Abstract
Chemogenomics methods seek to characterize the interaction between drugs and biological systems and are an important guide for the selection of screening compounds. The acid/base character of drugs has a profound influence on their affinity for the receptor, on their absorption, distribution, metabolism, excretion and toxicity (ADMET) profile and the way the drug can be formulated. In particular, the charge state of a molecule greatly influences its lipophilicity and biopharmaceutical characteristics.
This study investigates the acid/base profile of human small molecule drugs, chemogenomics datasets and screening compounds including a natural products set. We estimate the ionization constants (pKa values) of these compounds and determine the identity of the ionizable functional groups in each set. We find substantial differences in acid/base profiles of the chemogenomic classes. In many cases, these differences can be linked to the nature of the target binding site and the corresponding functional groups needed for recognition of the ligand. Clear differences are also observed between the acid/base characteristics of drugs and screening compounds. For example, the proportion of drugs containing a carboxylic acid was 20%, in stark contrast to a value of 2.4% for the screening set sample. The proportion of aliphatic amines was 27% for drugs and only 3.4% for screening compounds. This suggests that there is a mismatch between commercially available screening compounds and the compounds that are likely to interact with a given chemogenomic target family. Our analysis provides a guide for the selection of screening compounds to better target specific chemogenomic families with regard to the overall balance of acids, bases and pKa distributions.
Keywords: Acid, Acidity, Base, Basicity, Chemogenomics, Drug discovery, Functional groups, GPCR, Ion channels, Ionization constant, Kinases, pKa
Introduction
Chemogenomics is the study of the interaction between small molecules and biological systems.[1] To improve the efficiency of the drug discovery process, medicinal chemists must broadly apply chemogenomic principles to tailor synthesis or screening efforts so that the candidate molecules are matched to their target families. Correlating compound space with target space has fuelled many studies in this area[2] and in this current study we extend these studies by specifically exploring the influence of molecule acid/base properties on biological activity in a large number of chemogenomics datasets. Furthermore, we complement these analyses by investigating the acid/base properties of screening compounds (including a ‘natural product’ set) that are available for purchase and testing in medicinal chemistry settings. In all cases the data can be used to compare their profiles to drugs.
The acidic and basic functional groups present in a molecule are an important factor in chemogenomics because they strongly affect its physicochemical properties, such as lipophilicity and aqueous solubility and, in turn, these factors influence compound behaviour within biological systems; receptor interactions and absorption, distribution, metabolism, excretion and toxicology (ADMET).[3] Specific examples of the influence of acid/base properties on ADMET characteristics include the propensity of acids to exhibit higher plasma protein binding, affecting volumes of distribution,[4] while basic compounds tend to show greater toxicity through mechanisms such as phospholipidosis[5] and hERG channel binding.[6]
Attempts have been made to draw relationships between ligand promiscuity and the properties MW and logP.[7] Larger and more lipophilic molecules have the potential to interact with more than one target.[8] Charge states also may also play a role in off-target selectivity as basic compounds have been suggested to be more promiscuous than acids, ampholytes or neutral molecules.[8a,9] Ionization constants therefore, fundamentally relate to how compounds behave and their overall drug-like character.[3b] Moreover, pKa values and ionization states critically influence the formulation of drugs, especially in cases where compounds need to be administered in solution. Of the main contributors to aqueous solubility,[3a] polarity is a key parameter yet this needs to be balanced with suitable lipophilicity for a molecule to cross membranes and to interact with specific macromolecules. Receptor interactions likewise require that drug functional groups complement those in the binding site of the macromolecule. In energetic terms, electrostatic interactions may provide a boost to compound potency yet this needs to be considered alongside the concept of ligand efficiency in drug discovery.[10] Overall, charge states and ionization constants deeply influence the properties of drugs and further study of acid/base profiles is needed.
Despite the broad importance of acid/base properties in drug discovery, extensive surveys of this property have not been undertaken. To gain a greater understanding of the acid/base character of drugs we have previously examined the pKa distributions of marketed drugs.[11] This latter work[11b] however, only explored contemporary drugs and expanding this to broader sets of compounds will be of benefit.
This study examines the acid/base profile of drugs, chemogenomics datasets, and screening compounds and provides three levels of analysis. An overview of compound types will be provided that identifies compounds with ionizable functional groups and classifies these substances into categories. The pKa values of ionizable compounds are presented as histograms for both single acid and single base containing compounds. Finally, the proportion of ionizable functional groups in each dataset is listed. Overall, we wish to be able to ask questions regarding whether screening compounds or research chemicals match the acid/base character of drugs and to further our knowledge of chemogenomics. The research aims to improve our awareness of acid/base profiles and their fundamental influence on bioactivity.
Results and Discussion
The ionization state of a bioactive compound strongly influences its behaviour within a biological system; how the compound interacts with a target binding site, or how it partitions between aqueous and lipid environments. The actual ionization state of a compound is dependent on the pH of the local environment and while usually close to neutral (~7.4), this can cover a wide pH range. For example, within the gut pH values can range from 1 to 8 and, significantly, within the microenvironment of ligand binding sites the pKa of functional groups can be perturbed by two units or more.[12] Studies of drug formulation may also need to consider a wide pH range. The significance of acidic and basic behaviour is not therefore a simple study of ionization states, it must also embrace a range of biopharmaceutical scenarios encountered in the drug discovery process.
Contemporary drugs
The drugs dataset of human approved drugs (Drugs) was selected to represent contemporary therapeutic substances. This curated dataset consists of drugs approved by the United States, European and Asian regulatory agencies and can be considered to be internationally significant from a pharmaceutical and medicinal viewpoint.
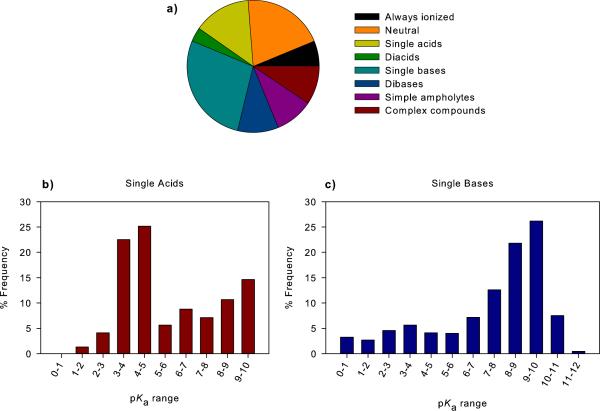
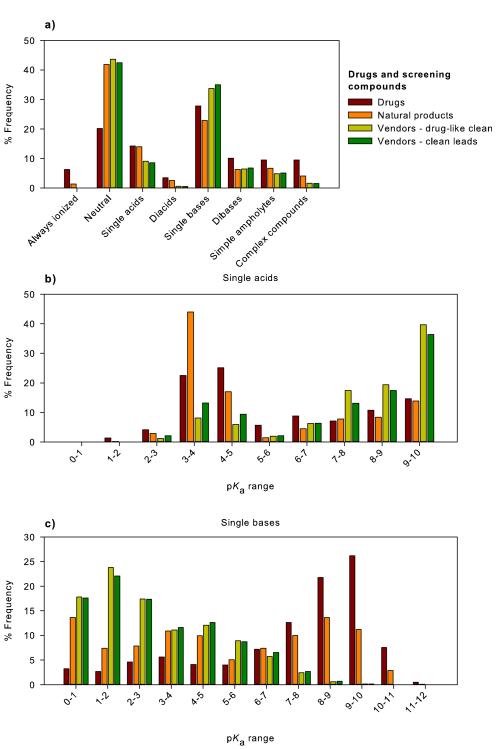
In this study, we filtered the drugs dataset to retain all carbon containing substances and remove compounds containing heavy metals, salts, mixtures, polymers and those with a molecular weight over 1000 to be consistent with previous work.[11b] The filtered dataset is comprised of a total of 3,766 compounds. pKa values were predicted for each molecule and compounds were classified into charge state categories; always ionized (6.3 %), neutral (20.2 %) single acids (14.2 %) diacids (3.5 %) single bases (27.8 %) dibases (10.1 %), simple ampholytes – compounds containing one acid and one base (9.5 %) and complex compounds (8.4 %) (Table 1, Figure 1A). Figures 1B and 1C show the pKa distributions of single acid and single base containing compounds in the drugs dataset.
Table 1.
