Abstract
The fission yeast Schizosaccharomyces pombe has more metazoan-like features than the budding yeast Saccharomyces cerevisiae, yet it has similarly facile genetics. Here, we present a large-scale verified binary protein-protein interactome network, “StressNet”, based on high-throughput yeast two-hybrid screens of interacting proteins classified as part of stress-response and signal transduction pathways in S. pombe. We performed systematic, cross-species interactome mapping using StressNet and a protein interactome network of orthologous proteins in S. cerevisiae. With cross-species comparative network studies, we detected a previously unidentified component (Snr1) of the S. pombe mitogen-activated protein kinase Sty1 pathway. Coimmunoprecipitation experiments showed that Snr1 interacted with Sty1 and that deletion of snr1 increased the sensitivity of S. pombe cells to stress. Comparison of StressNet with the interactome network of orthologous proteins in S. cerevisiae showed that the majority of interactions among these stress-response and signaling proteins are not conserved between species, but are “rewired;” orthologous proteins have different binding partners in both species. In particular, transient interactions connecting proteins in different functional modules were more likely to be rewired than conserved. By directly testing interactions between proteins in one yeast species and their corresponding binding partners in the other yeast species with yeast two-hybrid assays, we found that about half of the interactions traditionally considered “conserved” form modified interaction interfaces that may potentially accommodate novel functions.
INTRODUCTION
A crucial step towards understanding properties of cellular systems is to map networks of DNA-protein, RNA-protein, and protein-protein interactions, or the “interactome network,” of an organism. Over the last decade, large-scale binary protein-protein interactome datasets have been produced for several eukaryotes – Saccharomyces cerevisiae (1-3), Drosophila melanogaster (4, 5), Caenorhabditis elegans (6, 7), Arabidopsis thaliana (8), and human (9, 10), among which we produced a high-quality whole-proteome interactome network in S. cerevisiae using a high-throughput yeast two-hybrid (HT-Y2H) system (1). However, due to large evolutionary distances among these species [the last common ancestor of fungi and human is over 1 billion years ago (11, 12)] and extremely low coverage (most protein interactions are yet to be detected) of available interactome maps outside of S. cerevisiae, the overlap among these networks is sparse (13). This makes it difficult to extract meaningful information about evolutionary relationships from these interactomes. Thus, to bridge this gap, it is essential to construct a high-coverage interactome network for an intermediate species. The fission yeast, Schizosaccharomyces pombe, has an easily manipulatable genome and is estimated to have diverged from the budding yeast, S. cerevisiae, approximately 400 million years ago (11, 12). Furthermore, fission yeast is more similar to metazoans than is budding yeast, especially in its gene regulation by chromatin modification and RNA interference, mechanisms that are differently regulated and absent, respectively, in budding yeast (14). A high-quality map of the protein-protein interactome network of S. pombe will enable analysis of biological properties of many complex pathways common in metazoan species but missing in S. cerevisiae (15).
The two yeasts live in highly disparate ecological niches and have varied mechanisms of responding to external stimuli. Therefore, in this study, we focus on 658 S. pombe genes involved in key regulatory processes of stress response and cellular signaling. Because these pathways control how organisms sense and adapt to their immediate environments, they are likely to have diverged between the two species. Using our HT-Y2H pipeline (1), we obtained a binary interactome network among these 658 genes, which we named “StressNet”. All interactions were verified with two orthogonal assays to ensure their quality. By comparing with their S. cerevisiae counterparts, we measured the conservation rate of these StressNet interactions between fission and budding yeasts using a Bayesian method. We found species-specific wiring of stress-response and signaling pathways beyond what was expected by sequence orthology, indicating that rewiring of protein interactome networks in related species is likely to be a major factor for divergence. We also identified a previously unknown component Snr1 of the Sty1 mitogen-activated protein kinase (MAPK) pathway and experimentally validated that Snr1 has gained functions, through rewiring of its interactions, compared to the orthologous protein in S. cerevisiae. Furthermore, to better understand the evolution of proteins and their interactions, we developed a large-scale cross-species interactome mapping approach to directly test interactions between S. pombe proteins and the S. cerevisiae orthologs of their partners. Such analysis is only possible with the availability of two well-controlled high-coverage interactome maps generated with the same technology. We found that, for many conserved interactions, both partners had co-evolved to accommodate new interactions and functions, and their interaction interfaces can no longer be recognized by their S. cerevisiae counterparts.
RESULTS
Comparison of known interactions in S. cerevisiae and S. pombe
The number of known protein-protein interactions in S. pombe is disproportionately lower than in other model eukaryotic organisms and human. We estimated the number of all known interactions in S. cerevisiae and S. pombe by analyzing seven commonly-used databases – BioGRID (16), DIP (17), IntAct (18), iRefWeb (19), MINT (20), MIPS (21), and VisANT (22). There identified 110,443 interactions for budding yeast, but only 4,038 for fission yeast, from these databases. Furthermore, only those interactions or interaction sets that have been validated by at least two independent assays are reliable and defined as “high quality” (23, 24). Based on this criterion, 519 fission yeast interactions are of high quality, as opposed to 25,335 high-quality interactions known in budding yeast. Of these, only 160 S. pombe interactions are binary (a direct biophysical interaction between the two proteins), as opposed to 11,936 in S. cerevisiae. These numbers (table S1) indicate the extent to which the fission yeast interactions are underexplored and necessitate the systematic mapping of its interactome network.
StressNet: A large-scale high-quality protein interactome network for stress response and cellular signaling in S. pombe
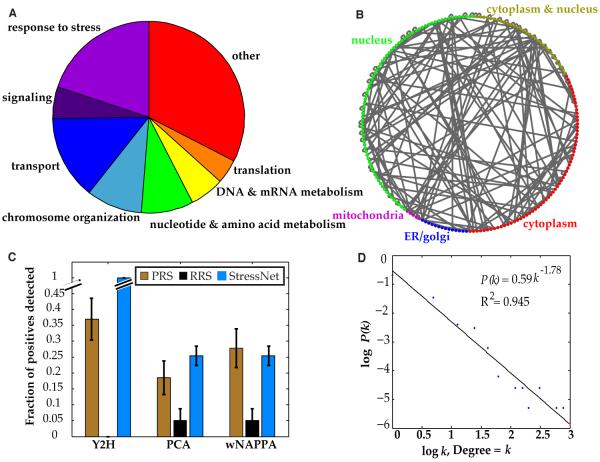
The subset of 658 genes for this study was selected using Gene Ontology (GO) (25) “Biological Process” (BP) functional annotations for fission yeast (Fig. 1A, table S2). To generate a high-quality high-coverage stress-response interactome map for S. pombe, we screened all possible protein pairs (>430,000) in this space three times using a high-quality HT-Y2H system, as we had done for S. cerevisiae (1). The resulting protein interactome network, StressNet (Fig. 1B), comprises 235 high-quality binary interactions among 200 proteins (table S3). Of these, 218 interactions were previously unknown. To validate our experimental pipeline and the quality of StressNet, from the 160 high-quality binary interactions we selected a set of 54 well-documented protein interactions from the literature and [“positive reference set” (PRS); table S4] and 43 random protein pairs that have never been reported or predicted to interact [“random reference set” (RRS); table S5]. 20 PRS interactions were successfully confirmed in our pipeline, whereas none of the RRS pairs were detected as positives (Fig. 1C). Therefore, the sensitivity [fraction of detected true positives among all possible true positives (1)] of our Y2H assay is 37.0%. To directly measure the quality of our Y2H-identified interactions (1, 26), we re-tested all 235 interactions detected in our HT-Y2H screen by two orthogonal assays: the protein complementation assay (PCA) (27) and the well-based nucleic acid programmable protein array (wNAPPA) (28), producing a fully-verified large-scale interactome map. The confirmation rates of our interactions with both orthogonal assays were similar to those of the PRS, further validating the high quality of StressNet (1, 26) (Fig. 1C). Using the results of the validating assays, we calculated the precision of StressNet as 95.3 ± 4.7% (Eq. 8 and 9 in Materials and Methods).
