Abstract
We performed a genome wide association scan in 1,461 patients with bipolar 1 disorder and 2,008 controls drawn from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD) and University College London sample collections with successful genotyping for 372,193 SNPs. Our strongest single SNP results are found in myosin5B (MYO5B; p=1.66 × 10−7) and tetraspanin-8 (TSPAN8; p=6.11 × 10−7). Haplotype analysis further supported single SNP results highlighting MYO5B, TSPAN8 and the epidermal growth factor receptor (MYO5B; p=2.04 × 10−8, TSPAN8; p=7.57 × 10−7 and EGFR; p=8.36 × 10−8). For replication, we genotyped 304 SNPs in a family-based NIMH sample (n=409 trios) and a University of Edinburgh case-control sample (n=365 cases, 351 controls) which do not provide independent replication after correction for multiple testing. A comparison of our strongest associations with the genome-wide scan of 1,868 patients with bipolar disorder and 2,938 controls completed as part of the Wellcome Trust Case-Control Consortium (1) indicates concordant signals for SNPs within the voltage-dependent calcium channel, L-type, alpha 1C subunit (CACNA1C) gene, but no other single SNP associations are highly significant in both studies. Given the heritability of bipolar disorder, the lack of agreement between studies emphasizes that susceptibility alleles are likely to be modest in effect size and require even larger samples for detection.
Keywords: Genetic, Schizophrenia, CACNA1C, MYO5B, TSPAN8, EGFR
Introduction
Bipolar disorder (BP) is characterized by profound mood symptoms that include episodes of mania, hypomania and depression. Although family and twin studies unequivocally demonstrate a strong contribution of inherited genetic variation to the risk for BP (2), traditional linkage mapping and prior candidate gene studies have failed to identify genes that increase risk in a consistent manner.
Advances in gene mapping created by the International HapMap Project (3, 4) as well as highly parallel genotyping technology, have enabled investigation of the hypothesis that common variation may play a role in the liability to BP. This strategy is supported by recent successes in other common, complex disease studies as well as previous lack of success in BP genetics. Rare, highly penetrant, Mendelian forms of bipolar disorder have not been identified. Linkage data has largely identified areas with modest evidence of linkage to bipolar disorder that are not well localized, even when data are pooled (5, 6). Candidate gene studies while implicating several genes (7), have only been able to focus on a minority of genes in the genome, and interpretation of replication of individual findings has been difficult. Furthermore, strong support for applying a whole genome approach to BP comes from the success in finding novel, strong and consistent susceptibility loci in type II diabetes (8-10), prostate cancer (11), Crohn’s disease (12), breast cancer (13), and coronary artery disease (14) using similar whole genome approaches. Like BP, these diseases have a clear genetic component. The underlying types of genetic susceptibility to these disorders has been variable. For example, age-related macular degeneration (AMD) is thus far found to have a modest number of risk genes and alleles that have relatively large effect sizes (15), while others are likely to have many smaller risk alleles. As yet there is no obvious relationship between the heritability of a particular disorder and the number or strength of the observed effects. For at least two disorders, type 2 diabetes (8) and breast cancer (13) while susceptibility alleles were identified, very large sample sizes (n > 10,000) were required to establish consistency of results.
In this paper, we present data from a whole genome scan of bipolar disorder, two independent replication samples and a comparison of our most associated results with with previous whole genome association studies (1, 16).
Materials and Methods
Study samples
Cases and controls included in the final analysis were obtained from several sources. DNA samples from Bipolar I (BPI) patients (n=955) were obtained from the Systematic Treatment Enhancement Program for Bipolar Disorder (STEP-BD). Additional BPI cases (n=506) were obtained from University College London, United Kingdom (UCL, UK). Control samples were obtained from two sources in the United States (US) through the NIMH Genetics Repository (n=1498) and from UCL (n=510, matched to the UCL cases). To achieve maximal power to detect common variants of modest effect, our primary analyses were of all samples combined, estimating a common odds ratio but controlling for any systematic differences between samples (see Association Analysis methods section below).
STEP-BD cases
STEP-BD was a national, longitudinal cohort study designed to examine the effectiveness of treatments and their impact on the course of bipolar disorder (17) that enrolled 4,361 participants across the US who met the Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) criteria for bipolar I, bipolar II, bipolar NOS, schizoaffective manic or bipolar type, or cyclothymic disorder based on diagnostic interviews. Assessment of DSM-IV psychopathology was performed using two semi-structured diagnostic interviews: 1) the Affective Disorders Evaluation (ADE) which includes mood and psychosis modules adapted from the Structured Clinical Interview for DSM-IV (SCID-IV) (18), was administered by treating psychiatrists who were trained and certified in administration of the interview, and 2) the Mini International Neuropsychiatric Interview (MINI-PLUS) (19), a validated structured diagnostic interview, was administered by trained Clinical Specialists. Both interviews use DSM-IV criteria to establish diagnoses.
From the parent study, 2,089 individuals who were over 18 years of age and consented to the collection of blood samples for DNA and cell lines for genetic studies were enrolled in the STEP Genetic Repository for Participants (STEP-GRP; Co-Directors: Jordan W. Smoller, Vishwajit Nimgaonkar). Of the 2,089 STEP-GRP participants 62% had aconsensus diagnosis of BPI on both the ADE and MINI. BPI was chosen for the phenotype in this study for the following reasons: 1) diagnostic accuracy is higher than other forms mood disorder with high inter-rater reliability (20-22), 2) heritability of BPI has been repeatedly demonstrated (2), and 3) psychosis, a common feature of bipolar disorder, has been shown to be familial (23-25).
UCL cases
The UCL sample comprised Caucasian individuals who were ascertained and received clinical diagnoses of BP disorder according to UK National Health Service (NHS) psychiatrists at interview using the categories of the International Classification of Disease version 10 (ICD10). In addition bipolar subjects were included only if both parents were of English, Irish, Welsh or Scottish descent and if three out of four grandparents were of the same descent. One grandparent was allowed to be of Caucasian European origin but not of Jewish or non-European Union (EU) ancestry, based on the EU countries before the 2004 enlargement. This data was recorded in an Ancestry Questionnaire. They were then interviewed by one of three research psychiatrists (JL, NJB, AA) using a structured interview and defined as having BP 1 disorder. Research subjects were volunteers from UK NHS psychiatric services and from the UK Manic Depression Fellowship, a self-help organization for patients with bipolar disorder. The interviews were conducted between October 1991 and June 2006 using the Schizophrenia and Affective Disorders Schedule – Life Time version (SADS-L) which provides diagnoses according to the probable level of the Research Diagnostic Criteria and also according the US DSMIIIR criteria (26, 27).
Further clinical information was collected using the 90-item Operational Criteria checklist (OPCRIT) (28). All volunteers read an information sheet approved by the Metropolitan Medical Research Ethics Committee who also approved the project for all NHS hospitals. Written informed consent was obtained from each volunteer.
Both the STEP-BD and UCL samples comprised Caucasian BPI individuals, and, as shown in Table S1, were similar in terms of other clinical characteristics except for a somewhat higher proportion of cases with psychotic symptoms in the UCL sample.
NIMH control samples
Two groups of control samples were obtained from the NIMH Genetics Initiative through the NIMH Center for Collaborative Studies (http://zork.wustl.edu/nimh/) from the Rutger’s Repository (http://rucdr.rutgers.edu/). The first comprised 454 DNA samples derived from US Caucasian anonymous cord blood donors (29). The second comprised 1,044 US Caucasian controls who completed an online self-administered psychiatric screen and were ascertained by Knowledge Networks (KN; http://www.knowledgenetwork.com), a survey and market research company whose panel contains approximately 60,000 households (>120,000 unrelated adults). Households were selected via random digit dialing. KN provides financial incentives to its panel members for participation in web-based surveys. The panel provides a weighted probability sample, representative of the US population. The online screen included questions regarding demographics, ethnic ancestry, and DSM-IV criteria for depression and anxiety disorders. In addition, participants were queried about any history of schizophrenia, psychosis or bipolar disorder using a three-part question: “Have you ever received treatment for, or been diagnosed with, any of the following conditions: a) schizophrenia or schizoaffective disorder; b) hearing voices others could not hear or believing things that others said were not true (such as that people were trying to harm you); c) bipolar disorder (manic-depression).” Controls were included only if they answered “no” to all three of these questions. In addition, controls who met lifetime criteria for recurrent major depressive disorder with functional impairment based on their responses to depression items were excluded. The second control sample and the control samples utilized in Baum et al. (16) were selected from the same larger group of controls collected by KN and may overlap considerably.
UCL controls
The UCL control subjects were recruited from London branches of the National Blood Service, from local NHS family doctor clinics and from university student volunteers. All control subjects were interviewed with the SADS-L to exclude all psychiatric disorders including alcohol dependence according to RDC/DSMIIIR criteria as well as drinking above the Royal College of Psychiatrists upper limit for safe drinking of 21 units per week for males and 14 units for females. The control subjects were further selected on the basis of not having a family history of bipolar disorder, schizophrenia or alcoholism. The UCL supernormal control sample was matched to the UCL case sample using the same ancestry criteria listed above for the BPI volunteers.
Replication study subjects
NIMH Genetics Initiative
Parent-proband trios (n=409 from 256 nuclear families) were obtained from samples previously collected for linkage studies from the NIMH Genetics Initiative (NIMH-GI). The offspring in all trios were affected with BPI
Edinburgh
This sample comprised Caucasian individuals contacted through the inpatient and outpatient services of hospitals in South East Scotland. A BPI diagnosis was based on an interview with the patient using the SADS-L supplemented by case note review and frequently by information from medical staff, relatives and care givers. Final diagnoses, based on DSM-IV criteria (30) were reached by consensus between two trained psychiatrists. Ethnically-matched controls from the same region were recruited through the South of Scotland Blood Transfusion Service. Controls were not directly screened to exclude those with a personal or family history of psychiatric illness; however the Blood Transfusion Service does not accept blood donations from subjects taking regular medication or with a history of a major illness. The study was approved by the Multi-Centre Research Ethics Committee for Scotland and patients gave written informed consent for the collection of DNA samples for use in genetic studies.
Genotyping
For all US samples, DNA was extracted from either whole blood, neonatal cord blood, or from lymphoblastoid cell lines at the Rutger’s Cell and DNA Repository. DNA samples were extracted from whole blood using standard protocols for all UCL samples. Case and control samples were randomized to 96-well plates. Individual plates did not contain mixtures of STEP-BD and UCL samples.
Genotyping was performed using the Affymetrix Gene Chip Human Mapping 500K Array Set. This set is comprised of two high-density arrays, NspI and StyI. All genotyping was performed by the Genetic Analysis Platform at the Broad Institute of Harvard and MIT using standard protocols (31) as previously described (8). Genotypes were called using the Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM) (32). A panel of 24 markers present on the whole genome product as well as 25 SNPs previously genotyped in the UCL samples were used as genetic fingerprints to detect sample switches.
Replication Genotyping
Genotyping of replication SNPs were performed in the NIMH and Edinburgh samples by Sequenom MassArray (33).
Technical Validation
Fifty-nine SNPs from among the most highly associated SNPs were selected for genotyping by an independent method (Sequenom MassArray (33)). For the 56 SNPs with Sequenom genotyping rates greater than 90%, concordance with the Affymetrix data was 99.7% (based on 187,960 genotypes).
Data analysis and quality control
We performed all data analysis and QC using the PLINK software package (http://pngu.mgh.harvard.edu/purcell/plink/) (34). Here we describe the individual and SNP exclusion criteria, methods to address population stratification, and the single SNP and haplotypic association analyses in the screening and replication samples.
Individual exclusions
Raw genotype data was available for 1,800 cases (STEP n= 1,247; UCL n=553) and 2,268 controls (cord blood n=546; NIMHGI 1,180; UCL n=547). Prior to evaluating detailed genotyping quality per individual, we removed individuals with overall call rates <85% (STEP cases n=36; UCL cases n=17; US controls n=56; UCL controls n=17), and SNPs with call rates <90% or which mapped to multiple locations in the genome or were monomorphic (n=10,298). All remaining individuals had call rates > 95%. Seventy-five individuals (cases n=50; controls n=25) were excluded based on heterozygosity. The heterozygosity screen excluded outliers approximately 3 standard deviations from the mean estimated inbreeding coefficient, removing 55 individuals with many more heterozygote calls than expected and 20 individuals with more homozygote calls.
To address potential population stratification, the genome-wide proportion of alleles shared identical-by-state (IBS) for each pair of individuals was calculated. Gross outliers were first removed including individual who appeared to be close relatives, sample duplications, or non-Caucasian (cases n=84, controls n=39). IBS clustering was then performed which indicated that the vast majority of individuals belonged to a single cluster; 203 individuals who did not were removed (cases n=113; controls n=90), Finally, an additional 72 individuals (cases n=39; controls n=33) were removed for low-level relatedness, consistent with some degree of sample contamination, resulting in a final dataset of 3,469 individuals (cases n=1,461; controls n=2,008).
Marker Exclusions
After excluding individuals as described above, SNPs were excluded for the following reasons: 1) call rate <95% (n=23,673), 2) minor allele frequency <1% (n=67,661), 3) Hardy-Weinberg equilibrium p < 1 × 10−6 in controls (n=11,671) and 4) differential rates of missing genotypes between cases and controls, using Fisher’s exact test, p <1 × 10−3 (n=388) (see Table S2). We also tested whether genotypes were non-randomly missing with respect to genotype (potentially unobserved), as indicated by a local haplotype strongly predicting genotyping failure; we excluded SNPs with a p value < 1 × 10 −10 (n=17,021). Finally, as genotypes were called on a per-plate basis, we identified “bad plates” where SNPs showed grossly different allele frequencies to all other plates (p < 1 × 10 −10 ) and removed those SNPs (n=2,397). Following the exclusion there remained 372,193 SNPs for further analysis.
Population Stratification
Since all primary analyses were based on the combined STEP-BD and UCL samples, to control for possible systematic differences reflecting differential ancestry and/or DNA collection conditions, the primary analyses condition on analysis panel (STEP-BD versus UCL). To further control for possible effects of population stratification within panel, we matched cases and controls based on the proportion of alleles shared identical-by-state (IBS), using complete linkage hierarchical clustering. Within panel, individuals were clustered to ensure a) at least 1 case and 1 control in clusters with 2 or more individuals; b) that no two individuals in the same cluster fail the population pairwise concordance test (PPC) with a p < 1 × 10−3; c) only individuals from the same sample (US or UK) were clustered with each other. For a given pair of individuals, the PPC test compares the ratio of IBS 0 (e.g. AA:BB pairs) versus IBS 2 SNPs (e.g. AB:AB pairs) conditional on observing two of each allele; if both individuals belong to the same random-mating population, these two classes should be observed in a 1 : 2 ratio . Finally, parallel to our cluster-based approach, we also applied classical multidimensional scaling (MDS) based on the matrix of IBS distances in order to visually represent any stratification (Figure S1).
Association Analyses
Primary Whole Genome Association Analysis
The primary analysis was of single SNPs using the Cochran-Mantel-Haenszel (CMH) test to assess allelic association with disease conditional on the strata as defined by the stratification analysis. For each SNP, we also calculated standard, allelic association tests not conditioning on strata, based on a chi-squared test for independence.
Multimarker and haplotype tests
We augmented the single SNP tests with multi-marker haplotype tests. We attempted 565,560 (345,288 unique tests) tests, of which 459,414 (340,925 unique) were examined (minor haplotype frequency > 0.01) (35) and tested for association (allowing for potential ambiguity in statistically-inferred haplotype phase and imputed genotype).
We used additional haplotype-based analyses to validate and refine the signals of the most associated SNPs (labeled ‘reference SNPs’ here). First, we used local haplotype information to probabilistically reconstruct missing genotypes for each reference SNP, to further ensure that the associations were not due to biased genotyping failure (labeled the pSNP test). Second, we scanned the local region for haplotypes (not including the reference SNP) which we call “proxies”, that were in linkage disequilibrium with the reference SNP (r2 > 0.2). All proxy haplotypes of up to 4 SNPs out of up to 6 SNPs on either side and within 250kb of the reference SNP are then tested for association with disease (the pHAP test): a positive result here is encouraging in that it shows that the original signal is not due to just a single SNP (and, therefore, not due to technical artifact that might influence a single SNP). As well as technical validation, if the reference SNP is in fact an imperfect proxy for some underlying haplotype, this approach could also help to refine the association signal.
In total, we performed 372,193 single SNP tests, 340,925 unique multi-marker tests, 200 pSNP tests and 183,513 pHAP tests (which represents approximately 10% of all possible haplotypes, i.e. if the constraint of correlation with the reference SNP was ignored), giving a total of 896,831 primary tests of association with bipolar disorder. Naturally, a high proportion of these tests will be strongly correlated with one or more other tests, particularly the pHAP tests which were selected for being highly correlated with only 200 reference SNPs.
Replication sample association analyses
We analyzed the combined NIMH-GI and Edinburgh replication samples using the DFAM test implemented in PLINK, which effectively combines a standard TDT with a Cochran-Mantel-Haenzel test (for a single stratum of all Edinburgh samples) into a single test statistic. When presenting results separately for the two samples, we used the TDT for the NIMH-GI family data and a standard allelic association test for the Edinburgh sample. All tests were one-sided (given the direction of effect in the screening sample). We report tests where p < 0.05 in one of the samples, or the combined sample.
We combined p-values obtained online for the WTCCC with the original STEP-BD/UCL sample using Fisher’s rule (36).
Results
We analyzed 372,193 SNPs genotyped on the Affymetrix GeneChip Human Mapping Array in 1,461 BPI patients and 2,008 controls following the data cleaning procedure described above. The overall genotyping call rate for analyzed SNPs was 99.4%. Including the multimarker predictors, our data set is estimated to capture 78.7% of common variation (SNPs with minor allele frequency >=5%) in the CEPH HapMap (CEU) samples with r2 >0.8.
Assessment and handling of population stratification
The IBS-based constrained hierarchical clustering resulted in 67 clusters (ranging from size 2 to 126 individuals per cluster, mean 51.8, median 48). The final results were not particularly sensitive to different PPC thresholds or clustering schemes, as expected given the initial removal of population outliers and the generally well-matched nature of the remaining samples. Figure S1 shows a series of multidimensional scaling (MDS) plots representing these samples and the HapMap samples. Relative to the entire HapMap, all current samples cluster quite closely with the CEU HapMap panel. There is greater dispersion among the US samples compared to the UK samples, which is to be expected given the ascertainment schemes and pre-matching for the UK samples, but importantly, within US and UK samples, the cases and controls show similar. In general, we did not observe obviously different patterns of results whether we conditioned on sample site and population strata or not.
Association analysis
The genome-wide association results for bipolar disorder from the CMH test are shown in Figure S2. Our most significant single SNP had a p value of 1.66 × 10−7. We did not observe any deviation in the extreme tail of the distribution of single SNP test statistics (see Q-Q plot Figure S3).
Table 1 lists the 20 most associated single SNPs ranked according to the CMH test. Five of these results are found for common SNPs (minor allele frequency in cases between 0.042 and 0.39) within the introns of brain-expressed genes (myosin5b, (MYO5B); tetraspanin-8 (TSPAN8); epidermal growth factor receptor (EGFR), ornithine transcarbamylase (OTC) and raft-linking protein (RFTN1)) conferring odds ratios up to 0.59 and 1.51, for protective and risk alleles respectively. Previous association to BP has not been reported for these genes. Four SNPs in strong LD are found within 142 kb on chromosome 9, in a region containing an open reading frame and ATP6V1G1. Two independent associations are found on 18q: in addition to MYO5B at 45.7 Mb, there are an additional 6 SNPs approximately 15.6Mb telomeric. One SNP is found near transmembrane protein 132E (TMEM132E) on chromosome 17. Chromosomes 12 and 13 each have a SNP in a region containing no annotated genes or transcripts.
Table 1.
Association results for top 20 SNPs in bipolar disorder
| CHR | SNP | Rank | Position (base pairs) |
Nearest gene or transcript |
Distance from |
Frequency Affected |
Frequency Unaffected |
Allele1/ Allele2 |
P value CMH |
P value allelic |
Odds ratio |
pSNP test P value |
pHAP test P value |
r2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | rs4939921 | 1 | 45716326 | MYO5B | Intron | 0.115 | 0.080 | G/A | 1.66E-07 | 5.02E-07 | 1.51 | 5.09E-07 | 2.04E-08 | 0.42 |
| 12 | rs1705236 | 2 | 69831825 | TSPAN8 | Intron | 0.042 | 0.070 | T/A | 6.11E-07 | 1.47E-06 | 0.58 | 1.46E-06 | 7.57E-07 | 0.87 |
| 9 | rs16931058 | 3 | 114547107 | C9orf91 | (−59589) | 0.055 | 0.032 | A/G | 3.57E-06 | 2.34E-06 | 1.76 | 2.34E-06 | 9.11E-06 | 0.88 |
| 13 | rs1341139 | 4 | 78439189 | C13orf7 | (+307874) | 0.025 | 0.043 | C/A | 7.23E-06 | 3.16E-05 | 0.56 | 2.15E-05 | 1.59E-04 | 0.45 |
| 12 | rs11050038 | 5 | 29149594 | MLSTD1 | +43806 | 0.167 | 0.207 | C/T | 1.47E-05 | 2.18E-05 | 0.77 | 2.18E-05 | 2.60E-05 | 0.98 |
| 9 | rs2274598 | 6 | 114499686 | C9orf91 | −12168 | 0.040 | 0.022 | C/T | 1.68E-05 | 1.28E-05 | 1.87 | 2.30E-05 | 7.04E-06 | 0.83 |
| 18 | rs1444067 | 7 | 61321666 | CDH7 | (+247471) | 0.470 | 0.419 | T/G | 1.83E-05 | 3.41E-05 | 1.23 | 7.26E-05 | 5.56E-06 | 0.61 |
| 18 | rs2850700 | 8 | 61268622 | CDH7 | (+300515) | 0.473 | 0.424 | T/C | 1.93E-05 | 4.36E-05 | 1.22 | 6.02E-05 | 2.50E-05 | 0.98 |
| 9 | rs2900591 | 9 | 114524833 | C9orf91 | −37315 | 0.048 | 0.029 | T/C | 2.06E-05 | 2.68E-05 | 1.71 | 8.05E-06 | 6.66E-06 | 0.55 |
| 18 | rs2658046 | 10 | 61273210 | CDH7 | (+295927) | 0.473 | 0.423 | G/A | 2.09E-05 | 4.31E-05 | 1.22 | 5.34E-05 | 2.31E-05 | 0.94 |
| 17 | rs10491113 | 11 | 30028246 | TMEM132E | −37797 | 0.144 | 0.185 | A/G | 2.69E-05 | 9.90E-06 | 0.74 | 1.03E-05 | 7.36E-02 | 0.21 |
| 18 | rs12970791 | 12 | 61316783 | CDH7 | (+252354) | 0.467 | 0.418 | A/C | 2.73E-05 | 5.30E-05 | 1.22 | 5.55E-05 | 1.22E-05 | 0.65 |
| 18 | rs2850699 | 13 | 61268473 | CDH7 | (+300664) | 0.472 | 0.424 | C/A | 2.78E-05 | 6.02E-05 | 1.22 | 6.02E-05 | 4.40E-05 | 0.95 |
| 18 | rs4455070 | 14 | 61305341 | CDH7 | (+263796) | 0.472 | 0.423 | T/A | 3.26E-05 | 5.60E-05 | 1.22 | 4.10E-05 | 1.34E-05 | 0.64 |
| 7 | rs17172438 | 15 | 54925746 | EGFR | Intron | 0.203 | 0.162 | C/T | 3.26E-05 | 1.26E-05 | 1.32 | 1.58E-05 | 9.28E-07 | 0.69 |
| 7 | rs729969 | 16 | 54902416 | EGFR | Intron | 0.163 | 0.125 | T/C | 3.30E-05 | 9.29E-06 | 1.36 | 8.67E-06 | 8.36E-08 | 0.68 |
| 9 | rs10982292 | 17 | 114404845 | ATP6V1G1 | +24703 | 0.039 | 0.022 | A/G | 3.32E-05 | 2.86E-05 | 1.82 | 2.52E-05 | 6.81E-06 | 0.77 |
| 12 | rs11049998 | 18 | 29085712 | MLSTD1 | (+107688) | 0.166 | 0.206 | G/A | 3.33E-05 | 3.93E-05 | 0.77 | 2.95E-05 | 2.05E-04 | 0.78 |
| X | rs5963419 | 19 | 38028410 | OTC | Intron | 0.386 | 0.331 | C/T | 3.44E-05 | 3.53E-05 | 1.27 | 4.22E-05 | 4.71E-05 | 0.99 |
| 3 | rs9817739 | 20 | 16455831 | RFTN1 | Intron | 0.184 | 0.223 | T/C | 3.47E-05 | 7.26E-05 | 0.79 | 7.30E-05 | 7.24E-03 | 0.28 |
Association results for the top 200 SNPs can be found in Table S3; a complete listing of all single SNP statistics is available at http://pngu/mgh/harvard/edu/purcell/bpwgas/. Of the two-marker haplotypes used in multimarker tests of SNPs not directly genotyped, no additional associations were identified at p < 5 × 10−5 except for within the region of EGFR already identified by single SNP analysis (Table S4). Analyzing STEP-BD and UCL individuals separately, no single SNP from either site exceeds those results listed in Table 1 (Table S5). There was no heterogeneity of odds ratios between study sites for the top 200 markers (Breslow-Day test for homogeneity of odds ratio, p > 0.01).
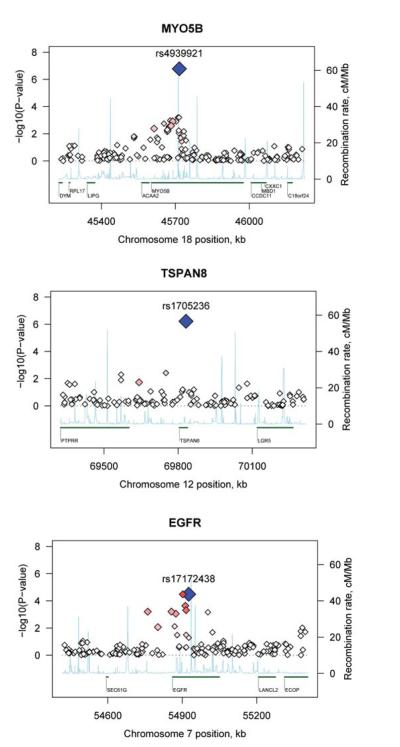
Reconstruction of missing genotypes for these top 200 SNPs, based on local haplotype information, indicated that none of these tests were substantially biased by missing genotype data (pSNP test; see Methods). Next, we established proxy alleles and haplotypes that were highly associated with the reference SNP, but did not contain the SNP (pHAP test). In general, this confirmed that the original signals were not driven by solely a single SNP. In particular, rs1705236 in TSPAN8 (ranked 2nd) does not have any flanking SNPs showing association, but does show a strong association with a number of proxy haplotypes (see Figure S4). For MYO5B, EGFR, and TSPAN8 (single SNP results shown in Figure 1) the pHAP tests showed haplotypic p values of 2.04 × 10−8, 8.4 × 10−8 and 7.5 × 10−7 respectively (Table 1). The MYO5B result withstands conservative correction for 1 million tests ( p= 5 × 10−8) and has an odds ratio of 1.51 (frequency in cases=0.115 and controls=0.08). Plots of gene and regions surrounding additional SNPs from Table 1 can be found in Figure S5.
Figure 1.
Plot of MYO5B, TSPAN8 and EGFR regions of association.
Association results (−log10 p) are plotted for all SNPs passing QC in the combined STEP-BD and UCL sample. A) MYO5B, B) TSPAN8, and C) EGFR. Chromosome position is plotted with reference to the NCBI build 35 and gene names are plotted with reference to the University of California at Santa Cruz Genome Browser. Recombination rate as estimated from the HapMap is plotted in light blue. Blue diamond: SNP with strongest evidence for association. Red diamonds: strength of LD with blue diamond SNP (darker red indicates stronger LD).
Testing in independent samples
We selected a total of 304 SNPs for genotyping in two independent samples: the family-based NIMH-GI sample and population-based Edinburgh sample. As well as genotyping of the 200 top hits (both samples), we report here additional SNPs that were genotyped earlier in the study only in the NIMH-GI sample, based on the results of interim analyses with less stringent QC metrics. We observed evidence for association, defined as same allele, same direction of effect, one-sided p <0.05, in the combined NIMH-GI and Edinburgh samples for 13 SNPs (2 SNPs at p<0.01) (Table S6). Of these, 9 were from our top 200 hits whereas 4 (rs12967023, rs9268853, rs749044 and rs6424785) were selected based on interim analyses (ranked 566th, 1220th, 662nd and 2405th out of the 372,193 SNPs in the final analysis, with CMH p-values of 0.001165, 0.002575, 0.00137 and 0.005191 respectively).
Four of these 13 SNPs are in strong LD in a region of chromosome 18q22 with no annotated genes, approximately 246 kb from cadherin 7 (CDH7); they were ranked 7th, 8th, 10th and 13th most significant in the whole genome scan and are marginally significant at p<0.05 (one-sided) in this follow-up. The next highest ranking SNP from the original scan that is also significant in the follow-up is rs10491113 (ranked 11th in the original scan), which is 38kb downstream of TMEM132E on chromosome 17. The strongest follow-up results are for rs12967023 (p=0.00091), which is in myelin basic protein (MBP) and rs9268853 (p=0.0011) on chromosome 6 near HLA-DRA; neither SNP was in the top 200 in the original scan however. We see modest evidence for association with rs4979416 in the gene calcium/calmodulin-dependent serine protein kinase-interacting protein (DFNB31) (ranked 172nd in the original screen, p=0.013 in follow-up) and an association for rs2237554, in the metabotropic glutamate receptor 3 gene (GRM3) (ranked 121st in the original screen, p=0.035 in follow-up). Importantly, though, given that 304 follow-up tests were performed, this number of significant results is consistent with that expected by chance.
Comparison to previous whole genome scan data
Two groups have recently published whole genome association studies of bipolar disorder. The Wellcome Trust Case Control Consortium (WTCCC) genotyped 1900 primarily BPI patients from the UK and 3000 controls using the same platform as described in this paper (1). The strongest signal observed in that study was at rs420259 near the partner and localizer of BRCA2 (PALB2) under a recessive genetic model. We do not observe evidence for association at this SNP under allelic, dominant or recessive models (p>0.05).
Looking at the WTCCC allelic p values (as available online), we do not observe any evidence for association with the 20 SNPs in Table 1 noted (all p>0.05). For our top 200 SNPs, we calculated combined p values for our study and the WTCCC (Table S7). Of our top 200 SNPs, the most significant result in the WTCCC consistent with our data was rs1006737 (OR=1.21 this study, OR=1.16 WTCCC) which is located in the third intron of the alpha subunit of the L-type, voltage-gated calcium channel (CACNA1C). Within CACNA1C, there are numerous other associated SNPs in both our sample and the WTCCC in an area of strong linkage disequilibrium (LD) as shown in Figure 2. However, we did not observe statistically significant association with this SNP in our replication samples (although the Edinburgh case-control sample displays broadly similar allele frequencies as well as an odds ratio of 1.15).
Figure 2.
Plot of CACNA1C region of association.
Association results (−log10 p) are plotted for all SNPs passing QC. A) Combined STEP-BD and UCL samples, and B) WTCCC. See Figure 1 for description of plots.
Baum et al. (2007) recently reported the results of a whole genome scan using a pooled DNA strategy on the Illumina HumanHap550 followed by individual genotyping of 37 SNPs (16). They identified SNPs in diacylglyceral kinase eta (DGKH) as associated with bipolar disorder in a US and German sample. As we expect some degree of overlap between our control sample and those used by Baum et al, we have excluded 1044 controls from the following analyses, to create a dataset of 1461 cases and 964 controls that is guaranteed to be independent of Baum et al. We do not see evidence for association with the 41 SNPs present on our arrays in DGKH (minimum p = 0.03). Furthermore, imputing the missing genotypes for the three SNPs reported as associated in DGKH (using the CEU HapMap samples as a reference panel and the pSNP test, as these SNPs were not directly genotyped in the current study) we do not see association for these SNPs, with pSNP tests imputing the missing genotype data showing p= 0.257, 0.744 and 0.381 respectively; the strongest haplotype-based proxies in our data (from the pHAP test) gave p = 0.066, 0.289 and 0.247 for r2 with the Baum et al SNPs of 0.40, 0.68 and 0.91 respectively).
The fifth most associated SNP in Baum et al. is rs942518 (p = 0.0001, not present in our genotyping platform) which is located near DFNB31. In our full sample including all controls, we observed an association with rs4979416 (located in DFNB31 approximately 10 kb from rs942518) which was ranked 172nd (p= 0.0003, odds ratio 1.45, MAF 7% in cases, 5% in controls, p=0.0027 in the non-overlapping subsample) and showed replication in our follow-up (p=0.013). Although, the NIMH-GI sample overlaps with the case samples used in Baum et al., we observe a marginal association in the Edinburgh sample (p=0.06, odds ratio 1.4, 7.4% in cases, 5.4% in cases) as well as the NIMH-GI trios (p= 0.06, odds ratio 1.4). This SNP is in moderate LD with rs942518, based on HapMap data (r2=0.35). The WTCCC also shows a number of SNPs (rs11787667, rs10982246, rs10982256, rs1535964 and rs998548) flanking and within ~20kb of rs4979416 that show association with BP (all p < 5 × 10−4 (Figure S6). However, as shown in Figure S7, the association in this region appears complex and there is no obvious haplotype spanning the region that can account for the entire signal. Although the current study, the WTCCC and Baum et al. all identify associations physically near each other in DFNB31, the WTCCC association would appear to be independent of the signal seen in our study and Baum et al.; further, the association in DFNB31 from our study appears to be at least partially representing indirect association with a stronger signal ~60kb downstream, to a region containing multiple SNPs that were ranked in our top 10 associations. Further work is warranted to confirm and refine the nature of association in this region.
Discussion
We report results from a large whole genome association study of bipolar disorder. For our strongest signals, we also report replication efforts using a combined family-based and case-control sample. Furthermore, we compared our top results with that of other whole genome association studies of bipolar disorder. While we did not detect single SNP signals that meet stringent criteria for genome-wide significance, a haplotype in the gene MYO5B achieved genome-wide significance and we identified several interesting loci that will require examination in much larger samples. In addition to its size and the genotyping of individual rather than pooled samples, specific strengths of this study include the rigor of the diagnostic assessments for both the US and UK samples, the use of psychiatrically screened controls for the majority of the control sample and the methodology implemented to limit the effects of population stratification.
Our strongest association signals were observed in three genes, MYO5B, TSPAN8, and EGFR that were supported by consideration of local haplotype information. MYO5B is located on chromosome 18 in a region initially reported as linked to bipolar disorder (37, 38), although a subsequent meta-analysis of linkage studies does not support this observation (6). MYO5B is a large gene (372 kb, 40 exons) encoding a multifunctional protein expressed in a variety of brain structures, including dendritic spines, at high levels (39, 40). Myosins are known to transport proteins in neurons through binding to their C-terminal globular domain (41) and MYO5B specifically regulates vesicle trafficking (42). Strikingly, MYO5B has been shown to regulate EGFR cycling in both canine (MDCK) and human cells (A431) (43). In light of the current associations with both MYO5B and EGFR, the functional relationship of these two genes may point to a possible molecular pathway (i.e. vesicle trafficking at the plasma membrane) in which converging susceptibility alleles of small effect size, in distinct component genes, may underlie the biological etiology of BP. EGFR, also known as ErbB1/HER1, belongs to the ErbB family of receptor tyrosine kinases that includes ErbB4, the Neuregulin 1 (NRG1) receptor, and heterodimeric complexes of EGFR, ErbB2, ErbB3 and ErbB4 have been shown to modulate downstream signaling of NRG1 and NRG2 (44).
Abnormalities in circadian rhythms have been hypothesized to lead to episodes of mania and depression in what has been termed the social zeitgeber theory (45, 46). The biology that connects mood symptoms and circadian rhythms is unknown. However, Kramer et al. have demonstrated that the actions TGF-α, transduced through the EGFR, inhibits locomotor activity on the running wheel in mice and disrupts circadian cycles when infused into the third ventricle (47). Waved-2 mice are a naturally occurring strain with an EGFR point mutation that reduced kinase activity more than 80% (48). In fact, this loss of function EGFR mutation, that would be predicted to result in excessive activity, demonstrates excessive daytime activity on the running wheel, phenotypically reminiscent of the prominent hyperactivity observed in manic patients. Conversely, Drosophila gain-of-function mutants that activate EGFR signaling demonstrate an opposite phenotype, excessive sleep (49). From this we would hypothesize that the SNPs and haplotypes associated with increased risk of bipolar disorder observed with EGFR would result in decreased tyrosine kinase activity. Through downstream interactions with known interacting proteins such as GAB1 and ErbB3, decreased PI3K activity could result in decreased phosphorylation (increased activity) of GSK3β. This is consistent with the effects of mood stabilizers which function as GSK3β inhibitors. However, these observations remain speculative in the absence of independent replication or knowledge of a causal polymorphism.
TSPAN8, located on chromosome 12, is a member of a large family of tetraspanin proteins that are involved in diverse cellular processes (50). Other tetraspanins are known to form tetraspanin-enriched microdomains (TERMs) and may be involved in clustering of receptors or cell signaling molecules (51).
We did not see evidence for replication of our strongest findings. We did however observe replication p values below 0.05 in a 53kb region of 18q22 with no annotated genes, approximately 246 kb from CDH7 a brain-expressed calcium dependent cell-cell adhesion molecule. Similarly, rs10491113 is found 38kb downstream of TMEM132E, an uncharacterized transmembrane protein. Two SNPs, one in myelin basic protein (MBP) and one in a metabotropic glutamate receptor (GRM3) followed based on results of an interim analyses show stronger results. Myelination and oligodendrocyte functions have been reported as abnormal in bipolar disorder patients (52). We have previously demonstrated upregulation the full length “golli” MBP mRNA transcription mice following treatment of lithium (53). Finally, two groups have tested SNPs in the GRM3 gene for association with bipolar disorder with varying results (54, 55). While we judge the evidence for association with GRM3 modest, we have previously reported lithium-induced changes in this gene (53).
Consistent association findings between our study and the WTCCC are observed within intron 3 of the alpha subunit of the L-type calcium channel (CACNA1C). L-type calcium channels mediate a variety of calcium dependent processes in neurons and are sensitive to dihydropyridine derivatives, such as verapamil. Treatment of mania with verapamil initially showed promise, but its efficacy remains ambiguous (56, 57). Mutations in CACNA1C have been shown to cause Timothy’s syndrome with severe prolongation of the QT interval on electrocardiogram, syndactyly, cognitive abnormalities and autism spectrum symptoms (58).
In summary, we have generated a list of genes and regions that warrant follow-up in more samples. These associations can be classified according to two categories: those with strong statistical significance in the primary scan but no supporting evidence, and those that show only moderate association in the primary scan but that also show replication in the follow-up, or some level of consistency with the WTCCC and/or Baum et al. study. In the first category are MYO5B, TSPAN8 and EGFR. In the second category are DFNB31, CACNA1C, MBP, GRM3 and HLA-DRA. This study, by itself, is unable to unambiguously conclude that any of these genes influence risk for BPI.
In contrast to a number of recent whole genome studies of other common complex disorders, the major findings from each bipolar disorder study are not obviously supported by any of the other studies. Possible explanations for this apparent non-replication, include inadequate power to detect alleles of modest effect sizes, population-specific disease alleles, phenotypic heterogeneity (or misspecification), epistasic interactions of multiple modest effect genes, and effects of copy number variants or other genetic variation not well captured by the panels of common SNPs, including multiple rare disease alleles. Although the current study is well-powered to detect large effects (odds ratios > 1.5 for common alleles), most rarer and more modest effects will have a substantial chance of not being detected, in both the whole genome screen and the replication stage. For example, if we define 5 × 10−4 as the significance threshold for declaring a “promising SNP” (i.e that we would expect to rank within the top 200 or so results) we can calculate joint power to detect a SNP in both our and the WTCCC samples. For common variants (40% minor allele frequency) the joint power is ~96% and ~48% for SNPs with multiplicative odds ratios of 1.3 and 1.2 respectively. However, for less common variants (10% minor allele frequency) joint power to detect the SNP in both studies is much lower, now at ~34% and ~3% for odds ratios of 1.3 and 1.2 respectively. And even if we were able to detect a rarer SNP in our study, for example one with a minor allele frequency of 5% and odds ratio of 1.2, then power to replicate in the WTCCC even at very modest significance thresholds (p<0.05 rather than p < 5 × 10−4) is still only approximately 50%. Given these estimates, and the modest effect sizes of replicated variants emerging in other disease areas, the lack of agreement between studies is perhaps less surprising. Joint analysis is expected to be more powerful than replication (59), and we are planning to embark on a combined analysis of the WTCCC bipolar data and our sample with the WTCCC investigators. As well as lack of power, phenotypic and genetic heterogeneity is another explanation for the lack of clear findings. It is unlikely, however, that population-specific alleles account for the observed difference between our study and the WTCCC: both samples are of European background and approximately 1/3 of the current sample was from the UK (as are the WTCCC samples). While phenotypic heterogeneity is always a possibility, the STEP-BD and UCL samples are selected from patients with BPI disorder and approximately 70% of the WTCCC sample fit DSM-IV criteria for BPI disorder. Ultimately, meta-analysis of diagnostic subphenotypes, ascertainment strategies and diagnostic conventions will be useful to investigate possible phenotypic heterogeneity between and within studies. In particular, the ADE, SADS-L and OPCRIT used in the diagnostic assessment of the STEP-BD and UCL samples also collects data on age of onset, prior episode frequency, prognosis, clinical features, medical history, family history, and mental status. One possibility to be explored is that the genetic influences on bipolar disorder act on these more specific aspects of the disease or in subtypes of the disorder characterized by age-of-onset or certain clinical symptoms. Future analyses will focus in particular on those aspects of the diagnosis that have been demonstrated to be inherited including psychosis, and age-at-onset. The diagnostic instruments used in this study and also by the WTCCC include additional phenotypic information.
Alternatively, the common variant hypothesis may not fit for bipolar disorder. Whole genome studies are not adequately powered to investigate the hypothesis that there are many rare private alleles (either in a small number of genes, or in many genes) leading to disease. Testing of this hypothesis may be possible in the near future as whole genome sequencing technologies become less costly. We are also pursuing alternate analytic approaches to the SNP data presented here, focusing on epistasis, copy number variation and rare variation as indexed by patterns of extended segmental sharing.
We were also unable to provide additional support for the most significant associations reported in the WTCCC and Baum et al studies. Despite this, the high heritability of BP suggests it should be possible to identify risk variants. Given the power and coverage of the present study, if common variants exist for bipolar disorder, they may be of very small effect and thus require very large samples to be reliably detected. This highlights the future need for meta-analysis of all whole genome bipolar disorder data already collected as well as for ongoing and larger sample collections.
Supplementary Material
Acknowledgements
We would like to thank the patients and families who have contributed their time and DNA to this study. The study was supported by grants from the NIMH (MH062137, PS; MH067288, PS; MH063445, JWS; MH63420, VLN), Charles A. King Trust Fellowship (JF); NARSAD Young Investigator Awards (RHP, SP, JF); NARSAD Independent Investigator Award (PS); Johnson & Johnson Pharmaceutical Research & Development (EMS); Johnson & Johnson Foundation (PS); the Sydney Herman Foundation (EMS); the Stanley Foundation for Medical Research (EMS); the Dauten Family (EMS; PS). The University College London clinical and control samples were collected with support from the MDF the Bipolar Organization (formerly the UK Manic Depression Fellowship), the Neuroscience Research Charitable Trust, the central London NHS Blood Transfusion Service and by a research lectureship from the Priory Hospitals. Processing and genetic analysis of the UCL cohort has been supported by UK Medical Research Council project grants G9623693N and G0500791. The collection of the Edinburgh cohort was supported by grants from The Wellcome Trust, London, The Chief Scientist Office of the Scottish Executive and the Translational Medicine Research Institute, Glasgow. The STEP-BD project was funded in whole or in part with Federal funds from the National Institute of Mental Health (NIMH), National Institutes of Health, under Contract N01MH80001 to Gary S. Sachs, M.D. (PI), Michael E. Thase M.D. (Co-PI), Mark S. Bauer, M.D. (Co-PI). Active STEP-BD Sites and Principal Investigators included Baylor College of Medicine (Lauren B. Marangell, M.D.); Case University (Joseph R. Calabrese, M.D.); Massachusetts General Hospital and Harvard Medical School (Andrew A. Nierenberg, M.D.); Portland VA Medical Center (Peter Hauser, M.D.); Stanford University School of Medicine (Terence A. Ketter, M.D.); University of Colorado Health Sciences Center (Marshall Thomas, M.D.); University of Massachusetts Medical Center (Jayendra Patel, M.D.); University of Oklahoma College of Medicine (Mark D. Fossey, M.D.); University of Pennsylvania Medical Center (Laszlo Gyulai, M.D.); University of Pittsburgh Western Psychiatric Institute and Clinic (Michael E. Thase, M.D.); University of Texas Health Science Center at San Antonio (Charles L. Bowden, M.D.). Collection of DNA from consenting participants in STEP-BD was supported by N01-MH-80001 (G Sachs, PI). We thank the following individuals for their assistance with this effort: Francine Molay, Beth Rosen-Sheidley, and Laurie Silfies. Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials were collected by the “Molecular Genetics of Schizophrenia II” (MGS-2) collaboration. The investigators and co-investigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA,MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA,MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA,MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson M.D. (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY,MH59586, Jeremy Silverman, Ph.D. (PI). NIMH Family samples were collected as part of ten projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1999-03, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Wayne State University), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, M.S., Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D., Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D,Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D.; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis M.D. , J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini M.D.,Ph.D. ; University of California at Irvine, CA, R01 MH60068, William Byerley M.D., and Mark Vawter M.D. ; University of Iowa, IA, R01 MH059548, William Coryell M.D. , and Raymond Crowe M.D. ; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner Ph.D. , Francis McMahon M.D. , Chunyu Liu Ph.D., Alan Sanders M.D., Maria Caserta, Steven Dinwiddie M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner M.D. , Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, PsyD, Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.
Data release policy: Genotypes for the NIMH control samples have been submitted to the NIMH Genetics Repository and are available under the usual data release policies. Genotypes for the STEP-BD case samples will be submitted to the NIMH repository and will be available for release using the same mechanism.
Footnotes
Author Contribution Statement Writing group: Sklar P, Fan J, Gurling HM, Smoller JW, Ogdie M, Nimgaonkar VL, Daly MJ, Purcell SM
Project management: Sklar P, Smoller JW, Nimgaonkar VL, Scolnick EM, Gurling HM
Clinical characterization and phenotypes: Smoller JW, Nimgaonkar VL, Perlis RH, Thase ME, Sachs GS, Faraone SV, Muir WJ, McGhee KA, MacIntyre DM, McLean A, VanBeck M, Blackwood DH, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Gurling HM
DNA sample QC and replication genotyping: Fan J, Chambert K, Franklin J, Ardlie KG
Whole genome genotyping: Gabriel SB, Chambert K, Gates C, Blumensteil B, Defelice M, Sougnez C
Analytic group: Purcell SM, Ferreira MAR, Fan J, Smoller JW, Perlis RH, McQueen MB, Todd-Brown K, De Bakker PIW, Daly MJ, Sklar P
Supplementary information is available at the Molecular Psychiatry website.