Charge state categories of compounds in the chemogenomics and screening compound datasets.
| Dataset | Number of compounds |
Proportion (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Always ionized |
Neutral | Single acids |
Diacids | Single bases |
Dibases | Simple ampholytes |
Complex comp- ounds |
||
| Drugs | 3766 | 6.3 | 20.2 | 14.2 | 3.5 | 27.8 | 10.1 | 9.5 | 8.4 |
|
| |||||||||
| Aspartyl proteases | 4300 | 0.1 | 25.9 | 11.1 | 6.0 | 24.3 | 18.5 | 5.0 | 9.1 |
| Cysteine proteases | 2503 | 0.7 | 45.2 | 10.4 | 3.5 | 22.5 | 8.6 | 4.4 | 4.7 |
| GPCR amines | 20712 | 2.7 | 0.3 | 0.0 | 0.0 | 54.1 | 24.5 | 7.4 | 11.1 |
| GPCR cannabinoid | 1510 | 0.1 | 35.5 | 13.8 | 0.5 | 39.9 | 7.6 | 2.4 | 0.1 |
| GPCR nucleotide | 5228 | 1.1 | 12.5 | 16.9 | 1.1 | 23.0 | 27.3 | 6.8 | 11.4 |
| GPCR class A other | 1972 | 1.5 | 12.8 | 2.9 | 1.0 | 55.7 | 15.5 | 5.6 | 4.9 |
| GPCR peptide | 19602 | 3.2 | 7.6 | 8.3 | 1.2 | 30.0 | 16.9 | 15.0 | 17.8 |
| GPCR prostaglandin | 1399 | 0.4 | 0.8 | 59.5 | 3.1 | 4.7 | 0.1 | 24.5 | 6.9 |
| GPCR class B | 1391 | 0.3 | 1.7 | 2.9 | 0.1 | 58.1 | 21.4 | 8.5 | 7.0 |
| GPCR class C | 1093 | 0.3 | 15.7 | 8.9 | 0.3 | 24.6 | 13.2 | 9.2 | 27.8 |
| Integrins | 3300 | 2.6 | 0.5 | 24.4 | 4.1 | 2.7 | 1.2 | 31.6 | 32.9 |
|
Ligand-gated ion
channel |
7357 | 3.8 | 9.2 | 7.3 | 1.7 | 37.8 | 22.0 | 8.5 | 9.8 |
|
Voltage-gated ion
channel |
2761 | 1.0 | 31.2 | 4.7 | 0.5 | 40.3 | 10.4 | 6.4 | 5.5 |
| Ion channel – other | 1263 | 9.8 | 10.3 | 22.8 | 1.0 | 19.2 | 17.7 | 4.1 | 15.1 |
| Tyrosine kinases | 4229 | 0.3 | 5.7 | 3.5 | 2.9 | 29.8 | 28.3 | 6.4 | 23.1 |
|
Kinases – non
tyrosine |
5635 | 0.7 | 12.4 | 7.6 | 1.6 | 24.2 | 24.2 | 9.4 | 19.8 |
| Metalloproteases | 2024 | 7.7 | 2.9 | 41.0 | 17.2 | 0.9 | 0.2 | 16.7 | 13.5 |
| Nuclear receptors | 6571 | 0.0 | 21.3 | 27.2 | 7.5 | 15.1 | 2.1 | 16.5 | 10.2 |
| Oxido reductases | 5744 | 3.7 | 9.9 | 14.6 | 2.9 | 27.7 | 20.3 | 7.4 | 13.4 |
| Oxygenases | 3890 | 0.1 | 31.2 | 15.7 | 1.9 | 27.1 | 5.2 | 13.0 | 5.9 |
| Phosphodiesterases | 2555 | 5.0 | 14.6 | 9.0 | 1.4 | 31.7 | 23.2 | 9.9 | 5.1 |
| Serine proteases | 5793 | 9.2 | 14.6 | 6.6 | 1.1 | 28.4 | 16.9 | 8.8 | 14.5 |
| Transporters | 5240 | 0.7 | 3.0 | 0.3 | 0.0 | 65.1 | 18.3 | 5.4 | 7.1 |
| Unknown targets | 22298 | 3.8 | 20.0 | 9.2 | 2.4 | 19.7 | 15.0 | 10.3 | 19.7 |
|
| |||||||||
| Vendors – drug-like clean |
6676776 | 0.0 | 43.6 | 9.1 | 0.6 | 33.7 | 6.5 | 4.8 | 1.6 |
| ‘Natural’ products | 89425 | 1.4 | 41.8 | 14.0 | 2.6 | 22.9 | 6.3 | 6.7 | 4.1 |
| Vendors – clean leads | 1716660 | 0.1 | 42.4 | 8.6 | 0.6 | 35.0 | 6.8 | 5.1 | 1.5 |
Figure 1.
(A) A breakdown of acid/base classes for small molecule drugs approved for use in humans (drugs). The acid/base classes are: always ionized (acids with pKa < 0, bases with pKa > 12 and compounds carrying a permanent charge), neutral compounds, single acid-containing substances, single base-containing substances, compounds with two acidic groups, compounds with two basic groups, simple ampholytes (one acidic and one basic group) and other complex combinations of acidic and basic groups. The pKa distribution of compounds containing (B) a single acidic group and (C) basic group. The pKa values were binned into single log unit ranges. The histogram column heights are expressed as a percentage of the total compounds.
The most common acidic groups are the carboxylic acids, heterocyclic nitrogen atoms, hydroxamic acids, phenols and sulfonamides which together make up 93.5% of acidic groups. Lower frequency acids were: phosphates, imides, sulfates, carbon acids, tetrazoles, carbamates, alcohols, acidic amides, acidic anilines, hydrazides and thiols. The most common basic groups were aliphatic amines, guanidines, amidines, anilines, basic amides and heterocyclic nitrogen atoms. Information on the range of pKa values for various functional groups is discussed in the following references.[13]
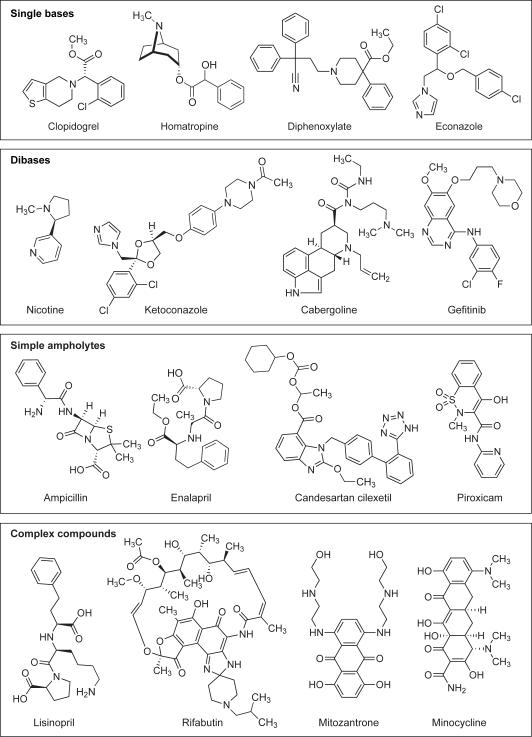
Examples of the drugs that fall into these classes are shown in Figure 2. Interestingly, over 60% of the drugs have only one ionizable functional group or no ionizable group. A review of the distribution of pKa values for compounds with one basic group (Figure 1C) shows that there are very few single bases with pKa values below 6.0 For the single acid containing substances (Figure 1B), there are very few acidic functional groups with pKa values in the range 5-8. This is linked to a lack of acidic functional groups that have pKa values in this range. This is also associated with the typical pKa ranges found for two of most common acidic groups, carboxylates (3.5-5) and phenols (7.5-10.5).[14]
Figure 2.
Examples of compounds in different acid/base categories.
Chemogenomic and screening compound datasets
The WOMBAT database comprises a collection of over 300,000 compounds with associated target-bioactivity data carefully curated from the peer reviewed literature.[15] From the database, we assembled 23 sets of compounds that are reported to interact with specific protein families and an additional set where the target macromolecule was unknown. Filters were applied to each set to select compounds exhibiting activity at concentrations of 1 μM or lower as well as a molecular weight cut-off. Following the estimation of pKa values, the compounds were placed in charge state categories. A chemogenomic analysis of ligand acid/base properties has not been conducted previously.
The Zinc database[16] is a virtual library of purchasable chemical structures from over 100 vendors which was developed to facilitate docking experimentation. In this study we employed three sets of compounds from the Zinc database[16] to represent compounds available for biological screening. These datasets have been prefiltered by the Zinc curators to remove compounds unsuitable for medicinal chemistry. The largest dataset consisted of 6.6 million commercially available screening compounds from the major suppliers (Vendors – drug-like clean). A smaller subset of these compounds, selected largely on the basis of lower molecular weight and lipophilicity,[16] is termed the ‘Vendors – clean leads’ set. Finally, a natural products collection was assessed and these compounds have been classified by the suppliers themselves as being natural products or derivatives of natural products.
Acid/base properties of chemogenomic datasets
Table 1 summarises the charge state categories of the drugs, chemogenomics and screening compound datasets. Tables 2 and 3 provide detailed information on the pKa distributions of the largest groups, the single acid and single base containing compounds. In our analysis, the smallest dataset contained just over 1,000 compounds while the majority of the datasets had over 2,000 substances. In this paper we have selected four chemogenomic families to contrast the differences in their acid/base properties. These targets are; the GPCRs (biogenic amine, nucleotide and peptide receptors), three protease families (aspartyl, cysteine and serine), kinases (tyrosine and non-tyrosine) and ion channels (ligand-gated, voltage-gated and other).
Table 2.
pKa distribution of compounds containing a single acidic functional group.
| Dataset | pKa range (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0-1 | 1-2 | 2-3 | 3-4 | 4-5 | 5-6 | 6-7 | 7-8 | 8-9 | 9-10 | |
| Drugs | 0.0 | 1.3 | 4.1 | 22.5 | 25.1 | 5.7 | 8.8 | 7.1 | 10.7 | 14.7 |
|
| ||||||||||
| Aspartyl proteases | 0.8 | 0.6 | 0.2 | 13.7 | 8.6 | 0.2 | 17.4 | 26.5 | 6.1 | 25.8 |
| Cysteine proteases | 0.0 | 0.0 | 3.8 | 11.1 | 29.5 | 13.8 | 0.0 | 2.7 | 13.4 | 25.7 |
| GPCR amines | 0.0 | 0.0 | 25.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 0.0 | 25.0 |
| GPCR cannabinoid | 0.0 | 0.0 | 0.0 | 0.5 | 1.9 | 3.8 | 1.4 | 1.4 | 2.4 | 88.5 |
| GPCR nucleotide | 0.0 | 0.0 | 0.1 | 1.2 | 0.5 | 0.8 | 2.0 | 35.2 | 39.8 | 20.4 |
| GPCR class A other | 0.0 | 0.0 | 0.0 | 13.8 | 6.9 | 0.0 | 3.4 | 8.6 | 20.7 | 46.6 |
| GPCR peptide | 0.1 | 0.4 | 8.4 | 30.3 | 37.3 | 2.5 | 5.9 | 5.6 | 2.1 | 7.5 |
| GPCR prostaglandin | 0.0 | 0.1 | 0.5 | 29.4 | 66.4 | 2.8 | 0.0 | 0.0 | 0.0 | 0.8 |
| GPCR class B | 0.0 | 0.0 | 0.0 | 95.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 |
| GPCR class C | 0.0 | 0.0 | 4.1 | 10.3 | 27.8 | 0.0 | 3.1 | 33.0 | 14.4 | 7.2 |
| Integrins | 0.0 | 0.0 | 5.2 | 86.5 | 7.4 | 0.1 | 0.0 | 0.0 | 0.0 | 0.7 |
| Ligand-gated ion channel | 0.0 | 0.4 | 8.3 | 20.0 | 8.5 | 6.5 | 14.7 | 6.9 | 3.7 | 31.0 |
| Voltage-gated ion channel | 0.0 | 0.0 | 0.0 | 5.4 | 26.4 | 0.0 | 2.3 | 31.8 | 7.0 | 27.1 |
| Ion channel - other | 0.0 | 0.3 | 0.0 | 0.7 | 0.3 | 0.0 | 1.4 | 14.2 | 7.3 | 75.7 |
| Tyrosine kinases | 0.0 | 2.0 | 4.8 | 3.4 | 22.4 | 1.4 | 0.0 | 6.1 | 20.4 | 39.5 |
| Kinases – non tyrosine | 0.0 | 0.0 | 0.2 | 0.5 | 0.7 | 0.2 | 2.6 | 6.1 | 29.3 | 60.4 |
| Metalloproteases | 0.0 | 0.0 | 0.7 | 5.4 | 18.8 | 0.0 | 0.0 | 0.0 | 71.0 | 4.1 |
| Nuclear receptors | 0.1 | 0.2 | 2.0 | 30.4 | 32.5 | 0.2 | 5.4 | 2.3 | 8.3 | 18.6 |
| Oxido reductases | 0.0 | 0.6 | 4.2 | 39.0 | 20.9 | 9.4 | 1.5 | 4.2 | 16.8 | 3.4 |
| Oxygenases | 0.0 | 0.2 | 0.0 | 14.4 | 12.3 | 1.5 | 4.8 | 8.9 | 37.2 | 20.8 |
| Phosphodiesterases | 0.0 | 5.2 | 1.3 | 22.7 | 7.0 | 0.9 | 7.9 | 6.1 | 44.5 | 4.4 |
| Serine proteases | 0.0 | 2.9 | 1.8 | 30.5 | 21.9 | 4.4 | 3.1 | 11.0 | 6.0 | 18.3 |
| Transporters | 0.0 | 0.0 | 0.0 | 0.0 | 18.8 | 0.0 | 0.0 | 12.5 | 12.5 | 56.3 |
| Unknown targets | 0.1 | 0.2 | 0.7 | 9.6 | 11.1 | 4.1 | 5.2 | 10.6 | 18.7 | 39.7 |
| Vendors – drug-like clean | 0.1 | 0.1 | 1.1 | 8.1 | 5.9 | 1.9 | 6.2 | 17.4 | 19.4 | 39.7 |
| ‘Natural’ products | 0.0 | 0.2 | 2.9 | 44.0 | 17.0 | 1.5 | 4.4 | 7.8 | 8.4 | 13.9 |
| Vendors – clean leads | 0.0 | 0.1 | 2.1 | 13.2 | 9.4 | 2.1 | 6.3 | 13.1 | 17.4 | 36.4 |
Table 3.
pKa distribution of compounds containing a single basic functional group.
| Dataset | pKa range (%) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0-1 | 1-2 | 2-3 | 3-4 | 4-5 | 5-6 | 6-7 | 7-8 | 8-9 | 9-10 | 10-11 | 11-12 | |
| Drugs | 3.2 | 2.7 | 4.6 | 5.6 | 4.1 | 4.0 | 7.2 | 12.6 | 21.8 | 26.2 | 7.5 | 0.5 |
|
| ||||||||||||
| Aspartyl proteases | 16.2 | 9.6 | 12.8 | 3.4 | 4.6 | 6.8 | 11.0 | 20.1 | 9.0 | 6.0 | 0.5 | 0.1 |
| Cysteine proteases | 12.3 | 28.5 | 4.3 | 11.0 | 11.4 | 17.8 | 2.1 | 3.4 | 4.8 | 2.0 | 0.4 | 2.1 |
| GPCR amines | 0.0 | 0.2 | 0.1 | 0.4 | 1.2 | 2.0 | 12.8 | 21.2 | 29.8 | 25.9 | 5.8 | 0.5 |
| GPCR cannabinoid | 16.8 | 32.3 | 4.6 | 4.5 | 3.0 | 1.0 | 29.0 | 2.3 | 5.6 | 0.5 | 0.3 | 0.0 |
| GPCR nucleotide | 15.3 | 13.8 | 14.9 | 19.7 | 26.2 | 5.0 | 3.5 | 1.0 | 0.3 | 0.3 | 0.1 | 0.0 |
| GPCR class A other | 4.5 | 2.1 | 1.3 | 0.9 | 1.3 | 0.4 | 2.1 | 10.4 | 24.1 | 36.4 | 16.3 | 0.4 |
| GPCR peptide | 5.0 | 1.4 | 4.3 | 2.4 | 5.4 | 3.2 | 7.2 | 11.2 | 25.4 | 25.1 | 5.8 | 3.6 |
| GPCR prostaglandin | 1.5 | 0.0 | 0.0 | 3.0 | 25.8 | 3.0 | 1.5 | 10.6 | 47.0 | 4.5 | 3.0 | 0.0 |
| GPCR class B | 4.2 | 7.7 | 11.5 | 22.6 | 21.9 | 12.3 | 9.9 | 3.1 | 6.3 | 0.5 | 0.0 | 0.0 |
| GPCR class C | 8.2 | 29.7 | 3.3 | 18.6 | 13.4 | 16.0 | 0.7 | 1.1 | 2.6 | 5.9 | 0.4 | 0.0 |
| Integrins | 19.1 | 57.3 | 4.5 | 4.5 | 3.4 | 1.1 | 0.0 | 1.1 | 2.2 | 0.0 | 4.5 | 2.2 |
| Ligand-gated ion channel | 3.6 | 8.0 | 7.9 | 13.2 | 3.5 | 4.0 | 4.6 | 12.7 | 13.4 | 17.1 | 10.3 | 1.8 |
| Voltage-gated ion channel | 2.9 | 5.1 | 3.8 | 5.8 | 7.5 | 5.8 | 4.8 | 9.2 | 33.6 | 16.8 | 4.8 | 0.0 |
| Ion channel - other | 2.5 | 14.0 | 11.5 | 6.2 | 35.8 | 11.9 | 3.7 | 0.4 | 0.8 | 11.5 | 1.6 | 0.0 |
| Tyrosine kinases | 2.4 | 7.8 | 8.5 | 18.7 | 22.7 | 20.7 | 6.7 | 7.9 | 2.1 | 2.3 | 0.2 | 0.1 |
| Kinases – non tyrosine | 12.7 | 25.0 | 16.6 | 13.9 | 10.5 | 6.4 | 8.4 | 1.2 | 0.9 | 3.4 | 0.7 | 0.2 |
| Metalloproteases | 11.1 | 0.0 | 11.1 | 16.7 | 16.7 | 11.1 | 5.6 | 5.6 | 16.7 | 5.6 | 0.0 | 0.0 |
| Nuclear receptors | 6.7 | 15.9 | 6.3 | 50.8 | 10.4 | 0.9 | 0.3 | 0.1 | 7.2 | 1.5 | 0.0 | 0.0 |
| Oxido reductases | 14.6 | 4.2 | 8.8 | 6.5 | 6.2 | 7.7 | 8.8 | 27.6 | 8.2 | 4.3 | 0.8 | 2.3 |
| Oxygenases | 18.0 | 10.7 | 13.8 | 14.3 | 13.5 | 4.2 | 13.0 | 4.1 | 6.0 | 2.3 | 0.2 | 0.0 |
| Phosphodiesterases | 9.1 | 26.3 | 13.8 | 9.1 | 11.5 | 16.9 | 5.9 | 2.3 | 2.5 | 1.6 | 0.7 | 0.0 |
| Serine proteases | 8.7 | 2.1 | 3.2 | 1.9 | 3.4 | 1.4 | 5.4 | 5.7 | 28.8 | 15.1 | 4.1 | 20.3 |
| Transporters | 0.1 | 0.1 | 0.7 | 2.0 | 1.3 | 0.3 | 0.8 | 5.9 | 26.2 | 51.1 | 11.1 | 0.4 |
| Unknown targets | 8.2 | 17.3 | 11.1 | 9.9 | 9.0 | 8.9 | 9.0 | 5.7 | 10.9 | 6.9 | 2.2 | 0.8 |
|
| ||||||||||||
| Vendors – drug-like clean | 17.8 | 23.8 | 17.4 | 11.1 | 12.1 | 8.9 | 5.7 | 2.4 | 0.6 | 0.1 | 0.0 | 0.0 |
| ‘Natural’ products | 13.6 | 7.4 | 7.8 | 10.9 | 9.9 | 5.1 | 7.4 | 10.0 | 13.6 | 11.2 | 2.9 | 0.1 |
| Vendors – clean leads | 17.6 | 22.1 | 17.3 | 11.6 | 12.6 | 8.7 | 6.5 | 2.7 | 0.7 | 0.1 | 0.0 | 0.0 |
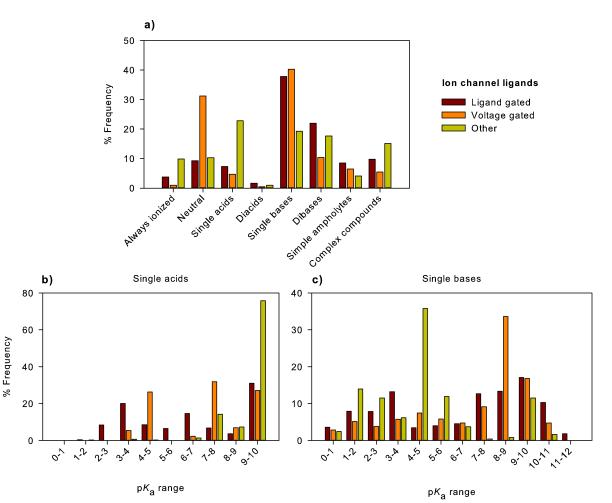
Figure 3A shows the charge state categories for the three GPCR families. Notably these targets are heavily weighted to recognising base containing compounds. The GPCR biogenic amine dataset contained 20712 compounds with over 50% possessing a single basic group comprising largely aliphatic amines and basic heterocyclic nitrogen atoms. In contrast, the same dataset had only four compounds containing a single acidic group. The high proportion of bases is explained by the key recognition of the basic amino group of their associated (neuro)transmitters[17] by a conserved aspartic acid residue. In contrast, the peptide and nucleotide receptors do recognise some neutral and acidic compounds. Figure 3B shows the profiles of single base containing compounds. Notably the nucleotide GPCR ligands show distinctly different properties from the peptide and biogenic amine GPCR ligands.
Figure 3.
Overview of compound types for (A) GPCR ligands and (B) the pKa distribution of compounds containing a single basic functional group.
Figure 4 shows the profiles of the protease examples illustrating the subtle differences between each class. Interestingly there are a larger number of compounds with basic groups above a pKa of 8 for the serine proteases. The presence of these basic groups (mostly amidine and basic heterocyclic nitrogen atoms) coincides with the need for functional groups to interact with the S1 pocket of proteins such as Factor Xa.[18]
Figure 4.
(A) Overview of charge state categories for protease ligands. (B) pKa distributions of compounds containing a single acidic functional group and (C) pKa distributions of compounds containing a single basic functional group.
The kinase classes are dominated by compounds that contain either one or two basic groups (54%). Figure 5 shows that for both the basic and acidic groups there are few compounds that would be fully ionized at physiological pH (i.e. acids with pKa values below 6.0 and bases with pKa values above 9.0). Once again, there is a paucity of functional groups that are charged at pH 7.4, in agreement with the hydrogen bonding requirements of the binding sites.[19]
Figure 5.
(A) Overview of charge state categories for kinase ligands. (B) pKa distributions of compounds containing a single acidic functional group and (C) pKa distributions of compounds containing a single basic functional group.
Finally, we compare ligand-gated, voltage-gated and other ion channels (Figure 6). These compounds are predominantly single or dibases but 31% of voltage-gated channel ligands are neutral and 23% of the ‘other’ class are single acids (predominantly weak acids such as phenols). The proportion of basic compounds with pKa values above 7.0 for the ligand and voltage-gated channels was 55.3 and 64.4%, respectively while the ‘other’ ion channels only had 14.4% of single bases in this range.
Figure 6.
(A) Overview of charge state categories for ion-channel ligands. (B) pKa distributions of compounds containing a single acidic functional group and (C) pKa distributions of compounds containing a single basic functional group (C).
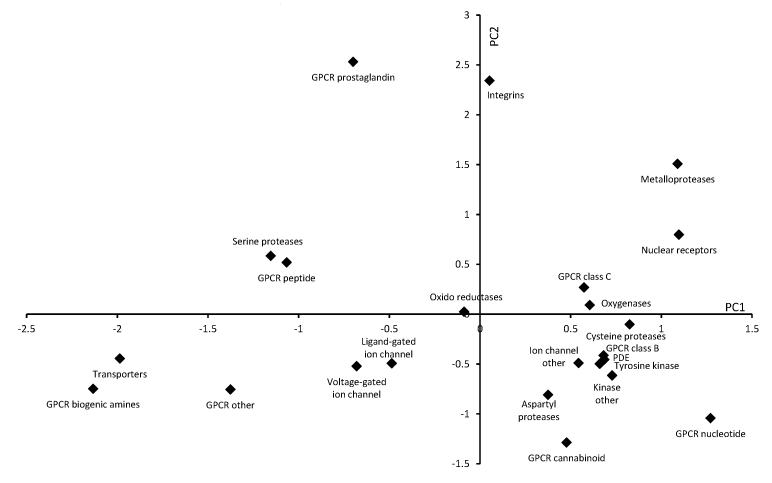
Chemogenomics seeks to add to our knowledge of how small organic compounds interact with and perturb biological systems. If this can be translated into tailoring compound sets for screening purposes then it is fulfilling the ambition to improve how we conduct drug discovery. It is clear from the analysis above that each ligand class possess distinct acid/base distributions. Figure 7 shows a principal component analysis (PCA) plot of the 23 chemogenomics datasets where the target family is known, employing the compound category, ampholyte proportion and pKa distribution data as input data revealing the clustering of target families. The loadings (appendix 1, supplementary material) for the PCA show that PC1 is associated with the base pKa distribution data and the proportion of ordinary and zwitterionic ampholytes. Acidic compounds are found in the upper region of the plot. PC2 is linked with the charge state category data as well as some of the acid pKa distribution data with basic compounds located in the lower left quadrant. Compound classes with fewer acid or base groups are found in the lower right quadrant.
Figure 7.
Scores plot of principal components (PC) 1 and 2 for the chemogenomics datasets. Loadings for the PCs are given in the supplementary information. PC1 and PC2 are associated with base and acid pKa values, respectively. Datasets with high proportions of acidic groups are generally located in the upper and right hand section of the plot while basic compounds dominate the datasets in the lower left hand portion of the plot.
In this analysis, GPCR biogenic amines are located close to transporters (predominantly monoamine transporter ligands) as a result of their similar single base pKa distributions. Ligand-gated and voltage-gated ion channel sets are also clustered. The kinase families group together and are joined by the PDE, ion channel ‘other’ and GPCR class B sets. PDE and kinase ligands often mimic purines and this grouping reflects the heterocyclic nature of these compounds. The GPCR prostaglandin, GPCR nucleotide, integrins, metalloproteases and nuclear receptor sets are well separated from other clusters reflecting high proportions of particular functional groups. For example, metalloprotease ligands contain specific functional groups (most often a hydroxamic acid) which can bind to an active-site metal (e.g. Zn). In a similar manner, GPCR prostaglandin ligands are dominated by carboxylic acids.
Acid/base properties of screening compounds
Current drug discovery relies heavily on screening technologies. Over the past decade, very large numbers of chemical compounds have become available from commercial suppliers and there has been a great deal of analysis of the physicochemical properties of drugs and lead compounds.[20] However, a systematic analysis of the acid/base properties of screening compounds has not been published previously. In this section of work, we analyse the acid/base properties of the vendors – drug-like, vendors – clean leads and natural products sets taken from the Zinc database.
As with the drugs and chemogenomics datasets, we estimated the pKa values of the screening compound sets (vendors – drug-like, vendors – clean leads and natural products) and classified them into charge state categories (Tables 1-3).
Figure 8 shows the charge state categories of the screening compound sets and compares them to the drugs dataset. Most clearly, all the screening sets have a greater proportion of neutral compounds and fewer always ionized or ionizable groups than drugs. The vendors – drug-like and vendors – clean leads are very similar to one another which is understandable as the vendors – clean leads is a subset of the larger group of commercially available compounds. The drugs dataset contains significant fraction (6%) of permanently ionized compounds which are essentially absent in the vendors sets and only make up 1.4% of the natural products set. Completely neutral compounds make up over 40% of all the screening compounds but only represent 20% of drugs. The mono- and particularly diacids are underrepresented in the vendors sets although these groups are quite well represented in the natural product set. Contrastingly, the proportions of single bases in the screening sets approached that found in drugs although the natural products had slightly fewer and the vendors sets had a slightly greater fraction. All the screening sets contained only about two-thirds of the number of dibases and ampholytes found in drugs. The remaining category of complex compounds constitutes about 8% of drugs but only 4.1% of natural products and 1.5% of screening compounds.
Figure 8.
(A) Contrasting differences in charge state categories between the drugs, compound suppliers (Vendors – drug-like clean, vendors – clean leads) and natural products datasets. pKa distributions of compounds containing a (B) single acidic or (C) single basic functional group; drugs, compound suppliers (Vendors – drug-like clean, vendors – clean leads) and the natural products dataset.
To further highlight the differences between the screening compound and drug sets, Figures 8B and 8C show the pKa distributions of single acid and single base-containing compounds (Table 2). Figure 8B reveals that the screening compound suppliers have more acids with pKa values above 6.0 relative to the drug and natural products sets. In this case, only 41% of drug single acids have pKa values above 6.0 in contrast to 82% of the vendors datasets. The drug and natural product sets have a considerable proportion of single acids with pKa values in the range 3-5 while these make up a smaller fraction of the vendor sets. Figure 8C illustrates major differences between the single base pKa distributions of contemporary drugs and the screening compound sets (Table 3). The vendor sets are dominated by weak bases with pKa values under 5 while the drugs distribution has a large fraction of bases with pKa values in the range 7-10. The natural products set lies between the two sets.
Simple ampholytes make up about 10% of the drugs dataset. Ampholytes can be defined as ordinary, where the acidic functional group has a pKa value higher than the basic group or zwitterionic in the reverse case. In ordinary ampholytes, the neutral species of the compound predominates at the isoelectric point while the doubly charged species is present in zwitterions. Table 4 lists the ratios of ordinary and zwitterionic ampholytes in drugs, the chemogenomic datasets and screening compounds. Zwitterions make up nearly half of drug simple ampholytes but the screening compounds were found to have more than 95% of ampholytes as ordinary in character (Table 4). The chemogenomic datasets display large variations in the number of ampholytes present and the ratio of ordinary and zwitterionic ampholytes. Most notably, over 30% of the integrin dataset are ampholytes and nearly all are zwitterions.
Table 4.
Fractions of ordinary and zwitterionic ampholytes present in drug, chemogenomic and screening datasets.
| Dataset | Proportion of total compounds (%) |
Fraction ordinary ampholytes (%) |
Fraction zwitterionic ampholytes (%) |
|---|---|---|---|
| Drugs | 9.5 | 53.9 | 46.1 |
|
| |||
| Aspartyl proteases | 5.0 | 90.8 | 9.2 |
| Cysteine proteases | 4.4 | 82.7 | 17.3 |
| GPCR amines | 7.4 | 42.9 | 57.1 |
| GPCR cannabinoid | 2.4 | 97.2 | 2.8 |
| GPCR nucleotide | 6.8 | 91.6 | 8.4 |
| GPCR class A other | 5.6 | 78.4 | 21.6 |
| GPCR peptide | 15.0 | 55.6 | 44.4 |
| GPCR prostaglandin | 24.5 | 28.0 | 72.0 |
| GPCR class B | 8.5 | 98.3 | 1.7 |
| GPCR class C | 9.2 | 62.0 | 38.0 |
| Integrins | 31.6 | 14.0 | 86.0 |
| Ligand-gated ion channel | 8.5 | 66.6 | 33.4 |
| Voltage-gated ion channel | 6.4 | 49.4 | 50.6 |
| Ion channel - other | 4.1 | 84.6 | 15.4 |
| Tyrosine kinases | 6.4 | 94.5 | 5.5 |
| Kinases – non tyrosine | 9.4 | 79.5 | 20.5 |
| Metalloproteases | 16.7 | 88.4 | 11.6 |
| Nuclear receptors | 16.5 | 85.4 | 14.6 |
| Oxido reductases | 7.4 | 79.4 | 20.6 |
| Oxygenases | 13.0 | 46.8 | 53.2 |
| Phosphodiesterases | 9.9 | 91.7 | 8.3 |
| Serine proteases | 8.8 | 26.8 | 73.2 |
| Transporters | 5.4 | 17.5 | 82.5 |
| Unknown targets | 10.3 | 52.3 | 47.7 |
|
| |||
| Vendors – drug-like clean | 4.8 | 95.8 | 4.2 |
| ‘Natural’ products | 6.7 | 59.3 | 40.7 |
| Vendors – clean leads | 5.1 | 94.1 | 5.9 |
This analysis reveals considerable differences in the acid/base makeup of the drug and screening datasets. It is reasonable to expect that much of the variation arises from the processes involved in screening compound synthesis and supply. Supplier companies are constrained by synthetic accessibility and cost of the compounds. As such, the physicochemical properties of the compounds are strongly influenced by the necessity of expedient synthesis. A study of medicinal chemists observed that there is a tendency to prepare largely neutral molecules that are soluble in organic solvents, and which may be readily crystallized, in bulk, from organic solvent as part of the purification process.[21] We expect that a similar situation applies to compound suppliers. High throughput encourages fewer synthetic steps (i.e., avoiding the protection and deprotection of chemically reactive, ionizable groups) and compounds that are easy to purify (e.g., zwitterions are notoriously difficult to purify on silica columns). Other factors, including stability may reduce the number of certain functional groups. For example, amines are prone to oxidation on storage. With these factors in mind, it is understandable that the screening compounds have: (a) more neutral compounds; (b) more ordinary ampholytes; (c) fewer acids with pKa < 7; and (d) fewer bases with pKa > 7. Indeed, the profile conforms to compound types that are preferred by organic chemists with regard to their properties for convenient synthesis, isolation and purification. Recent offerings from some compound suppliers are beginning to encompass a greater variety of medicinal chemist-friendly substances and target-based collections have also emerged.
Natural products have historically been an important source of drugs and drug leads[22] and are often claimed to be more drug-like than purely synthetic screening compounds.[23] Ganesan found that properties of a natural product set to be similar to drugs, however, they did have more rotatable bonds, stereogenic centres and higher molecular weights.[24] Despite any advantages they may bring, the pharmaceutical industry has generally decreased its use of natural products in screening due to the complexities of working with these compounds. In our analysis, we find the natural products set to be clearly different from the other screening compounds and that they share some of the characteristics of drugs. Although the Zinc natural product set does contain a large fraction of neutral compounds, it also includes higher proportions of complex ionizable compounds (always ionized, simple and complex compounds) and a greater number of zwitterionic compounds compared to screening compounds. The pKa distributions of single acids and single bases more closely resemble drugs than screening compounds. Overall, the natural products dataset could not be considered to closely match the profile of drugs but it does contain some features of the drug pKa distributions. This analysis can also be used alongside the data generated by Feher and Schmidt, who examined a wide range of physicochemical properties for drugs, natural products and combinatorial chemistry compounds.[25]
Functional groups
What functional groups are responsible for the acid/base properties of drugs and screening compounds? An analysis of the drug, chemogenomic and screening datasets is shown in Tables 5 and 6. This reveals that for drugs, the predominant acidic groups are carboxylic acids (17%), phenols (8%), heterocycles (4%) and sulfonamides (3%) while the most important bases are aliphatic amines (29%), heterocyclic amines (28%) and anilines (5%).
Table 5.
Acidic functional groups present in chemogenomics datasets and screening compound libraries.
| Dataset | Number of Compounds |
Percentage of dataset |
|||||
|---|---|---|---|---|---|---|---|
| Carboxylate | Heterocyclic nitrogen |
Hydrox- amate |
Phenol | Sulfonamide | Others | ||
| Drugs | 3766 | 16.8 | 3.7 | 0.3 | 8.3 | 3.3 | 6.5 |
|
| |||||||
| Aspartyl proteases | 4300 | 4.2 | 0.4 | 0.1 | 12.5 | 8.3 | 10.2 |
| Cysteine proteases | 2503 | 13.3 | 2.7 | 0.5 | 1.7 | 3.5 | 3.2 |
| GPCR amines | 20712 | 1.2 | 1.2 | 0.1 | 6.3 | 3.5 | 0.8 |
| GPCR cannabinoid | 1510 | 0.3 | 0.4 | 0.1 | 13.6 | 0.1 | 2.7 |
| GPCR nucleotide | 5228 | 2.1 | 16.3 | 0.1 | 1.5 | 0.2 | 15.6 |
| GPCR class A other | 1972 | 3.2 | 3.5 | 0.8 | 4.9 | 1.3 | 4.6 |
| GPCR peptide | 19602 | 19.4 | 1.5 | 0.1 | 10.9 | 6.8 | 7.3 |
| GPCR prostaglandin | 1399 | 83.8 | 0.1 | 0.1 | 4.0 | 9.7 | 2.6 |
| GPCR class B | 1391 | 3.5 | 0.0 | 0.0 | 8.6 | 0.0 | 3.2 |
| GPCR class C | 1093 | 27.5 | 1.4 | 0.0 | 4.5 | 0.2 | 14.0 |
| Integrins | 3300 | 91.8 | 1.0 | 0.0 | 3.8 | 3.5 | 7.7 |
| Ligand-gated ion channel |
7357 | 9.1 | 7.2 | 0.1 | 7.8 | 0.5 | 4.4 |
| Voltage-gated ion channel |
2761 | 3.5 | 2.9 | 0.0 | 2.1 | 1.6 | 3.0 |
| Ion channel - other | 1263 | 6.8 | 1.8 | 0.4 | 15.6 | 8.0 | 9.5 |
| Tyrosine kinases | 4229 | 2.8 | 3.9 | 1.6 | 7.1 | 0.2 | 9.6 |
| Kinases – non tyrosine |
5635 | 2.6 | 16.9 | 0.4 | 7.2 | 0.9 | 4.7 |
| Metalloproteases | 2024 | 35.7 | 0.9 | 56.8 | 4.5 | 5.8 | 15.3 |
| Nuclear receptors | 6571 | 28.9 | 3.1 | 0.0 | 29.2 | 0.9 | 2.2 |
| Oxido reductases | 5744 | 23.7 | 4.0 | 0.1 | 4.5 | 0.2 | 5.3 |
| Oxygenases | 3890 | 16.5 | 0.6 | 7.2 | 7.1 | 3.5 | 1.2 |
| Phosphodiesterases | 2555 | 4.9 | 8.9 | 3.1 | 2.2 | 0.4 | 8.8 |
| Serine proteases | 5793 | 13.2 | 1.5 | 0.1 | 6.4 | 6.4 | 9.1 |
| Transporters | 5240 | 4.7 | 0.0 | 0.0 | 1.8 | 0.1 | 1.0 |
| Unknown targets | 22298 | 14.4 | 6.7 | 1.2 | 8.7 | 1.7 | 7.1 |
|
| |||||||
| Vendors – drug-like clean |
6676776 | 2.4 | 2.6 | 0.0 | 1.8 | 4.5 | 5.6 |
| Vendors – clean leads |
1716660 | 3.3 | 2.7 | 0.0 | 1.9 | 3.2 | 5.0 |
|
| |||||||
| ‘Natural’ products | 89425 | 13.8 | 2.8 | 0.0 | 8.4 | 0.7 | 3.4 |
Values are calculated by dividing the number of times the functional group is encountered by the total number of compounds and expressed as a percentage.
Table 6.
Basic functional groups present in chemogenomics datasets and screening compound libraries.
| Dataset | Number of Compounds |
Percentage of dataset |
||||||
|---|---|---|---|---|---|---|---|---|
| Amidine | Anilinic | Guanidine | Heterocyclic nitrogen |
amines | Amidic | Other | ||
| Drugs | 3766 | 1.2 | 5.3 | 1.5 | 27.9 | 29.2 | 0.7 | 2.0 |
|
| ||||||||
| Aspartyl proteases | 4300 | 0.5 | 6.0 | 0.7 | 37.3 | 19.7 | 5.7 | 1.9 |
| Cysteine proteases | 2503 | 0.0 | 9.4 | 0.8 | 26.8 | 9.4 | 1.1 | 3.1 |
| GPCR amines | 20712 | 2.5 | 4.3 | 0.7 | 75.2 | 45.7 | 3.0 | 2.2 |
| GPCR cannabinoid | 1510 | 0.0 | 1.3 | 2.5 | 28.1 | 18.5 | 6.6 | 0.9 |
| GPCR nucleotide | 5228 | 0.2 | 2.5 | 0.5 | 101.5 | 2.4 | 0.4 | 1.4 |
| GPCR class A other | 1972 | 4.8 | 3.0 | 2.3 | 38.1 | 44.4 | 0.4 | 0.9 |
| GPCR peptide | 19602 | 0.7 | 2.9 | 4.9 | 59.8 | 38.0 | 5.0 | 2.0 |
| GPCR prostaglandin | 1399 | 0.2 | 0.4 | 3.6 | 28.2 | 0.9 | 2.7 | 3.4 |
| GPCR class B | 1391 | 0.0 | 4.7 | 0.4 | 102.1 | 9.3 | 0.1 | 7.6 |
| GPCR class C | 1093 | 0.1 | 1.1 | 0.2 | 56.6 | 30.7 | 0.0 | 0.4 |
| Integrins | 3300 | 16.6 | 1.6 | 12.9 | 42.4 | 12.2 | 8.9 | 0.2 |
| Ligand-gated ion channel |
7357 | 1.7 | 2.7 | 3.1 | 70.9 | 26.4 | 1.0 | 0.6 |
| Voltage-gated ion channel |
2761 | 1.1 | 2.9 | 1.5 | 38.7 | 33.1 | 1.5 | 0.5 |
| Ion channel - other | 1263 | 0.6 | 4.3 | 3.6 | 54.9 | 9.8 | 1.0 | 2.5 |
| Tyrosine kinases | 4229 | 0.0 | 5.2 | 0.5 | 114.7 | 21.9 | 0.1 | 0.8 |
| Kinases – non tyrosine | 5635 | 0.1 | 7.3 | 0.5 | 95.0 | 19.2 | 0.6 | 2.9 |
| Metalloproteases | 2024 | 0.1 | 1.2 | 1.4 | 21.2 | 14.8 | 2.7 | 0.1 |
| Nuclear receptors | 6571 | 0.0 | 4.7 | 0.0 | 31.7 | 8.5 | 0.7 | 1.2 |
| Oxido reductases | 5744 | 2.5 | 8.6 | 7.6 | 52.3 | 9.0 | 5.4 | 1.7 |
| Oxygenases | 3890 | 0.0 | 1.3 | 0.5 | 48.9 | 5.3 | 0.6 | 3.9 |
| Phosphodiesterases | 2555 | 0.2 | 4.1 | 0.2 | 86.7 | 4.7 | 1.3 | 4.1 |
| Serine proteases | 5793 | 29.0 | 3.7 | 5.5 | 41.5 | 23.5 | 2.0 | 0.6 |
| Transporters | 5240 | 0.9 | 4.4 | 0.0 | 36.4 | 71.1 | 0.4 | 1.3 |
| Unknown targets | 22298 | 1.1 | 7.2 | 0.9 | 53.0 | 30.6 | 2.0 | 3.2 |
|
| ||||||||
| Vendors – drug-like clean |
6676776 | 0.1 | 4.4 | 0.1 | 44.1 | 3.4 | 3.1 | 0.5 |
| Vendors – clean leads | 1716660 | 0.0 | 3.4 | 0.1 | 45.0 | 3.8 | 3.2 | 0.4 |
| ‘Natural’ products | 89425 | 0.2 | 2.9 | 0.1 | 24.0 | 13.4 | 4.6 | 3.0 |
Values are calculated by dividing the number of times the functional group is encountered by the total number of compounds and expressed as a percentage. Where the frequency is high this can lead to percentages over 100.
The chemogenomic datasets reveal a great diversity in acid base character reflecting the influence of the binding site on the acid/base properties of the ligand. For example, the integrin and GPCR prostanoid sets have extremely high proportions of carboxylic acids; the GPCR nucleotide and kinase non tyrosine sets have high proportions of acidic heterocyclic nitrogen atoms and, strikingly, over half the metalloprotease set contain hydroxamic acids (Table 1). Finally, the nuclear receptor dataset showed a higher proportion of phenolic acids which are largely estrogenic ligands. In all these cases, the high proportions of specific functional groups can be rationalized by considering the nature of their macromolecular binding sites. For example, the high proportion of hydroxamic acids for the metalloprotease set is associated with the need for this group to bind to the zinc atom within the enzyme. It is noteworthy that relative to the drug dataset, the vendors screening collections are deficient in both carboxylic acids and phenols having only ~3% and 2% respectively.
The analysis of the basic groups also provided similar associations between the ligands and their binding sites. Integrin ligands have a high proportion of basic amidine groups and guanidines (Table 6). The serine protease set favours amidine groups while the cysteine protease dataset is rich in anilines. Tyrosine kinase ligands contain a high number of basic heterocyclic nitrogen atoms. Several groups contained a high proportion of aliphatic amines; the GPCR amines, GPCR peptides, GPCR class A other and transporters. In these latter cases, there is a clear association between the basic group of the natural ligand for these macromolecules and the aliphatic amines present in the chemogenomic sets. Once again there is a significant disparity between the proportion of compounds containing aliphatic amines in the drug and screening sets; being 29 and 2% respectively.