Figure 1.
S. pombe stress-response binary interactome network, StressNet. (A) Functional classification of the proteins included in our high-quality high-coverage HT-Y2H screen. (B) Network view of the stress-response binary interactome network in S. pombe. (C) Fraction of protein pairs in PRS, RRS, and StressNet that tested positive using Y2H, PCA, and wNAPPA. Data are shown as measurements + statistical error (SE). (D) Degree distribution of StressNet. P(k) is the probability that a protein has a degree = k.
To assign a confidence score to each interaction in StressNet, we implemented a random forest algorithm to integrate results from the three orthogonal assays (figs. S1 and S2 and Materials and Methods). Every detected interaction had a confidence score >0.76 (table S3). This value represents a normalized probability on a scale of 0 to 1 and indicated that all the interactions in StressNet were of high quality. Finally, to evaluate the topological properties of our network, we plotted the degree (number of interactions each protein has) distribution of StressNet (Fig. 1D and table S6). Protein interactomes are small-world scale-free networks (29, 30) and our stress-response interactome for S. pombe exhibited similar topological properties to other large-scale biological networks.
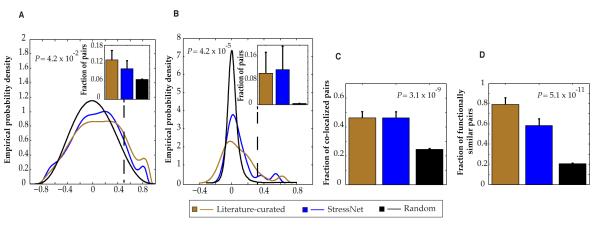
To assess the biological relevance of this network, we investigated overall relationships between protein pairs using expression and genetic interaction profile similarities (14, 31), subcellular colocalization (32), and GO functional similarities (25). We found significant enrichment of interactions in StressNet of protein pairs that colocalized or were functionally similar, and that were encoded by coexpressed genes or genes that exhibited similar genetic interaction profiles [calculated using the Pearson Correlation Coefficient (PCC)], relative to random expectation (Fig. 2A-D). Furthermore, the enrichment of StressNet in all four categories was similar to that of high-quality literature-curated binary interactions. These results confirmed the high quality of StressNet and indicated that these interactions are likely to be functionally relevant.
Figure 2.
Biological properties of StressNet interactions. (A) Pearson correlation coefficient (PCC) distribution of expression profiles of interacting and random protein pairs (dashed line corresponds to PCC cutoff above which pairs are considered to be significantly coexpressed; inset shows the fraction of significantly coexpressed pairs). (B) PCC distribution of genetic interaction profiles of interacting and random protein pairs (dashed line corresponds to PCC cutoff above which pairs are considered to be significantly similar; inset shows the fraction of pairs with significantly similar interaction profiles). (C) Enrichment of colocalized protein pairs. (D) Enrichment of protein pairs sharing similar functions. For each panel, the random set is constructed by considering all pairwise combinations of genes or proteins in the corresponding space. All P values represent comparisons between StressNet interactions and random pairs using a cumulative binomial test. Inset graphs and data in C and D are shown as measurements + SE.
Evolutionary relationships in StressNet
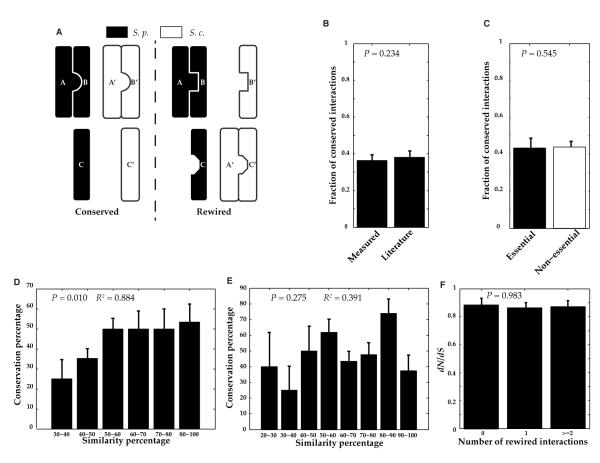
For biological networks, evolutionary relationships are commonly measured in terms of conservation and rewiring: If a pair of interacting proteins in one species has corresponding orthologs in another that also interact, then the interaction is considered to be conserved (an interolog); otherwise, the interaction is considered to be rewired (33-35) (Fig. 3A). To understand key principles governing the evolution of protein-protein interactions, especially for those in stress-response and signaling pathways, we compared the interactions in StressNet to their corresponding ortholog pairs in S. cerevisiae. We experimentally tested all corresponding S. cerevisiae protein pairs of the 235 interactions in StressNet and found that for 35 interactions, the corresponding budding yeast ortholog pairs were detected as interacting by our Y2H experiments. We developed a Bayesian framework to calculate the percentage of conserved interactions based on three parameters – the proportion of observed conserved interactions (35/235 = 14.9%), the precision (95.3 ± 4.7%), and the sensitivity (37.0% ± 4.4%) of our Y2H assay (see Eq. 12 and 13 in Materials and Methods). Substituting appropriate values, the percentage of conserved interactions between S. pombe and S. cerevisiae is calculated as 36.3 ± 2.9% (Fig. 3B).
Figure 3.
Evolutionary analysis of interactions. (A) Schematic of conserved and rewired interactions between the two yeast species. S.p., S. pombe; S.c., S. cerevisiae (B) Conservation rate (fraction of conserved interactions) in our interactome calculated in two different ways. Measured represents the value calculated using a Bayesian framework that incorporates the precision and recall of our assay. Literature represents the value estimated using budding yeast interactions reported in the literature. (C) Fraction of conserved interactions involving essential and non-essential proteins. The differences in B and C are not significant based on a cumulative binomial test. (D) Distribution of the fraction of conserved interactions as a function of overall sequence similarity. (E) Distribution of the fraction of conserved interactions as a function of sequence similarity of interaction interfaces. For D and E, P values are used to test whether there is a significant difference (using a cumulative binomial test) in conservation percentage between the groups corresponding to the lowest and highest similarity percentages. R2 (coefficient of determination) represents the significance of the correlation between conservation and similarity percentages. (F) Distribution of dN/dS [ratio of the number of non-synonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS)] as a function of number of rewired interactions. The differences are not significant based on a two-sided Kolmogorov-Smirnov test. Data are shown as the measurements + SE.
Using an orthogonal approach, we supplemented S. cerevisiae interactions detected in our Y2H experiments with high-quality known S. cerevisiae interactions curated from the literature to obtain 55 more StressNet interactions for which the corresponding budding yeast orthologs were reported to interact in the literature (24). There are 90 (35 + 55) conserved interactions in total (table S7) and the conservation is 38.3% ± 3.2%, consistent with the conservation calculated using the Bayesian framework (Fig. 3B). Furthermore, this agreement shows that after combining our Y2H experimental results with high-quality literature-curated interactions, the number of known interactions in our search space in S. cerevisiae is nearly complete, because if there were still a large number of unidentified interactions, the observed proportion of conserved interactions based on literature-curated interactions would have been much lower. Because it is always difficult to determine a negative interaction (1, 36), to ensure the set of rewired interactions is of high quality, we used a stringent set of criteria to define them (table S7) as those StressNet interactions without corresponding S. cerevisiae ortholog pairs and those interactions whose corresponding S. cerevisiae ortholog pairs have other high-quality interactions but have never been reported as interacting in the literature or tested positive in our Y2H experiments, and these ortholog pairs are known to have different cellular localizations (37).
Proteins encoded by essential genes, those when deleted cause lethality, tend to have more interacting partners (hubs) and also evolve more slowly than non-essential ones (38, 39). We found that essential and non-essential genes (40) in our interactome were equally likely to be involved in conserved interactions (Fig. 3C), contrary to previous studies (39). Stress-response and signal-transduction pathways play a crucial role in the process of adaptation to distinct ecological environments. As measured by the ratio of nonsynonymous to synonymous substitution rates (dN/dS) (41, 42), we found that the essential genes in these pathways evolve at the same rate as the non-essential genes in the pathways evolve, although on average all essential genes in the genome evolve significantly slower (fig. S3) than non-essential genes. To ensure that this is not an artifact of the calculation method, we also calculated dN/dS values for all essential and non-essential genes. Consistent with earlier findings (24), we observed that overall, the essential genes had a significantly lower average dN/dS (fig. S3). The average dN/dS for all stress-response genes is not significantly different from that for the entire genome (fig. S4). The dN/dS distributions for these two species are highly similar (fig. S5). This finding is consistent with analyses that suggest that these species are at comparable evolutionary distances from S. pombe (21, 22) and confirm that there are no inherent biases in our dN/dS calculations. Thus, our findings suggest that essential genes in stress-response and signal transduction pathways are under less negative selection such that their interactions are rewired for adaptive advantages through evolution.
To better understand the mechanisms underlying conservation and rewiring of interactions, we examined the relationship between sequence similarity of orthologous pairs and interaction conservation rates. Consistent with expectation (33), interactions involving proteins with higher overall sequence similarity or identity were more likely to be conserved (Fig. 3D, fig. S6). However, proteins interact through specific domains (43); therefore, we examined the role of sequence similarity of these interfaces in determining the conservation of corresponding interactions. Previous studies have established a homology modeling approach (44, 45) to locate interaction interfaces using co-crystal structures in PDB (46) and have found that analysis of these interfaces provides insights into their evolutionary rate (44). The conservation of an interaction depends on the conservation of the interfaces involved (47). Using a similar approach, we inferred interaction interfaces for proteins involved in 161 interactions in our network (Materials and Methods). We found no significant correlation between the similarity or identity of interaction interfaces and the conservation of the corresponding interactions (Fig. 3E, fig. S6). Examination of the average dN/dS ratios for proteins with different numbers of rewired interactions showed that the selection pressure on the gene did not affect the degree to which the interactions of the orresponding protein were rewired (Fig. 3F), further indicating that the rewiring of interactome networks and the divergence of related species are not completely dictated by evolution detected at the sequence level.
Functional profile of conserved and rewired interactions
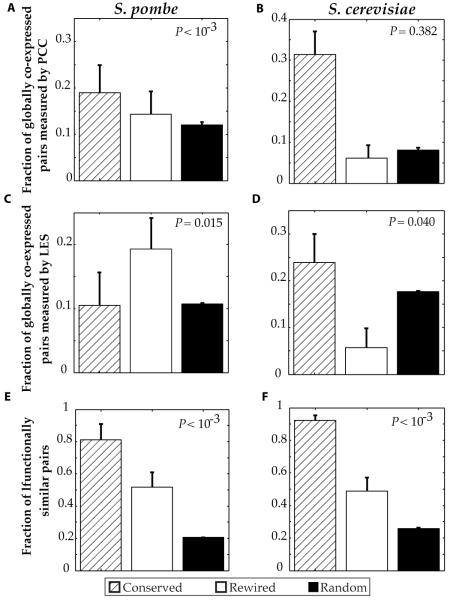
To investigate whether gene pairs encoding proteins involved in conserved and rewired interactions are differently regulated at the transcriptional level, we measured global coexpression between these pairs using the PCC. Global coexpression means that the pattern of gene expression of both genes is the same (fig. S7). Whereas conserved interactions had the highest fraction of coexpressed pairs,gene pairs encoding proteins involved in rewired interactions were also significantly more coexpressed than random in S. pombe (Fig. 4A). We also calculated coexpression relationships for the corresponding budding yeast pairs. By definition, the conserved pairs also interact in budding yeast, but the rewired pairs do not. The enrichment in gene expression is consistent with this distinction: Gene pairs encoding proteins involved in conserved interactions were coexpressed, genes encoding rewired pairs were not significantly enriched than random expectation in S. cerevisiae (Fig. 4A).
Figure 4.
Functional analysis of conserved and rewired interactions in S. pombe and S. cerevisiae. (A) Fraction of globally coexpressed pairs (as measured by PCC) among conserved and rewired interactions. (B) Fraction of locally coexpressed pairs (as measured by LES) among conserved and rewired interactions (C) Fraction of functionally similar pairs among conserved and rewired interactions. For each panel, the random set is constructed by considering all pairwise combinations of genes/proteins in the corresponding space. All P values represent comparisons between rewired interactions and random pairs using a cumulative binomial test. Data are shown as measurements + SE.
PCC captures only global coexpression relationships, but cannot capture local or transient coexpression that occurs only under certain conditions (fig. S7). Furthermore, gene pairs encoding proteins involved in stable interactions tend to be globally coexpressed, whereas those in transient interactions are often only locally coexpressed without significant PCC values (48). Stable and transient interactions both have important biological functions – the former constitute tightly connected modules, whereas the latter form key links between modules, especially in signal transduction pathways, and are more important than the stable ones or random interactions in maintaining the integrity of cellular networks (48). To detect transient interactions, we used the Local Expression-correlation Scores (LES) (48, 49). Rewired interactions in fission yeast had significantly higher LES values (Fig. 4B) than both conserved interactions and random expectation, suggesting that transient interactions are more likely to be rewired through evolution. Rewired pairs in budding yeast had LES values lower than random pairs (Fig. 4B), indicating that gene regulation for these pairs is also rewired.
Next, we examined GO functional similarities between interacting proteins involved in conserved and rewired interactions. Whereas conserved interactions had higher functional similarity than rewired interactions in fission and budding yeast, interacting protein pairs in both categories were significantly more functionally similar than random (Fig. 4C). This is in agreement with previous findings that conserved interactions tend to be in modules with specific functions, whereas rewired interactions tend to be inter-modular and have greater diversity in function (48).
In our analysis of rewired interactions above, we focused on those that are present in fission yeast but lost in budding yeast. Because the S. pombe interactome is still considerably underexplored in the literature and the sensitivity of our Y2H assay is 37.0%, it is not yet possible to determine non-interacting pairs in S. pombe reliably. Therefore, although it is possible to define lost interactions in S. cerevisiae by combining literature-curated interactions with our Y2H-detected ones, the same cannot be done to define lost interactions in S. pombe. However, there are 1,638 S. cerevisiae interactions where one protein has a corresponding S. pombe ortholog in the space of the 658 open reading frames (ORFs) that we explored and another protein has no S. pombe ortholog. Thus, there can be no corresponding S. pombe interactions and these are rewired interactions in S. cerevisiae by definition. We found that these interactions had significantly higher PCC, LES, and functional similarity as compared to random (fig. S8). The trend is comparable to that of rewired interactions in S. pombe (Fig. 4), further confirming the robustness of our results. We performed PCC and LES analysis of coexpression (fig. S9) and functional similarity (fig. S10) of conserved and rewired interactions defined at different confidence levels and obtained similar results, indicating that the analysis is robust and reliable.
Modes of rewiring uncovered by cross-species interactome mapping
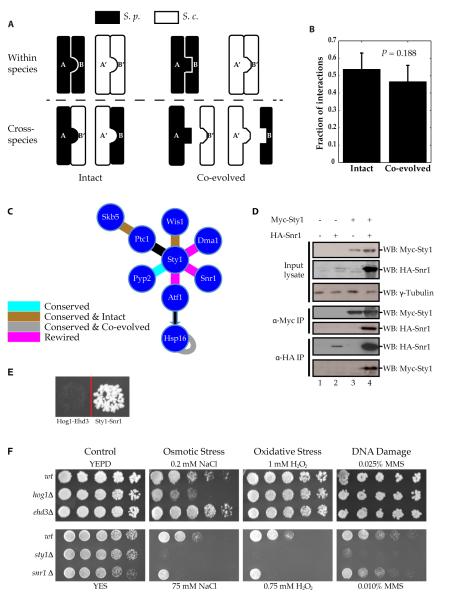
To further understand the meaning of “conservation” of interactions and experimentally explore the molecular mechanisms through which interaction interfaces evolve, we performed a systematic cross-species interactome mapping by testing all conserved interactions between corresponding S. cerevisiae and S. pombe proteins. Using orthologous pairs of interacting proteins in the two yeast species, we examined whether a protein in one species interacted with the ortholog of its partner in the other (Fig. 5A). Because we could detect the original interacting pairs from the same species with our Y2H experiments, we know that all four proteins are correctly expressed, folded, and are amenable to detection by our Y2H approach, thereby avoiding technical false negatives. The traditional definition of “conservation” implies the notion of conserved interfaces across different species. However, there are many examples where proteins with conserved interactions form new interactions and carry out new functions that are not conserved. The interface of a conserved interaction in fission yeast is considered “intact” if the proteins involved could also interact with the corresponding orthologs of their partners in budding yeast; otherwise, the interface is considered “co-evolved” (Fig. 5A). We found that these conserved interactions were equally likely result from an intact interface or co-evolved interface that formed new interaction interfaces that were unrecognizable by their orthologous counterparts in the other species (Fig. 5B). Earlier studies have suggested that interacting proteins may co-evolve to maintain structural complementarity and binding specificity (50-52). In this calculation, we used a lenient definition for an intact interface: We considered the interface intact if one or both of the cross-species interactions was positive, which provides a lower bound estimation of co-evolution between interacting proteins.
Figure 5.
Analysis of intact and co-evolved interactions. (A) Schematic of intact and co-evolved interactions. (B) Fraction of intact and co-evolved interactions in our interactome. Data are shown as measurements + SE. No significant difference detected using a cumulative binomial test. (C) The MAPK Sty1 stress response pathway. All undirected lines represent interactions detected in our interactome. The black arrow represents transcriptional regulation. (D) Sty1-Snr1 interaction validated in S. pombe using co-immunoprecipitation (N = 3 blots). (E) Y2H analysis of the ability of Hog1 and Ehd3 to interact and Sty1 and Snr1 to interact (N = 3 experiments). (F) Sensitivity assays for different deletion strains of S. cerevisiae and S. pombe under various stress conditions (N = 3 experiments).
Divergence of the Sty1 stress-response pathway through interaction conservation and rewiring
In S. pombe, Sty1 is activated in response to various stresses, including oxidative and osmotic stress, starvation, and other conditions (53, 54). Sty1 has orthologs in S. cerevisiae (Hog1, with 89% sequence similarity) and human (p38, with 69% sequence similarity). Both p38 and Sty1 respond to a wide range of stresses and both are different from Hog1 in terms of function (55). With our stress-response interactome, we detected key interactions at every step of the MAPK signal transduction pathway and, therefore, completely recapitulated the entire Sty1 pathway (fig. S11). This confirmed the sensitivity and accuracy of our HT-Y2H method, especially for discovering transient interactions in signaling pathways. Among all Sty1 interactions in StressNet, those with its activator (Wis1) and inhibitor (Pyp2) were both conserved between the two yeast species, and the Sty1-Wis1 interaction interface was intact. By contrast, the interaction between Sty1 and its known target in fission yeast, Atf1, represented a rewired interaction (Fig. 5C). We also identified a previously unknown interactor of Sty1: SPBC2D10.09, a protein that we named Snr1 (Sty1-interacting stress-response protein). To confirm this interaction in vivo, we performed co-immunoprecipitation of tagged proteins expressed in S. pombe (Fig. 5D, table S8). The amount of Snr1 pulled down in the presence of Sty1 was greater than that pulled down in the absence of Sty1, indicating that the interaction with Sty1 stabilizes Snr1 (Fig. 5D). The corresponding orthologous pair of Hog1 and Ehd3 in S. cerevisiae did not interact by Y2H (Fig. 5E). Cells lacking snr1 (snr1Δ cells) grew slower under stress, similar to sty1Δ cells (Fig. 5F and fig. S12), whereas growth of ehd3Δ cells was not compromised. These results suggested that Snr1 is a component of the Sty1 pathway and that its functions diverged from its budding yeast counterpart. Moreover, snr1 also has a human ortholog, HIBCH, further investigation of which may expand our knowledge of the human p38 MAPK pathway.
DISCUSSION
We generated StressNet – a high-quality high-coverage binary interactome for stress-response and signal-transduction pathways in the fission yeast, S. pombe. All interactions were verified by three orthogonal assays and assigned probabilistic confidence scores. We performed comparative network analysis to study the evolution of protein interactomes between the fission and budding yeast species. Even though 84% of StressNet interactions have corresponding orthologous pairs in S. cerevisiae, only about 40% of these interactions are conserved, indicating considerable evolutionary changes beyond simple sequence orthology. Thus, the interolog concept should be used with caution to infer interactions across species, especially if the two are not closely related. Furthermore, our results suggested that rewiring of protein interactome networks in related species is likely a major factor for divergence. Surprisingly, we found no significant correlation between the similarity of interaction interfaces and the conservation of corresponding interactions. This demonstrates that conservation of interactions is more complex than previously expected – domains that are not part of the interaction interface also play some indirect role in making the interaction possible. Even if the interface is conserved, the corresponding interaction could still be rewired because of steric hindrance due to altered overall structure or loss of nearby structural scaffolds that make the interaction thermodynamically favorable (56). We also experimentally explored the evolution of interaction interfaces and our analysis indicated that interactions traditionally considered “conserved” are equally likely to have intact interfaces as to have co-evolved ones that are different from their orthologous counterparts. These results suggest a molecular mechanism by which the interactome network is rewired through evolution: Many proteins have co-evolved with their partners to form modified interfaces that can, therefore, accommodate new interactions and functions.
Our results indicated that conserved interactions tended to be stable and rewired ones were more likely to be transient. Therefore, our finding provides a molecular-level mechanistic explanation for previous studies showing that genetic cross talk between functional modules can differ substantially (14, 57, 58). However, our results also suggest that, overall, proteins tend not to rewire all of their interactions; thus, even if they acquire novel interactions, they still generally conserve at least some of the original functions.
Our results indicate that substantial evolutionary changes, both rewiring and co-evolution, of stress-response pathways could be a major mechanism by which different organisms adapt to diverse living environments. Conservation of interactions in other pathways might be different from what we observed here. Therefore, similar cross-species interactome mapping and comparative network analyses of more pathways and species will provide a more comprehensive understanding of underlying principles that help shape distinct characteristics of individual organisms through evolution.
MATERIALS AND METHODS
Selection of genes for the study
This study focused on stress response and signal transduction proteins (based on GO Biological Process annotations) and their known interactors in S. pombe. We also include S. pombe orthologs of S. cerevisiae proteins that are known to interact with orthologs of fission yeast stress-response and signal transduction proteins. While selecting the 658 ORFs (table S2), we also ensured that a set of PRS interactions in S. pombe could be constructed with genes from our space, a limiting criterion because there are only 160 binary high-quality S. pombe interactions reported in the literature.
Yeast two-hybrid (Y2H)
Y2H experiments were carried out as described (59). Briefly, 658 S. pombe ORFs in Gateway entry vectors were transferred into AD and DB vectors using Gateway LR reactions. After bacterial transformation, plasmids of all AD-Y and DB-X clones were transformed into yeast two-hybrid strains MATa Y8800 and MATα Y8930 (genotype: leu2-3, 112 trp1-901 his3Δ200 ura3-52 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ cyh2R), respectively. The MATa Y8800 strain was obtained from the MATa Y550 strain after mutating CYH2 to introduce cycloheximide resistance. MATα Y8930 was generated by crossing MATa Y8800 with MATα Y1541 (3), followed by sporulation and identification of the MATα cycloheximide-resistant yeast strain by tetrad analysis. After AD-Y and DB-X were transformed into Y8800 and Y8930, respectively, autoactivators were screened by spotting onto synthetic complete media (SC) lacking histidine and tryptophan (AD-Y) or histidine and leucine (DB-X). These autoactivators were excluded from all further screenings. Each unique DB-X was mated with pools of ~188 unique AD-Y by co-spotting onto yeast extract peptone dextrose (YEPD) plates. Diploids were selected by replica plating onto SC plates without leucine and tryptophan (SC–Leu–Trp). To select for positive interactions, Y2H screening was performed by replica plating the diploids onto SC plates with 1 mM 3-amino-1,2,4-triazole (3-AT) and without leucine, tryptophan, and histidine (SC–Leu–Trp–His+3-AT). SC-Leu-Trp-His plates were used for the HT-Y2H screen in S. cerevisiae (1). We used 1 mM 3-AT, because this concentration greatly reduces background and improves the quality of the screens (8, 59, 60). Newly occurring autoactivators were determined by concurrently replica plating the diploids onto SC media with cyclohexamide (CHX) and 1 mM 3-AT and lacking leucine and histidine (SC–Leu–His+3-AT+CHX). Screening for these autoactivators relies on CHX to select for cells that do not have the AD plasmid, due to plasmid shuffling. Thus, growth on the latter plate identifies spontaneous autoactivators; these were removed from further analyses. All plates were replica cleaned the following day and scored after three additional days. The space was screened three times.
Y2H positives were grown two to three days at 30°C and then spotted onto four plates for secondary phenotype confirmation (phenotyping II) (SC–Leu–Trp–His+3-AT; SC–Leu–His+3-AT+CHX; SC–Leu–Trp–adenine; SC–Leu–adenine+CHX). Colonies that either grew on SC–Leu–Trp–His+3-AT but not on SC–Leu–His+3-AT+CHX or on SC–Leu–Trp–adenine but not on SC–Leu–adenine+CHX were identified as positives.
For colonies that scored positive in phenotyping II, the identities of DB-X and AD-Y were determined by the Stitch-seq approach (59) using Illumina sequencing. All identified interacting pairs were retested by pairwise Y2H.
Construction of PRS and RRS
The PRS and RRS are representatives of true positive interactions and negative pairs, respectively, and we used the PRS and the RRS to optimize the assay performance and they may be interpreted as positive and negative controls. The PRS comprises a set of 54 protein interactions from the literature, each of which is supported by at least two independent assays from two different publications (table S4). RRS pairs were generated from a random selection out of all possible protein pairs within our search space for which no interaction has yet been detected by any method (table S5). Because fission yeast interactions are underexplored, we also required that their corresponding budding yeast ortholog pairs have never been reported to interact.
Another way to construct the RRS is to consider protein pairs with different cellular localizations because these are unlikely to interact. 31 out of the 43 RRS pairs are indeed localized in different cell compartments. Using the whole RRS (Fig. 1C), we estimate the false positive rate for Y2H, PCA, and wNAPPA are 0/43, 2/43 (4.7% ± 3.2%), and 2/43 (4.7% ± 3.2%), respectively. If we only use the 31 RRS pairs localized in different cell compartments (named “RRS_DiffLocal”), the false positive rates for the three assays are 0/31, 2/31 (6.5% ± 4.4%), and 1/31 (3.2% ± 3.2%). Therefore, the false positive rates for all three assays used in our experiments do not change whether we use the complete RRS or RRS_DiffLocal.
With these controls, we found that 20 of the 54 PRS were detected in our screen and none of the RRS set. We calculated the sensitivity of our assay as 20/54 (37.0% ± 4.4%) .
Protein Complementation Assay (PCA)
S. pombe ORFs available in Gateway entry vectors were transferred by Gateway LR reactions into vectors encoding the two fragments of YFP (Venus variant) fused to the N-terminus of the tested proteins. Baits were fused to the F1 fragment (amino acids 1-158 of YFP) and preys to the F2 fragment (amino acids 159-239 of YFP). After bacterial transformation, plasmid DNA was prepared on a Tecan Freedom Evo bio-robot, and DNA concentrations are determined by OD260nm with a Tecan M1000 in a 96-well format. A 50 ng aliquot of each vector encoding the two proteins was used for transfection into HEK 293T cells in 96-well plates, using Lipofectamine 2000 (Invitrogen) reagent according to the instructions of the manufacturer. At approximately 48 hrs post-transfection, cells were processed with a Tecan M1000. A pair is considered interacting if the YFP fluorescence intensity was ≥2 fold higher over background.
Well-based nucleic-acid programmable protein array (wNAPPA)
ORFs encoding the interacting proteins were cloned into Gateway-compatible pCITE-HA and pCITE-GST vectors by LR reactions. After bacterial transformation, growth, DNA minipreps, and determination of DNA concentration, ~0.5μg of each plasmid were added to Promega TnT coupled transcription-translation mix (catalog number: L4610) and incubated for 90 minutes at 30°C to express proteins. During this time anti-GST antibody-coated 96-well plates (Amersham 96-well GST detection module, catalog number: 27-4592-01) were blocked at room temperature with PBS containing 5% dry milk powder. After protein expression, the expression mix was diluted in 100μl blocking solution, and added to the emptied pre-blocked 96-well plates. Expression mix was incubated in the 96-well plates for two hours at 15°C with agitation to allow for protein capture. After capture, plates were washed three times and developed by incubation with primary and secondary antibodies. Signal was visualized using chemiluminescence (Amersham ECL Reagents, catalog number: RPN2106) with a Tecan M1000. Wells with ≥3 fold higher intensity over background in either configuration were considered positives.
Measuring the precision of our assay
The precision of the Y2H assay was calculated using PCA and wNAPPA as orthogonal validation assays. Using Bayes’ rule we can build relationships between true and false positive rates of Y2H and observed positive interactions by a validating assay as:
| (Eq.1) |
where A+ corresponds to observing a positive interaction using the validating assay, Y+ corresponds to observing a positive interaction using Y2H, and T+ (T−) corresponds to an interaction being a real positive (negative) interaction. The precision of the Y2H is the term Pr(T+∣Y+) [which is also equal to 1−Pr(T −∣Y +)].
Assuming conditional independence between the validating assay and Y2H based on previously defined reasons (1), we can write:
| (Eq.2) |
Solving for the precision of the Y2H assay yields:
| (Eq.3) |
Pr(A +∣T+) and Pr(A+∣T−) were measured in the PRS and RRS experiments. So, for our Y2H assay we can write precision as:
| (Eq.4) |
where FStressNet is the fraction positive by an assay for StressNet, which is the best estimator for Pr(A+∣Y+). FPRS is the fraction positive by the assay for the PRS, which is an estimator for Pr(A+∣T+). FRRS is the fraction positive by the assay for the RRS, which is an estimator for Pr(A+∣T−).
The standard errors of FStressNet, FPRS, and FRRS are calculated using the standard error for binomial distributions:
| (Eq.5) |
where F is the fraction positive by the assay (FStressNet, FPRS, or FRRS and N is the total number of pairs tested.
To estimate the standard error for the precision, we used the standard delta method:
| (Eq.6) |
where X = f(A, B, C, …). A, B, C, … are independent random variables.
Here, the standard error of the precision is calculated as:
| (Eq.7) |
We have two validating assays, and we can incorporate the precision rates from these assays by calculating the average precision:
| (Eq.8) |
The standard error for the average precision is calculated by the delta method as:
| (9) |
Using this framework, we estimate the precision of our Y2H assay to be 95.3 ± 4.7%.
Calculating confidence scores for interactions
Using the random forest algorithm (61), we integrate results from Y2H, PCA, and wNAPPA and calculated confidence scores for interactions. Random forest is an ensemble classifier that constructs multiple decision trees by stochastic discrimination (62) and predicts a final class based on a weighted combination of the output class of each decision tree. It is considered to be a robust and accurate classifier for noisy datasets (61). We evaluated the performance of our classifier by five-fold cross validation on our reference set (union of PRS and RRS) and obtained moderately good performance (AUC = 0.64; fig. S2).
Determination of orthologs between S. pombe and S. cerevisiae
We use the list of orthologs provided by PomBase (63). The genome of S. cerevisiae underwent a duplication event (64). Thus, many S. pombe genes have two corresponding S. cerevisiae orthologous genes. Moreover, in a number of cases, the same S. cerevisiae gene has multiple S. pombe orthologs. Thus, the mapping considered for the study is “many-to-many”.
Estimation of the conservation of interactions
To estimate the conservation of protein-protein interactions between S. pombe and S. cerevisiae, we used a Bayesian framework that incorporates the precision and sensitivity of our Y2H assay:
| (Eq.10) |
where Pr(Cons+) corresponds to the conservation of protein-protein interactions between S. cerevisiae and S. pombe. The best estimator for Pr(Det) (the probability of detecting a S. cerevisiae interaction among proteins pairs that are orthologous to an interacting protein pair in StressNet) is Fdet, the fraction of the 235 StressNet interactions in S. pombe with corresponding Y2H-detected interactions in S. cerevisiae (35/235). Pr(Det∣Cons+) and Pr(Det∣Cons−) are estimated by FPRS and FRRS, the fractions of PRS and RRS interactions detected by our Y2H assay (20/54 and 0/43, respectively). By definition:
| (Eq.11) |
We can simplify the earlier equation to obtain an expression for Pr(Cons+):
| (Eq.12) |
To estimate the error for the conservation percentage, we used the standard delta method as described earlier. The standard deviation of Cons+ is given by:
| (Eq.13) |
Using the Y2H data, we calculated a conservation of 36.3 ± 2.9% interactions.
Another approach for measuring the conservation is to calculate fraction of S. cerevisiae interactions conserved in S. pombe. We mapped all S. pombe proteins in our space to their corresponding S. cerevisiae orthologs. We calculated the number of interactions in this S. cerevisiae space detected by our Y2H assay. We then mapped all the observed S. cerevisiae interactions to their corresponding S. pombe ortholog pairs and calculate the number of pairs detected as interacting in StressNet. We find that for 48/386 (12.4%) S. cerevisiae interactions, the corresponding S. pombe ortholog pairs also interact. Using the Bayesian framework described above, we calculate the conservation between S. pombe and S. cerevisiae interactions as 34.7 ± 2.0%, which is statistically the same (P = 0.708 using a cumulative binomial test) as the conservation calculated using the Y2H results (36.3 ± 2.9%).
Interaction conservation and confidence scores
After supplementing our Y2H experiments with high-quality interactions from the literature, we find that 90/235 (38.3%) interactions are conserved in StressNet. The statistical error associated with this measurement is related to the sample size and is calculated as the standard error [standard error = standard deviation / square root (N), where N is the number of samples]. The standard deviation is calculated based on the underlying probability distribution. The conservation percentage is obtained by a simple division (90/235 = 38.3%) and the underlying probability distribution is binomial (since each interaction can either be conserved or not, it corresponds to a Bernoulli event, the ensemble of which is modeled by a binomial distribution). The standard error is calculated using the appropriate formula for a binomial distribution: square root [p × (1-p) / N] = 3.2%, p = fraction of interactions that are conserved (90/235) and N = sample size (235)].
To test whether interactions with higher confidence scores were more likely to be conserved, we divided all StressNet interactions into two groups. The first group comprises interactions with confidence scores in the lower two quartiles and the second group comprises interactions with confidence scores in the upper two quartiles. We then compared the conservation for these two groups. We find that there is no significant difference (P = 0.37 using a two-sided Fisher exact test) in conservation rate between the two groups. This validates that the observed conservation rate is robust and not correlated with the confidence score associated with each interaction.
Evolutionary rates of genes and protein interactions
The evolutionary rate of genes is commonly measured in terms of the ratio of asynchronous nucleotide substitutions per asynchronous site to synchronous substitutions per synchronous site or dN/dS. This quantifies the selective evolutionary pressure on certain protein-coding genes to diverge faster, as opposed to others that may almost remain unchanged across species (20). To calculate the dN/dS values for all S. pombe genes, used two sequenced species in the Schizosaccharomyces genus – S. cryophilus and S. octosporus (21, 22). To determine orthology relationships, we used BLAST-x with default parameters (23) on all S. pombe genes. The top BLAST hit for each S. pombe gene against the indexed database of proteins for each of the two species was designated to be an ortholog, provided the E-value of the hit was < 0.05. Although the E-value cutoff is relatively high, it ensures that no potential pairs are missed. For pairs that have been incorrectly estimated to be orthologs, there is a correction step in downstream calculations that will return a dN/dS value of NaN (not a number), because of too high divergence. For all orthologous pairs, the Nei-Gojobori algorithm (20), which uses the Jukes-Cantor substitution model, was used to calculate dN/dS values.
Conservation of interactions and sequence similarity
Sequence similarity between S. pombe ORFs and their S. cerevisiae orthologs was measured by performing pairwise sequence alignment between all known ortholog pairs using the Needle program in the EMBOSS suite (65). It uses the Needleman-Wunsch alignment algorithm (66) to find the optimum alignment of two sequences along their entire length. The recommended default parameters – an affine gap penalty model (67) with an opening penalty of 10 and an extension penalty of 0.5 and the BLOSUM62 scoring matrix (68) – were used for the alignment. Because the lengths of orthologs may be dissimilar, we calculated the overall similarity percentage (OPS) with reference to the length of the S. pombe ORFs:
| (Eq.14) |
where, Nst is the total number of similar residues and L_Spt is the total length of the S. pombe ORF.
We then examined the relationship between the similarity percentage and the percentage of conserved interactions. Because the number of interactions varies considerably across different groups corresponding to different similarity percentages, we required each group to have at least 5 interactions. If any group had less than 5 interactions, it was merged with the next (higher) group. This ensured that our results were robust to outlier effects. We found that there was an increase in the degree of conservation with an increase in overall sequence similarity. To examine if the primary cause of this trend is the similarity of conserved domains, we identified domains on ortholog pairs that interact (69, 70). We defined the percentage similarity of interacting domains (PSID) as:
| (Eq.15) |
where Nsi is the number of similar residues in interacting domains and L_Spi is the sum of the lengths of the interacting domains in S. pombe.
We also repeated our analysis using sequence identity instead of similarity (fig. S6).
Inferring interaction interfaces from 3did and iPfam
In this study, we use interacting domains identified by 3did (69) and iPfam (70) to define interaction interface. To verify the reliability of inferring these domain-domain interactions, we performed three-fold cross-validation for 1,456 interaction pairs that have co-crystal structures. Because there are few co-crystal structures for S. pombe, this approach allowed us to obtain a meaningful estimate of the quality of the domain-domain predictions in these two databases. We split the pairs into three subsets such that two subsets were used for training and the third one was the test set. For each interaction pair in the test dataset, we scored a successful structural prediction when the predicted domain-domain interaction(s) had at least one co-crystal structure in support of it. We repeated the procedure thrice with each of the three subsets as the test set. Among the 1,456 PPI pairs, over 90% were correctly predicted with corresponding interacting domains, indicating that the predicted interaction interfaces used for our calculations were accurate (45).