References
- 1.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123(1):48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 3.The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 4.Altshuler D, Brooks LD, Chakravarti A, Collin FS, Daly MJ, Donnelly P, et al. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI, Jr., et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. Am J Hum Genet. 2003;73(1):49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77(4):582–95. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craddock N, Forty L. Genetics of affective (mood) disorders. Eur J Hum Genet. 2006;14(6):660–8. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- 8.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 9.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 12.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9):1055–9. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 16.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2007 doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachs GS, Thase ME, Otto MW, Bauer M, Miklowitz D, Wisniewski SR, et al. Rationale, design, and methods of the systematic treatment enhancement program for bipolar disorder (STEP-BD) Biol Psychiatry. 2003;53(11):1028–42. doi: 10.1016/s0006-3223(03)00165-3. [DOI] [PubMed] [Google Scholar]
- 18.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders. Patient Edition (SCID-P) New York State Psyciatric Institute; New York: 1996. [Google Scholar]
- 19.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 20):22–33. quiz 34-57. [PubMed] [Google Scholar]
- 20.Faraone SV, Blehar M, Pepple J, Moldin SO, Norton J, Nurnberger JI, et al. Diagnostic accuracy and confusability analyses: an application to the Diagnostic Interview for Genetic Studies. Psychol Med. 1996;26(2):401–10. doi: 10.1017/s0033291700034796. [DOI] [PubMed] [Google Scholar]
- 21.Simpson SG, McMahon FJ, McInnis MG, MacKinnon DF, Edwin D, Folstein SE, et al. Diagnostic reliability of bipolar II disorder. Arch Gen Psychiatry. 2002;59(8):736–40. doi: 10.1001/archpsyc.59.8.736. [DOI] [PubMed] [Google Scholar]
- 22.Nurnberger JI, Jr., Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51(11):849–59. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863-4. [DOI] [PubMed] [Google Scholar]
- 23.Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, et al. The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry. 2001;158(8):1258–64. doi: 10.1176/appi.ajp.158.8.1258. [DOI] [PubMed] [Google Scholar]
- 24.Potash JB, Chiu YF, MacKinnon DF, Miller EB, Simpson SG, McMahon FJ, et al. Familial aggregation of psychotic symptoms in a replication set of 69 bipolar disorder pedigrees. Am J Med Genet. 2003;116B(1):90–97. doi: 10.1002/ajmg.b.10761. [DOI] [PubMed] [Google Scholar]
- 25.Schulze TG, Hedeker D, Zandi P, Rietschel M, McMahon FJ. What is familial about familial bipolar disorder? Resemblance among relatives across a broad spectrum of phenotypic characteristics. Arch Gen Psychiatry. 2006;63(12):1368–76. doi: 10.1001/archpsyc.63.12.1368. [DOI] [PubMed] [Google Scholar]
- 26.Spitzer RL, Endicott J. The Schedule for Affective Disorder and Schizophrenia, Lifetime Version. 3rd ed New York State Psychiatric Institute; New York: 1977. [Google Scholar]
- 27.Spitzer RL, Endicott J. Research Diagnostic Criteria for a selected group of functional disorders. 3rd ed New York State Psychiatric Institute; New York: 1978. [Google Scholar]
- 28.McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48(8):764–70. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- 29.Mansour HA, Talkowski ME, Wood J, Pless L, Bamne M, Chowdari KV, et al. Serotonin gene polymorphisms and bipolar I disorder: focus on the serotonin transporter. Ann Med. 2005;37(8):590–602. doi: 10.1080/07853890500357428. [DOI] [PubMed] [Google Scholar]
- 30.Association AP. Diagnostic and statistical manual of mental disorders (DSM-IV) Fourth Edition American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- 31.Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14(3):414–25. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22(1):7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 33.Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, et al. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Brain-derived neutrophic factor. Mol Psychiatry. 2002;7(6):579–93. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pe’er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38(6):663–7. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 36.Fisher RA. Combining independent tests of significance. Am Stat. 1948;2:30. [Google Scholar]
- 37.Stine OC, Xu J, Koskela R, McMahon FJ, Gschwend M, Friddle C, et al. Evidence for linkage of bipolar disorder to chromosome 18 with a parent- of-origin effect. Am J Hum Genet. 1995;57(6):1384–94. [PMC free article] [PubMed] [Google Scholar]
- 38.De bruyn A, Souery D, Mendelbaum K, Mendlewicz J, Van Broeckhoven C. Linkage analysis of families with bipolar illness and chromosome 18 markers. Biol Psychiatry. 1996;39(8):679–88. doi: 10.1016/0006-3223(95)00293-6. [DOI] [PubMed] [Google Scholar]
- 39.Zhao LP, Koslovsky JS, Reinhard J, Bahler M, Witt AE, Provance DW, Jr., et al. Cloning and characterization of myr 6, an unconventional myosin of the dilute/myosin-V family. Proc Natl Acad Sci U S A. 1996;93(20):10826–31. doi: 10.1073/pnas.93.20.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lise MF, Wong TP, Trinh A, Hines RM, Liu L, Kang R, et al. Involvement of myosin Vb in glutamate receptor trafficking. J Biol Chem. 2006;281(6):3669–78. doi: 10.1074/jbc.M511725200. [DOI] [PubMed] [Google Scholar]
- 41.Bridgman PC. Myosin-dependent transport in neurons. J Neurobiol. 2004;58(2):164–74. doi: 10.1002/neu.10320. [DOI] [PubMed] [Google Scholar]
- 42.Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, et al. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12(6):1843–57. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapierre LA, Goldenring JR. Interactions of myosin vb with rab11 family members and cargoes traversing the plasma membrane recycling system. Methods Enzymol. 2005;403:715–23. doi: 10.1016/S0076-6879(05)03062-4. [DOI] [PubMed] [Google Scholar]
- 44.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. Embo J. 1996;15(10):2452–67. [PMC free article] [PubMed] [Google Scholar]
- 45.Ehlers CL, Frank E, Kupfer DJ. Social zeitgebers and biological rhythms. A unified approach to understanding the etiology of depression. Arch Gen Psychiatry. 1988;45(10):948–52. doi: 10.1001/archpsyc.1988.01800340076012. [DOI] [PubMed] [Google Scholar]
- 46.Grandin LD, Alloy LB, Abramson LY. The social zeitgeber theory, circadian rhythms, and mood disorders: review and evaluation. Clin Psychol Rev. 2006;26(6):679–94. doi: 10.1016/j.cpr.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294(5551):2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 48.Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, et al. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8(4):399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- 49.Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10(9):1160–7. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 50.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 51.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8(2):89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 52.Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362(9386):798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 53.McQuillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenet Genomics. 2007;17(8):605–17. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- 54.Marti SB, Cichon S, Propping P, Nothen M. Metabotropic glutamate receptor 3 (GRM3) gene variation is not associated with schizophrenia or bipolar affective disorder in the German population. Am J Med Genet. 2002;114(1):46–50. doi: 10.1002/ajmg.1624. [DOI] [PubMed] [Google Scholar]
- 55.Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77(6):918–36. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy NA, Janicak PG. Calcium channel antagonists for the treatment of bipolar disorder. Bipolar Disord. 2000;2(2):108–19. doi: 10.1034/j.1399-5618.2000.020204.x. [DOI] [PubMed] [Google Scholar]
- 57.Wisner KL, Peindl KS, Perel JM, Hanusa BH, Piontek CM, Baab S. Verapamil treatment for women with bipolar disorder. Biol Psychiatry. 2002;51(9):745–52. doi: 10.1016/s0006-3223(01)01338-5. [DOI] [PubMed] [Google Scholar]
- 58.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 59.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 60.Stirling WD. Enhancements to Aid Interpretation of Probability Plots. The Statistician. 1982;31(3):211–220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.