Conclusions
Acidic and basic groups of drugs are the primary recognition elements in a large proportion of drug/receptor interactions. This is made evident by clustering of particular acidic and basic functional groups observed within chemogenomic classes, for example the preponderance of carboxylic acid groups in GPCR-prostanoid ligands. Acid/base complementarity between ligands and their binding sites is responsible for much of the affinity desired by drug designers. The acid/base properties of a ligand are also fundamental determinants of lipophilicity which greatly affects both biopharmaceutical properties[3b,26] (ADMET) and off-target activity.[20] Drug designers also need to be aware of strongly ionized compounds that may result in the following development problems: poor absorption and distribution; excessive plasma protein binding; significant hERG channel affinity, phospholipidosis; and poor formulation prospects.[3b,20] Taken together, ionization states and pKa values are key considerations in the development of candidate drugs from screening hits. Acid/base properties also deeply influence formulation considerations and chemical stability. As such, greater understanding of charge states and pKa values in the field of drug discovery has the potential to enhance efficiency.
The primary focus of this study was to compare the acid/base profiles of drugs, chemogenomics datasets and screening compounds. Specifically, we have detailed charge state categories, pKa distributions of single acid and single base containing compounds, proportions of ordinary and zwitterionic ampholytes as well as identifying the ionizable functional groups for each dataset. We have identified differences in the profiles of drugs and screening compounds, for example there is a paucity of basic compounds with pKa values above 7.0 in the screening compound sets. In addition, neutral compounds dominate the screening compounds set having twice as many as the drugs dataset. As drug discovery is target oriented, we have also looked at 23 chemogenomics datasets to provide acid/base profiles for these protein families. These datasets demonstrated great diversity in acid/base profiles reflecting the requirements of their binding sites. This work builds substantially on our previous studies examining drug pKa profiles.[11]
Given the profound impact of acid/base character on drug behaviour, there are deep implications for considering charge states and pKa values within drug discovery. In particular, a great deal of care is taken over the selection of screening compounds as their choice affects hit rate efficiency and subsequent optimisation processes. Filtering of compounds prior to their selection is aimed at avoiding the inclusion of molecules that are unsuitable for testing or further chemistry manipulation.[27] Compound filtering usually considers the physicochemical properties of the compounds, their structural diversity and toxic or other undesirable functional groups.[28] Less attention has been applied to the consideration of pKa distributions in screening compounds although there is an awareness of balancing the ratio of acids and bases. For example, Blomberg et al. described two libraries used at AstraZeneca for fragment-based screening.[29] Both libraries were selected to contain a 1:1:4 cation:anion:neutral ratio that is dominated by neutral compounds. Looking specifically at our drugs dataset at pH 7.0 and making the simple assumption that acids are defined as having a pKa below 7.0 while bases are defined as having a pKa above 7.0 results in a cation:anion:neutral ratio of 2:1:2.5. This is in contrast to Blomberg et al.[29] however their requirements were generic in nature and were oriented to a small fragment library.
We believe that there is a need to consider pKa distributions and the diversity of acidic and basic functional groups when selecting screening compounds. If care is not taken, then compounds will reflect the acid/base profiles of the vendor libraries and will deviate significantly from having a drug-like profile or more importantly, to the general needs of a specific chemogenomic family.[30] It can be argued that, by mimicking the profile of already known drugs and targets, discovery will be biased to previously-explored chemical biology. However, by including specific acid/base information in filtering protocols, there is the potential to select compounds that have a greater probability of success than simple property filters alone, which may improve the success rate of chemogenomic primary screens.
Our prime intention in performing this study was to provide information that extends the breadth of physicochemical properties that are considered by drug designers. We envisage that it will assist the selection of high quality screening compounds and serve to highlight the general impact of acid/base character in drug development. In addition, this work can be used in conjunction with other data, such as that generated by Chuprina et al.,[23b] who determined the physicochemical properties (MW, ClogP, number of hydrogen bond acceptors/donors, PSA, rotatable bonds, calculated solubility and calculated Caco-2 membrane permeability) of a similar collection of screening compounds. Certainly, acid-base profiling should be considered by drug designers, as well as organisations that provide compounds for screening purposes.
Experimental
The drugs dataset was obtained from an in-house collection at UNM. Compounds not containing carbon, containing heavy metals, salts, polymers, mixtures and substances with a molecular weight greater than 1000 Da were removed, leaving a set of 3766 compounds. These compounds represent contemporary, clinically used single substances that have been approved for use in Europe, Asia and the U.S.A. This dataset is available on request from Tudor Oprea (toprea@salud.unm.edu).
A database of commercially available chemical compounds was selected from the Zinc database[16] (Oct 2011) (http://zinc.docking.org/) which contains information on over 107 commercially available compounds. The Zinc collection provides curated subsets derived from the full database based on calculated properties such as; MW, logP, number of hydrogen bond donors/acceptors, number of rotatable bonds and PSA.
The following Zinc subsets were examined
- A ‘vendor – drug-like clean’ subset comprising 6,676,776 compounds that are available for purchase. This subset was selected by the curators of the Zinc database using filters to reduce the number of compounds. These filters combined various ‘drug-like’ criteria included the following restrictions: logP <= 5, MW <= 500 and MW > 150, rotatable bonds < 8, PSA < 150 and number of H-bond acceptors < 10. A set of substructure identifiers was used to exclude certain unwanted functional groups from a medicinal chemistry perspective (e.g. acid halides, phosphoranes, thiocyanates, peroxides, aldehydes, etc.). For a full list see appendix 2 (supplementary material).
- The ‘vendor – clean leads’ subset contains 1,716,660 compounds that might be used in early discovery research to identify useful lead compounds. These compounds have the following characteristics[16]: ClogP < 3.5, MW < 350 and rotatable bonds <= 7, however this did not include compounds with a ClogP <=2.5 and MW <=250 and rotatable bonds <= 5.
- The ‘natural products’ dataset contained 89,398 compounds from seven vendors. These compounds have been specified by the vendors as being of natural origin or as derivatives of natural products. The organizations whose compounds comprise the natural products dataset were: InterBioScreen, Molecular Diversity Preservation International, TimTec, AmbInter, Indofine, Specs & BioSpecs and AnalytiCon Discovery. Some thought should be given to the definition of ‘natural’ as this has been determined by the supplier.
The chemogenomics database, WOMBAT[15] (v2010.01) was used to generate lists of bioactive compounds acting at a range of macromolecular target classes. Instant JChem (InstantJChem 5.7.0, 2011, ChemAxon (http://www.chemaxon.com)) was used for compound searching and file manipulation. The following target classes were investigated: aspartyl proteases, cysteine proteases, GPCR – biogenic amines, GPCR – cannabinoids, GPCR – nucleotide-like, GPCR – peptide, GPCR –prostanoid, GPCR – other (class A), GPCR – class B, GPCR – class C, integrins, ligand-gated ion channels, voltage-gated ion channels, ion channels – other, metalloproteases, nuclear hormone receptors, tyrosine kinases, kinases (non-tyrosine), oxidoreductases, oxygenases, phosphodiesterases, serine proteases and transporters. In addition, a set of compounds with unspecified target were placed in a group titled ‘unknown’. In all, 24 chemogenomics datasets were examined. For each dataset, filters were applied to reduce the size of the lists. A molecular weight cut-off of 1000 was applied to remove large compounds and compounds that were less potent than 1 μM for the specific target class were discarded.
For each of the datasets the following procedure was followed
Compounds containing functional groups that were permanently ionized (e.g., quaternary nitrogen atoms) were identified. In this analysis these molecules were classified as ‘always ionized’ and pKa values were not estimated for these compounds.
The pKa values for each compound were predicted using the Calculator Plugin within the Marvin software package [Marvin 5.7.0, 2011, ChemAxon (http://www.chemaxon.com)] and the pKa data was used to classify compounds as ‘always ionized’, ‘ionizable’ and ‘neutral’. Our classification system was based on the following: compounds possessing acidic groups with pKa values below 0 or basic groups with pKa values above 12 were added to the list of always ionized substances. Acidic groups with pKa values above 10 or basic functional groups with pKa values below zero were considered neutral in character. Acids with pKa values in the range 0 – 10 and bases with pKa values in the range 0 – 12 were classified as ionizable. In our hands using a carefully curated set of accurately measured pK values,[13b] a the Marvin software was able to estimate these pKas to within a single log unit (unpublished data). The pKa ranges used to define ionizable groups for acids and bases were chosen following discussions with experienced medicinal chemists and drug developers. This decision coincided with our consideration of pKa values that fall close to pH values encountered throughout the body. In addition, the pKa ranges also have relevance to pH values encountered in the field of pharmaceutical formulation.
A further structural search was conducted using an in-house algorithm in conjunction with the pKa calculations to determine the identity of each ionizable group within each entire dataset.
Simple ampholytes are compounds that contain one acidic and one basic functional group. These compounds can be further classified as ordinary or zwitterionic ampholytes by considering the pKa values of the functional groups. If the pKa value of the acidic group is greater than the pKa value of the basic group then this is considered to be an ordinary ampholyte. Likewise, if the pKa value of the acidic group is lower than the pKa value of the basic group then this defines zwitterionic ampholytes. The proportion of ordinary and zwitterionic ampholytes was determined from the pKa values for each simple ampholyte.
Principal components analysis
The IBM SPSS statistical package was used to perform principal components analyses. The input data comprised the compound category percentages, ampholyte proportions and pKa distribution data for single acids and single bases. Each variable was standardized and the first two principal components were extracted and plotted. The loadings for each variable were also extracted and are presented in Appendix 1 in the supplementary information.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Peter Kenny for his valuable comments on this manuscript. This work was supported in part by NIH grants GM-095952 and MH-084690 (OU, TIO).
Contributor Information
Dr. David T. Manallack, Monash Institute of Pharmaceutical Sciences Monash University (Parkville Campus) 381 Royal Parade, Parkville VIC 3052, Australia..
Dr. Richard J. Prankerd, Monash Institute of Pharmaceutical Sciences Monash University (Parkville Campus) 381 Royal Parade, Parkville VIC 3052, Australia.
Ms. Gemma C. Nassta, Monash Institute of Pharmaceutical Sciences Monash University (Parkville Campus) 381 Royal Parade, Parkville VIC 3052, Australia.
Dr. Oleg Ursu, The University of New Mexico School of Medicine Department of Internal Medicine Translational Informatics Division Innovation Discovery & Training Complex, MSC10 5550 Albuquerque NM 87131, USA
Prof. Tudor I. Oprea, The University of New Mexico School of Medicine Department of Internal Medicine Translational Informatics Division Innovation Discovery & Training Complex, MSC10 5550 Albuquerque NM 87131, USA
Dr. David K. Chalmers, Monash Institute of Pharmaceutical Sciences Monash University (Parkville Campus) 381 Royal Parade, Parkville VIC 3052, Australia.
References
- [1].Müller G, Kubinyi H. In: Chemogenomics in Drug Discovery: A Medicinal Chemistry Perspective. Müller G, Kubinyi H, editors. Wiley-VCH; Weinheim: 2004. pp. 1–4. [Google Scholar]
- [2].Kubinyi H. Ernst Schering Res Found Workshop. 2006:1–19. doi: 10.1007/978-3-540-37635-4_1. [DOI] [PubMed] [Google Scholar]
- [3] a).Avdeef A. Wiley; Hoboken: 2003. [Google Scholar]; b) Manallack DT, Prankerd RJ, Yuriev E, Oprea TI, Chalmers DK. Chem. Soc. Rev. 2012 doi: 10.1039/c2cs35348b. DOI: 10.1039/C2CS35348B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4] a).Jusko WJ, Gretch M. Drug Metab. Rev. 1976;5:43–140. doi: 10.3109/03602537608995839. [DOI] [PubMed] [Google Scholar]; b) Piafsky KM. Clin. Pharmacokinet. 1980;5:246–262. doi: 10.2165/00003088-198005030-00004. [DOI] [PubMed] [Google Scholar]
- [5].Pelletier DJ, Gehlhaar D, Tilloy-Ellul A, Johnson TO, Greene N. J. Chem. Inf. Model. 2007;47:1196–1205. doi: 10.1021/ci6004542. [DOI] [PubMed] [Google Scholar]
- [6] a).Raschi E, Ceccarini L, De Ponti F, Recanatini M. Expert. Opin. Drug Metab. Toxicol. 2009;5:1005–1021. doi: 10.1517/17425250903055070. [DOI] [PubMed] [Google Scholar]; b) Vaz RJ, Li Y, Rampe D. Prog. Med. Chem. 2005;43:1–18. doi: 10.1016/S0079-6468(05)43001-5. [DOI] [PubMed] [Google Scholar]
- [7] a).Hopkins AL, Mason JS, Overington JP. Curr. Opin. Struct. Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]; b) Morphy R, Rankovic Z. Drug Discov. Today. 2007;12:156–160. doi: 10.1016/j.drudis.2006.12.006. [DOI] [PubMed] [Google Scholar]; c) Azzaoui K, Hamon J, Faller B, Whitebread S, Jacoby E, Bender A, Jenkins JL, Urban L. ChemMedChem. 2007;2:874–880. doi: 10.1002/cmdc.200700036. [DOI] [PubMed] [Google Scholar]; d) Leeson PD, Springthorpe B. Nat. Rev. Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]; e) Peters JU, Schnider P, Mattei P, Kansy M. ChemMedChem. 2009;4:680–686. doi: 10.1002/cmdc.200800411. [DOI] [PubMed] [Google Scholar]
- [8] a).Gleeson MP, Hersey A, Montanari D, Overington J. Nat. Rev. Drug Discov. 2011;10:197–208. doi: 10.1038/nrd3367. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hann MM. Med. Chem. Commun. 2011;2:349–355. [Google Scholar]
- [9].Gleeson MP. J. Med. Chem. 2008;51:817–834. doi: 10.1021/jm701122q. [DOI] [PubMed] [Google Scholar]
- [10] a).Hopkins AL, Groom CR, Alex A. Drug Discov. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]; b) Perola E. J. Med. Chem. 2010;53:2986–2997. doi: 10.1021/jm100118x. [DOI] [PubMed] [Google Scholar]
- [11] a).Manallack DT. Perspect. Medicin. Chem. 2008;1:25–38. [PMC free article] [PubMed] [Google Scholar]; b) Manallack DT. SAR QSAR Environ. Res. 2009;20:611–655. doi: 10.1080/10629360903438313. [DOI] [PubMed] [Google Scholar]
- [12].Harris TK, Turner GJ. IUBMB life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- [13] a).Albert A, Serjeant EP. The determination of ionization constants: a laboratory manual. 3rd ed Chapman & Hall; New York: 1984. [Google Scholar]; b) Prankerd RJ. A critical compilation of pKa values for pharmaceutical substances. Vol. 33. Elsevier Academic Press; Amsterdam: 2007. [DOI] [PubMed] [Google Scholar]; c) Kortum G, Vogel W, Andrussow K. Dissociation constants of organic acids in aqueous solution. 1st ed Butterworth; London: 1961. [Google Scholar]; d) Perrin DD. Dissociation constants of organic bases in aqueous solution. 1st ed Butterworth; London: 1965. [Google Scholar]; e) Perrin DD. Dissociation constants of weak bases in aqueous solution. 1st ed Butterworths; London: 1972. [Google Scholar]; f) Serjeant E, Dempsey B. Ionization constants of organic acids in aqueous solution. 1st ed Pergamon Press; Oxford: 1979. [Google Scholar]
- [14].Lemke TL. Review of organic functional groups: introduction to medicinal organic chemistry. 4th ed Lippincott Williams & Wilkins; Baltimore: 2003. [Google Scholar]
- [15] a).Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, Oprea TI. In: Chemoinformatics in Drug Discovery. Oprea TI, editor. Wiley-VCH; New York: 2004. pp. 223–239. [Google Scholar]; b) Olah M, Oprea TI. In: Comprehensive Medicinal Chemistry II. Taylor JB, Triggle DJ, editors. Vol. 3. Elsevier; Oxford: 2006. pp. 293–313. [Google Scholar]; c) Olah M, Rad R, Ostopovici L, Bora A, Hadaruga N, Hadaruga D, Moldovan R, Fulias A, Racec M, Oprea TI. In: Chemical Biology: From Small Molecules to Systems Biology and Drug Design. Schreiber TKSL, Wess G, editors. Wiley-VCH; New York: 2007. pp. 760–786. [Google Scholar]
- [16].Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang ES. Protein Sci. 2003;12:1360–1367. doi: 10.1110/ps.0305603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pinto DJ, Smallheer JM, Cheney DL, Knabb RM, Wexler RR. J. Med. Chem. 2010;53:6243–6274. doi: 10.1021/jm100146h. [DOI] [PubMed] [Google Scholar]
- [19].Liao JJ. J. Med. Chem. 2007;50:409–424. doi: 10.1021/jm0608107. [DOI] [PubMed] [Google Scholar]
- [20].Meanwell NA. Chem. Res. Toxicol. 2011;24:1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- [21].Cooper TW, Campbell IB, Macdonald SJ. Angew. Chem. Int. Ed. Engl. 2010;49:2–12. doi: 10.1002/anie.201002238. [DOI] [PubMed] [Google Scholar]
- [22].Harvey AL. Drug Discov. Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- [23] a).Grabowski K, Schneider G. Curr. Chem. Biol. 2007;1:115–127. [Google Scholar]; b) Chuprina A, Lukin O, Demoiseaux R, Buzko A, Shivanyuk A. J. Chem. Inf. Model. 2010;50:470–479. doi: 10.1021/ci900464s. [DOI] [PubMed] [Google Scholar]
- [24].Ganesan A. Curr. Opin. Chem. Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- [25].Feher M, Schmidt JM. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- [26].Leeson PD, St-Gallay SA. Nat. Rev. Drug Discov. 2011;10:749–765. doi: 10.1038/nrd3552. [DOI] [PubMed] [Google Scholar]
- [27] a).Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv. Drug Del. Rev. 1997;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]; b) Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]; c) Wager TT, Hou X, Verhoest PR, Villalobos A. ACS Chem. Neurosci. 2010;1:435–449. doi: 10.1021/cn100008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28] a).Baell JB, Holloway GA. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]; b) Hann M, Hudson B, Lewell X, Lifely R, Miller L, Ramsden N. J. Chem. Inf. Comput. Sci. 1999;39:897–902. doi: 10.1021/ci990423o. [DOI] [PubMed] [Google Scholar]
- [29].Blomberg N, Cosgrove DA, Kenny PW, Kolmodin K. J. Comput. Aided Mol. Des. 2009 doi: 10.1007/s10822-009-9264-5. [DOI] [PubMed] [Google Scholar]
- [30].Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.