Robustness of differences between sets of conserved and rewired interactions
To assess the robustness of the differences between sets of conserved and rewired interactions, we constructed different sets of conserved and rewired interactions corresponding to different confidence levels.
We constructed two sets of conserved interactions at different confidence levels – Conserved_HQ and Conserved_All. Conserved_HQ comprises only those interactions with corresponding S. cerevisiae ortholog pairs that tested positive in our Y2H experiments or were confirmed by two or more independent orthogonal assays in the literature. Conserved_All comprises all interactions in Conserved_HQ and those S. cerevisiae ortholog pairs that have been reported as interacting in the literature by only one assay.
We constructed five sets of rewired interactions at different confidence levels – Rewired_ByDefn, Rewired_HQ, Rewired_LC, Rewired_All_DiffLocal, and Rewired_All. Rewired_ByDefn comprises only those StressNet interactions for which at least one of the interacting proteins does not have a S. cerevisiae ortholog and, therefore, no corresponding interaction can exist in S. cerevisiae. Thus, these interactions are rewired by definition. Rewired_HQ comprises all interactions in Rewired_ByDefn and those interactions for which the corresponding S. cerevisiae ortholog pairs have other high-quality interactions but have never been reported as interacting in the literature or tested positive in our Y2H experiments, and these ortholog pairs are known to have different cellular localizations. Thus, these correspond to S. pombe interactions with corresponding budding yeast ortholog pairs that are in principle non-interacting, because they have different cellular localizations (1, 71) and they participate in well-validated interactions with other proteins but have never been reported to interact in the literature. Rewired_LC comprises all interactions in Rewired_ByDefn and those interactions with corresponding ortholog pairs that have other high-quality interactions but have never been reported as interacting in the literature or tested positive in our Y2H experiments. Rewired_All_DiffLocal corresponds to all interactions in Rewired_ByDefn and those interactions with corresponding ortholog pairs that have different cellular localizations. Rewired_All comprises all interactions that are not in Conserved_All.
Construction of myc-sty1 and HA-snr1 expression clones
S. pombe sty1 and snr1 genes were PCR amplified using the following primers – sty1-pNCH1472-Forward, sty1-pNCH1472-Reverse, snr1-pSGP73-Forward, and snr1-pSGP73-Reverse (table S8). The sty1 PCR product was cloned into a pNCH1472-myc vector using the NotI and SalI restriction sites. The snr1 PCR product was cloned into a pSGP73-HA vector using the NotI and BglII restriction sites. pNCH1472-myc-sty1 and pSGP73-HA-snr1 were single or double transformed into S. pombe KGY553 (ATCC). Transformed yeast was selected on Edinburgh minimal medium (EMM)–Ura plates for pNCH1472-myc-sty1, EMM–Leu for pSGP73-HA-snr1, and EMM–Ura–Leu for double transformation.
Coimmunoprecipitation and Western blotting
Transformed yeast (KGY553) containing pNCH1472-myc-sty1 or pSGP73-HA-snr1 or both were cultured overnight in 10 mL EMM selection medium. Yeast pellets were washed in 5 mL of cold TE buffer before protein extraction. To lyse cells, 1 mL of lysis buffer (50mM Tris-HCl pH 7.5, 0.2% Tergitol, 150 mM NaCl, 5 mM EDTA, Complete Protease Inhibitor tablet) and 600 μL glass beads were added to each tube and mixed in a beater for two rounds of 10 minutes each. Protein extracts were centrifuged for 10 minutes at 13,200 rpm at 4°C in an Eppendorf 5415R centrifuge.. Then, 500 μL of supernatant was immunoprecipitated overnight using 20 μL of EZview™ Red Anti-c-Myc Affinity Gel (Sigma-Aldrich E6654) or EZview™ Red Anti-HA Affinity Gel (Sigma-Aldrich E6779). The next morning, beads were washed three times using cold lysis buffer before being subjected to SDS-PAGE and Western blotting analysis. Primary antibodies used in our analysis were anti-c-Myc (Santa Cruz sc-789), anti-HA (Roche 12CA5), and anti-γ-tubulin (Sigma-Aldrich T5192).
Construction of yeast deletion strains
The snr1Δ strain was obtained from the Bioneer Schizosaccharomyces pombe Genome-wide Deletion Library. The deletion strain was verified by PCR using primers SpEhd3_Up_Fwd and Sp_Dn_Rev spanning the 3′ end of snr1 and the region immediately downstream. Primers specific for KanMX4 (KanMX4-Fwd and KanMX4-Rev) were used to detect the deletion cassette. A PCR-based strategy was used to construct the sty1Δ strain. Briefly, in the first round of PCR, primers (PFA6a_Sty1_Fwd and PFA6a_Sty1_Rev) with 20 base pairs (bp) homology to the regions upstream and downstream of sty1, respectively, were synthesized for PCR of the pFA6a-KanMX6 cassette. Primers with 20 bp homology to the pFA6a-KanMX6 were synthesized to PCR 290 bp upstream (Sty1Del-Up_Fwd and Sty1Del-Up_Rev) and 290 bp downstream (Sty1Del-Dn_Fwd and Sty1Del-Dn_Rev) of sty1, not including the sty1 gene. The three PCR products were stitched together sequentially with a second round of PCR. Stitch PCR of the upstream region and pFA6a-KanMX6 and of the downstream region and pFA6a-KanMX6 were carried out separately. In the third round of PCR, both upstream and downstream stitched PCR products were further stitched together to produce a final product of pFA6a-KanMX6 flanked on the 5′ and 3′ ends by 290 bp that are homologous to the upstream and downstream regions of chromosomal sty1 (Sty1Del-Up_Fwd and Sty1Del-Dn_Rev). The final PCR product was transformed into S. pombe 972h-canonical wild-type (ATCC). Transformed yeast was selected on yeast extract-sucrose (YES) media plates containing 150 mg/L G418. Insertion of the pFA6a-KanMX6 cassette by homologous recombination at the sty1 locus was verified by PCR using primers to target the entire cassette (Sty1Del-Up_Fwd and Sty1Del-Dn_Rev) and to target a sty1 internal region of 401 bp (Sty1_Fwd and Sty1_Rev). In addition, sty1 and snr1 deletion were performed in S. pombe KGY553 (ATCC) wild-type (h- his3-D1 leu1-32 ura4-D18 ade6-M216) background using a similar PCR strategy. Sequences of primers used for deletion and verification of strains in this study are listed in table S8.
Stress Sensitivity Assays
S. cerevisiae was grown in YEPD and S. pombe was grown in YES medium. All yeast strains were initially grown as a starter culture overnight at 30°C. From the starter culture, yeast cells were diluted into fresh medium to an initial OD600nm = 0.2. The cultures were grown to mid-log phase (OD600nm = 0.7). The S. cerevisiae and S. pombe strains were serially diluted 4-fold in sterile water and spotted onto YEPD and YES plates, respectively, containing various stressors. Spotted plates were incubated at 30°C and yeast growth was assessed after 3 days.
Supplementary Material
Acknowledgements
We thank Dr. Anthony Bretscher for providing budding yeast deletion strains, Dr. Susan Forsburg for providing fission yeast expression vectors, Dr. Marcus Smolka for experimental advice, and Drs. Andrew Clark and Eric Alani for critical reading of our manuscript.
Funding: J.D. was supported by the Tata Graduate Fellowship. A.M. was supported by Grant-in-Aid for Scientific Research (C). J.P. was supported by NIGMS grant GM098634. F.R. was supported by NIH Grant HG001715, by the Canada Excellence Research Chairs Program, and by the Canadian Institute for Advanced Research. This work was funded by US National Institute of General Medical Sciences grant R01 GM097358 to H.Y.
Footnotes
Author Contributions: H.Y. and F.R. conceived the study. H.Y. designed all experiments and analyses, and oversaw all aspects of the project. J.D. carried out upstream experimental design and all downstream computational analyses. A.M. and M.Y. were involved in Y2H library construction. T.V.V., V.T., A.G.D., L.W., and N.A.C performed interactome screens using HT-Y2H. T.V.V. and N.A.C. performed PCR stitching. J.M. and F.R. performed Illumina sequencing. X.Wang performed sequence alignment for Stitch-seq. T.V.V. performed pairwise re-tests using HT-Y2H. H.Y. performed PCA and wNAPPA experiments. J.P. provided S. pombe strains. J.P., S.M.L., and H.Y. designed co-immunoprecipitation and stress-response assays. X.Wei performed coimmunoprecipitation experiments. T.V.V. and N.Z.K. performed stress-response assays. J.D. and H.Y. wrote the manuscript with contributions from all authors.
Conflict of interest: None.
Data and materials availability: The Y2H data are available from HINT (http://hint.yulab.org), BioGRID (http://thebiogrid.org/) and IntAct (http://www.ebi.ac.uk/intact/).
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
REFERENCES AND NOTES
- 1.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet AS, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabasi AL, Tavernier J, Hill DE, Vidal M. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 4.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de Gunzburg J, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr., White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 6.Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, Cevik S, Klitgord N, Fan C, Braun P, Li N, Ayivi-Guedehoussou N, Dann E, Bertin N, Szeto D, Dricot A, Yildirim MA, Lin C, de Smet AS, Kao HL, Simon C, Smolyar A, Ahn JS, Tewari M, Boxem M, Milstein S, Yu H, Dreze M, Vandenhaute J, Gunsalus KC, Cusick ME, Hill DE, Tavernier J, Roth FP, Vidal M. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium AIM. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. REVIEWS1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 13.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, Mishra G, Nandakumar K, Shen B, Deshpande N, Nayak R, Sarker M, Boeke JD, Parmigiani G, Schultz J, Bader JS, Pandey A. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 14.Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, Hoe KL, Kim DU, Ideker T, Grewal SI, Weissman JS, Krogan NJ. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan NJ, Stewart AF. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust JM, Winter A, Dolinski K, Tyers M. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner B, Razick S, Turinsky AL, Vlasblom J, Crowdy EK, Cho E, Morrison K, Donaldson IM, Wodak SJ. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database (Oxford) 2010;2010:baq023. doi: 10.1093/database/baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mewes HW, Ruepp A, Theis F, Rattei T, Walter M, Frishman D, Suhre K, Spannagl M, Mayer KF, Stumpflen V, Antonov A. MIPS: curated databases and comprehensive secondary data resources in 2010. Nucleic Acids Res. 2011;39:D220. doi: 10.1093/nar/gkq1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, Ng DM, Yamada T, Chen C, Kawashima S, Mellor J, Linghu B, Kanehisa M, Stuart JM, DeLisi C. VisANT 3.0: new modules for pathway visualization, editing, prediction and construction. Nucleic Acids Res. 2007;35:W625. doi: 10.1093/nar/gkm295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, Vandenhaute J, Galli M, Yazaki J, Hill DE, Ecker JR, Roth FP, Vidal M. Literature-curated protein interaction datasets. Nat Methods. 2009;6:39. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das J, Yu H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst Biol. 2012;6:92. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, Venkatesan K, Rual JF, Vandenhaute J, Cusick ME, Pawson T, Hill DE, Tavernier J, Wrana JL, Roth FP, Vidal M. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 29.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 30.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 31.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 33.Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shou C, Bhardwaj N, Lam HY, Yan KK, Kim PM, Snyder M, Gerstein MB. Measuring the evolutionary rewiring of biological networks. PLoS Comput Biol. 2011;7:e1001050. doi: 10.1371/journal.pcbi.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Hur A, Noble WS. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformatics. 2006;7 Suppl 1:S2. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 38.Hirsh AE, Fraser HB. Protein dispensability and rate of evolution. Nature. 2001;411:1046. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 39.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 40.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 42.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Muller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. Comparative functional genomics of the fission yeasts. Science. 2011;332:930. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotech. 2012;30:159. doi: 10.1038/nbt.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Espadaler J, Romero-Isart O, Jackson RM, Oliva B. Prediction of protein-protein interactions using distant conservation of sequence patterns and structure relationships. Bioinformatics. 2005;21:3360. doi: 10.1093/bioinformatics/bti522. [DOI] [PubMed] [Google Scholar]
- 48.Das J, Mohammed J, Yu H. Genome scale analysis of interaction dynamics reveals organization of biological networks. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J Mol Biol. 2001;314:1053. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 50.Hakes L, Lovell SC, Oliver SG, Robertson DL. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc Natl Acad Sci U S A. 2007;104:7999. doi: 10.1073/pnas.0609962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim WK, Bolser DM, Park JH. Large-scale co-evolution analysis of protein structural interlogues using the global protein structural interactome map (PSIMAP) Bioinformatics. 2004;20:1138. doi: 10.1093/bioinformatics/bth053. [DOI] [PubMed] [Google Scholar]
- 52.Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. J Mol Biol. 2000;299:283. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- 53.Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24:961. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 54.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 55.Bone N, Millar JB, Toda T, Armstrong J. Regulated vacuole fusion and fission in Schizosaccharomyces pombe: an osmotic response dependent on MAP kinases. Curr Biol. 1998;8:135. doi: 10.1016/s0960-9822(98)00060-8. [DOI] [PubMed] [Google Scholar]
- 56.Kastritis PL, Moal IH, Hwang H, Weng Z, Bates PA, Bonvin AM, Janin J. A structure-based benchmark for protein-protein binding affinity. Protein Sci. 2011;20:482. doi: 10.1002/pro.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, Kliegman JI, Jiang L, Kuo D, Tosti E, Kim HS, Edelmann W, Keogh MC, Greene D, Tang C, Cunningham P, Shokat KM, Cagney G, Svensson JP, Guthrie C, Espenshade PJ, Ideker T, Krogan NJ. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frost A, Elgort MG, Brandman O, Ives C, Collins SR, Miller-Vedam L, Weibezahn J, Hein MY, Poser I, Mann M, Hyman AA, Weissman JS. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu H, Tardivo L, Tam S, Weiner E, Gebreab F, Fan C, Svrzikapa N, Hirozane-Kishikawa T, Rietman E, Yang X, Sahalie J, Salehi-Ashtiani K, Hao T, Cusick ME, Hill DE, Roth FP, Braun P, Vidal M. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, Yildirim MA, Simonis N, Heinzmann K, Gebreab F, Sahalie JM, Cevik S, Simon C, de Smet AS, Dann E, Smolyar A, Vinayagam A, Yu H, Szeto D, Borick H, Dricot A, Klitgord N, Murray RR, Lin C, Lalowski M, Timm J, Rau K, Boone C, Braun P, Cusick ME, Roth FP, Hill DE, Tavernier J, Wanker EE, Barabasi AL, Vidal M. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breiman L. Random forests. Machine Learning. 2001;45:5. [Google Scholar]
- 62.Kleinberg EM. An overtraining-resistant stochastic modeling method for pattern recognition. Annals of Statistics. 1996;24:2319. [Google Scholar]
- 63.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, Oliver SG. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 65.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 66.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 67.Vingron M, Waterman MS. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J Mol Biol. 1994;235:1. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
- 68.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stein A, Ceol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn RD, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 71.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.