S., Carter C.S. Multivariate pattern analysis of functional magnetic resonance imaging data reveals deficits in distributed representations in schizophrenia. Biological Psychiatry. 2008;64:1035–1041. doi: 10.1016/j.biopsych.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.S.L., Johnston S.J., Martino F., Sorger B., Formisano E., Linden D.E.J., Goebel R. Pattern classification predicts individuals' responses to affective stimuli. Translational Neuroscience. 2012;3:278–287. [Google Scholar]
- Zhang L., Samaras D., Brook S., Tomasi D., Volkow N., Goldstein R. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition CVPR. 2005. Machine Learning for Clinical Diagnosis from Functional Magnetic Resonance Imaging. [Google Scholar]
- Zhu C.Z., Zang Y.F., Liang M., Tian L.X., He Y., Li X.B., Sui M.Q., Wang Y.F., Jiang T.Z. Medical Image Computing and Computer-assisted Intervention: MICCAI … International Conference on Medical Image Computing and Computer-Assisted Intervention 8. 2005. Discriminative analysis of brain function at resting-state for attention-deficit/hyperactivity disorder; pp. 468–475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials


