Abstract
Purpose
To investigate the relationship between chromosomal radiosensitivity and early-onset cancer under age 35 years and to examine the heritability of chromosomal radiosensitivity.
Materials and methods
Peripheral blood lymphocytes were cultured for 72 hours prior to being irradiated with 0.5 Gy, 300 kV X-rays. Colcemid was added to cultures 30 minutes post-irradiation. Cultures were harvested 90 minutes post-irradiation and analysed for chromatid gaps and breaks. Heritability was estimated using Sequential Oligogenic Linkage Analysis Routines (SOLAR) software and by segregation analysis.
Results
Elevated radiosensitivity was seen for 7 out of 29 (24.1%) cancer survivors, 3 out of 29 (10.3%) partners and 10 out of 53 (20.8%) offspring. Although the proportion of individuals displaying enhanced radiosensitivity was twice as high in both the cancer survivor and offspring groups than the partner controls, neither reached statistical significance. Heritability analysis of the radiosensitive phenotype suggested 57.9 – 78.0% of the variance could be attributed to genetic factors.
Conclusions
An association between G2 chromosomal radiosensitivity and childhood and young adult cancer is suggested but was not statistically significant. In contrast, there is strong evidence for heritability of the radiosensitive phenotype. The cancer survivors included a broad range of malignancies and future studies should focus on specific cancers with known or likely faults in deoxyribonucleic acid (DNA) damage recognition and repair mechanisms.
Keywords: Chromosomal radiosensitivity, heritability, cancer
Introduction
The association of G2 chromosomal radiosensitivity with cancer predisposition has been demonstrated for a number of common cancers (Scott et al. 1994, Scott et al. 1996, Scott et al. 1999, Scott 2000, Baria et al. 2001, Riches et al. 2001, Baeyens et al. 2002, Baria et al. 2002, Howe et al. 2005, Lisowska et al. 2006, De Ruyck et al. 2008) with many studies concluding that G2 chromosomal radiosensitivity may be a marker of inherited, low-penetrance cancer predisposition genes (Scott et al. 1999, Scott 2000, Baria et al. 2001, Scott 2004, Howe et al. 2005). Heritability of the radiosensitive phenotype has been demonstrated in healthy first-degree relatives of cancer patients (Roberts et al. 1999). Furthermore, enhanced G2 chromosomal radiosensitivity has been shown to be associated with early-onset cancers (Papworth et al. 2001, Baria et al. 2002), thus supporting the premise that such cancers have a genetic inherited component. Consequently, it has been postulated that inheritance of low-penetrance genes may indeed function to predispose young individuals to cancer. We previously reported on the relationship between chromosomal radiosensitivity and early-onset cancer in a population of 23 Danish childhood and adolescent cancer survivors, their partners and offspring using the G2 assay of radiation-induced chromatid breakage (Curwen et al. 2005). Failure to distinguish between the G2 aberration profiles of the apparently normal partners and the cancer survivors suggested the lack of an association between chromosomal radiosensitivity and cancer predisposition, although questions were raised as to the suitability of the partner group as controls. Moreover, analysis of the Danish families did suggest that the radiosensitive phenotype was a heritable trait. In the current study, we examined an additional 30 Danish families for G2 chromosomal radiosensitivity in an attempt to clarify the relationship with predisposition to early-onset cancer and to investigate heritability of the radiosensitive phenotype.
Methods
Study group
Blood samples were obtained from 30 Danish survivors of childhood and young adult cancer who had been treated with radiotherapy under age 35 years, their partners and 55 out of a potential of 57 offspring born after treatment. This work forms part of an ongoing international study of trans-generational effects of cancer treatment in children and young adults (Boice et al. 2003) (www.gcct.org). Patients were invited to participate on the basis of their young age at diagnosis and having received a high gonadal radiation dose. This resulted in the group having a range of malignancies, including Hodgkin lymphoma, testicular cancer and Wilms’ tumour. Selection criteria were the same as previously described (Curwen et al. 2005) with the exception that age at diagnosis was increased from under 20 years to under 35 years. To ensure anonymity, each family was assigned a study number (T29 – T58) and samples were further coded to avoid identification of cancer survivor, partner and offspring within each family group. A total of 115 blood samples were collected from the families and sent to the United Kingdom (UK) in eight shipments between June and December 2006. In addition, blood samples were collected at every shipment from two healthy adult Danish volunteers as controls for the transportation system. Repeat blood samples from one healthy adult in-house volunteer were obtained in parallel to the eight shipments as the internal assay control. The culture failure of T36 meant that only 29 of the Danish families were available for chromosome analysis giving a total of 111 family samples. Approval for the study was obtained from the Danish Scientific Ethical Committee and the Danish Data Protection Agency.
Sample identity was confirmed using the Applied Biosystems AmpFlSTR COfiler polymerase chain reaction (PCR) amplification kit (Applied Biosystems, Warrington, UK). The kit provides human identification by amplifying six tetranucleotide short tandem repeat loci (D3S1358, D16S539, TH01, TPOX, CSF1PO, D7S820) plus a segment of the sex-specific amelogenin locus. PCR products were analysed on a semi-automated ABI 310 genetic analyser platform (Applied Biosystems), the data files generated were then run through Genotyper 2.X (Applied Biosystems) software to automate the genotyping process. A mismatch between parent and offspring at two or more loci was considered to be a non-paternity or non-maternity. No cases of non-paternity or non-maternity were observed.
The G2 assay
The G2 assay was performed as previously described (Curwen et al. 2005) and is based on a modified version of that reported by Scott et al. (1999). Peripheral blood was drawn into lithium heparin vacutainers (Fisher Scientific, Leicestershire, UK) and cultures (2ml whole blood) were set up in pre-warmed (37°C) and pre-gassed (5% CO2/95% air) Roswell Park Memorial Institute (RPMI-1640) medium (Sigma Aldrich, Dorset, UK) supplemented with 15% foetal calf serum (Invitrogen Limited, Paisley, UK), 1% L-glutamine (Invitrogen Limited) and 1% phytohaemagglutinin (M-form) (Invitrogen Limited) to a total volume of 20ml. Each culture was set up in duplicate, one flask (VWR International, Leicestershire, UK) for the determination of the spontaneous aberration frequency and one to be irradiated for the determination of the induced aberration frequency. A single foetal calf serum batch was used for the entire period of the study. Flasks were placed upright in a humidified, CO2-gassed incubator (Binder, Tuttlingen, Germany). After 48 hours, 15ml of medium was carefully removed and replaced with 15ml of fresh pre-gassed, pre-warmed medium. At 72 hours the cells were transported in a portable incubator (M-Tech Diagnostics, Warrington, UK) at 37°C and irradiated (or sham irradiated) with 0.5 Gy, 300 kV X-rays using a Siefert Isovolt 320 X-ray set (Aegleteq Ltd, Bucks, UK). Following a 30 minute recovery time, 0.2ml colcemid (10µg/ml) (Invitrogen Limited) was added and at 90 minutes after irradiation the contents of the culture flasks were transferred to 13.5ml conical base centrifuge tubes (Sterilin Ltd, Carerphilly, UK) and placed on ice. Subsequent centrifugation, hypotonic treatment (0.075M potassium chloride (KCl)) (VWR International) and fixation (methanol:glacial acetic acid, 3:1) (VWR International) were carried out at 4°C. Fixed cells were stored at −20°C overnight, or longer, prior to slide making.
Metaphase slides were made according to standard procedures and stained with Giemsa (VWR International). Slides from the family blood samples together with those from the transport and internal assay control samples were further coded prior to cytogenetic analysis. For each irradiated sample, 100 well-spread metaphases were analysed and the total number of chromatid gaps and breaks determined to give a G2 aberration yield. Chromatid-type aberrations were scored according to previously outlined criteria (Smart et al. 2003), with breaks being defined as mis-aligned discontinuities and gaps as single aligned discontinuities wider than the width of a chromatid. Two scorers were used to reduce scorer bias, with each scorer analysing 50 metaphases per sample. Previous studies using the G2 assay in our laboratory have shown the spontaneous aberration yield to be at a minimal level (<1.0 aberration per 100 cells). On this basis, and due to the additional time required to score control samples, it was decided that it was not necessary to determine spontaneous aberration yields. However, control sample slides were available for each individual to investigate any spurious results. For comparisons with the previous study (Curwen et al. 2005), data were re-analysed using induced aberration yields from the irradiated samples only.
Statistical methods
The distributions of chromatid type aberrations amongst cells in each population (cancer survivors, partner controls, offspring, internal assay control, transport control 1 and transport control 2) were analysed for approximation to the Poisson distribution using the dispersion index test of Papworth, as previously described by Savage (1970). If the observed distributions followed Poisson statistics, the variance and the mean (number of aberrations divided by the number of metaphase cells studied) of the observed distributions would be equal and a ratio value of 1.0 would be expected. If the variance is greater than the mean, this indicates that aberrations are overdispersed and a Poisson distribution does not apply. Ratios of variance to mean for each sample provided by a donor were calculated and population ratios were determined using the average value.
Chi-squared (χ2) analysis was used to investigate intra-individual variation in the transport and internal assay controls, adopting the formula χ2 = Σ(O–E)2/E, where O is the observed value of aberrations per 100 cells and E is the expected value of total aberrations per 100 cells. Overdispersion of distributions of aberrations amongst metaphase cells was corrected for in each population by multiplying the expected value of total aberrations per 100 cells (E) in the denominator of the equation for chi-squared by a compensation factor (Z = average value of ratio of variance to mean) and so the formula χ2 = Σ(O – E)2/(EZ) was adopted. Chi-squared analysis was also used to assess inter-individual variation within each population with overdispersion taken into account as above.
In all cases, standard errors (SE) were calculated by adjusting for overdispersion of chromatid aberrations using the appropriate compensation factor (Z) equated for each population (Z = average value of ratio of variance to mean, calculated for each population). In addition, for the internal assay and transport controls (where repeat sampling had occurred), any additional intra-individual variation introduced was considered. This was estimated by adding all the values of chi-squared for those individuals who were sampled more than once and dividing by the total degrees of freedom and is termed Y. Standard errors were thus calculated according to the formula √(Number of aberrations x Z x Y), and normalised to 100 cells scored.
The non-parametric Mann-Whitney U-test was used to compare median induced aberration frequencies between controls and the other donor groups. The 90th percentile value of the aberration yield for the partner control group was determined and used as the cut-off point for determining the proportions of radiosensitive individuals in the other groups. This method was introduced by Scott et al. (1999) and, although the 90th percentile is an arbitrary value, it has resulted in good discrimination and been adopted in the majority of subsequent G2 chromosomal radiosensitivity studies. Fisher’s exact tests were used to compare the proportion of radiosensitive individuals in each of the three family groups.
Heritability was estimated using Sequential Oligogenic Linkage Analysis Routines (SOLAR) software, which is an extensive, flexible software package for genetic variance components analysis (Almasy and Blangero 1998). SOLAR assumes a normal distribution for the trait, given genotype. We also performed segregation analysis using a regressive model as implemented in the Statistical Analysis for Genetic Epidemiology software (S.A.G.E. v6.0.1) as previously described (Curwen et al. 2005). In this approach, the residuals from the genotype means are assumed to be normally distributed, after Box-Cox transformation. Different disease transmission models (parameterised by τg, the probability that a parent of genotype g transmits allele A to his/her offspring) and different models for the genotype means (unrestricted, dominant, additive) were compared by maximum likelihood and by use of the Akaike information criterion (AIC). Gender was included as a covariate although this was not statistically significant in any model. A parameter representing residual polygenic correlation between the family members (in addition to correlation induced by the putative major gene) was also included. For the most parsimonious model, we estimated the heritability as the ratio of the genetic variance (the variability of genotype means across the putative genotypes) to the total phenotypic variance.
Results
Internal assay and transport controls
Table I illustrates the characteristics and G2 aberration frequencies for the internal assay and transport control samples. The ratio of variance to mean indicated overdispersion of aberration yields compared with Poisson expectations and adjustment was made for this as described in the Methods section. The mean aberration yield for the internal assay control was 113.6 ± 3.4 per 100 cells and a coefficient of variation (CV) of 20.7% was determined for intra-individual variability. Chi-squared analysis revealed a statistically significant difference between the samples (χ26 = 18.74, P = 0.005). Analysis of the samples collected from the two individuals as controls for the transportation system also indicated significant intra-individual variation {(χ26 = 42.95, P < 0.001), (χ24 = 30.85, P < 0.001)}. Mean induced aberration yields for these individuals were 131.3 ± 5.9 and 124.8 ± 7.5, respectively. Moreover, sequential sampling of these individuals produced CV of 31.2% and 29.9%, respectively.
Table I.
Characteristics of internal assay and transport control donors and radiation-induced G2 chromatid aberration frequencies
| Aberrations per 100 cells at sampling shipment |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor | Sex | Age at first sampling (years) |
1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Ratio of variance to mean (average value) |
Mean aberration frequency ± SE per 100 cells |
CV (%) |
| Internal assay control | F | 41 | 132 | 126 | 150 | 85 | - | 97 | 111 | 94 | 1.55 | 113.6 ± 3.4 | 20.7 |
| Transport control 1 | F | 41 | 149 | 123 | - | 75 | 160 | 143 | 82 | 187 | 1.78 | 131.3 ± 5.9 | 31.2 |
| Transport control 2 | F | 56 | 148 | 152 | - | 61 | - | - | 123 | 140 | 1.45 | 124.8 ± 7.5 | 29.9 |
SE = standard error, CV = coefficient of variation, F = female, - = sample failed/not available.
Cancer survivors, partners and offspring
Details of the childhood and young adult cancer survivors, their partners and offspring are presented in Table II, together with their respective G2 aberration yields. Again the test for Poisson distribution indicated overdispersion in all groups. Results were considered as groups (cancer survivors, partner controls and offspring) with information on group characteristics being provided in Table III. Comparison of median aberration frequencies revealed no statistically significant difference between the partner control group and the cancer survivor group (P = 0.576) nor between the partner control group and the offspring group (P = 0.497). Furthermore, mean aberration frequencies for the three groups were similar.
Table II.
Individual characteristics of childhood and young adult cancer survivors, their partners and offspring and radiation-induced G2 aberration yields
| Survivor | Partner | Offspringa | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Family code |
Gender | Primary cancer | Age at time of treatment (years) |
Age at time of sampling (years) |
Aberrations per 100 cells |
Age at time of sampling (years) |
Aberrations per 100 cells |
Age at time of sampling (years) |
Aberrations per 100 cells |
| T29 | M | Hodgkin | 27 | 41 | 147 | 44 | 130 | 14 | 162 |
| 12 | 202 | ||||||||
| T30 | M | Hodgkin | 14 | 45 | 102 | 43 | 131 | 18 | 136 |
| 15 | 125 | ||||||||
| T31 | F | Hodgkin | 18 | 47 | 153 | 50 | 130 | 23 | 137 |
| 19 | 148 | ||||||||
| T32 | F | Neuroblastoma | 2 | 32 | 123 | 37 | 103 | 6 | 171 |
| T33 | F | Lymphosarcoma | 10 | 62 | 123 | 75 | 167 | 30 | 118 |
| T34 | F | Lymphocytic lymphosarcoma | 9 | 41 | 74 | 40 | 113 | 12 | 101 |
| T35 | F | Reticulosarcoma | 9 | 44 | 110 | 43 | 143 | 18 | 82 |
| 14 | 105 | ||||||||
| T37 | F | Hodgkin | 14 | 34 | 170 | 34 | 130 | 12 | 127 |
| 9 | 167 | ||||||||
| T38 | F | Hodgkin | 15 | 41 | 142 | 53 | 144 | 8 | 114 |
| 5 | 125 | ||||||||
| T39 | F | Lymphosarcoma | 10 | 61 | 95 | 62 | 95 | 34 | 98 |
| T40 | M | Lymphosarcoma | 10 | 43 | 172 | 41 | 75 | 16 | 89 |
| 11 | 110 | ||||||||
| T41 | F | Hodgkin | 19 | 61 | 114 | 63 | 122 | 41 | 66 |
| 38 | 114 | ||||||||
| T42 | M | Hodgkin | 27 | 48 | 135 | 48 | 166 | 14 | 170 |
| 12 | 167 | ||||||||
| T43 | M | Hodgkin | 29 | 41 | 127 | 38 | 86 | 2 | 86 |
| T44 | M | Reticulosarcoma | 17 | 56 | 107 | 52 | 121 | 28 | 152 |
| 28 | 155 | ||||||||
| T45 | F | Reticulosarcoma | 16 | 55 | 105 | 63 | 87 | 22 | 101 |
| T46 | F | Wilms’ | 3 | 32 | 183 | 32 | 79 | 7 | 128 |
| 1 | 160 | ||||||||
| T47 | M | Hodgkin | 31 | 43 | 98 | 42 | 101 | 8 | 101 |
| T48 | M | Hodgkin | 20 | 52 | 204 | 45 | 204 | 18 | 160 |
| 15 | 207 | ||||||||
| 10 | 203 | ||||||||
| T49 | M | Hodgkin | 29 | 47 | 187 | 40 | 167 | 11 | 178 |
| 9 | 156 | ||||||||
| T50 | M | Hodgkin | 5 | 38 | 124 | 38 | 171 | 9 | 180 |
| 2 | 197 | ||||||||
| T51 | M | Lymphosarcoma | 11 | 60 | 180 | 54 | 199 | 33 | 173 |
| 31 | 133 | ||||||||
| T52 | M | Hodgkin | 14 | 50 | 206 | 47 | 122 | 26 | 115 |
| 23 | 168 | ||||||||
| T53 | M | Testis (seminoma) | 33 | 68 | 101 | 63 | 137 | 32 | 111 |
| 29 | 108 | ||||||||
| T54 | F | Hodgkin | 16 | 54 | 133 | 57 | 141 | 29 | 124 |
| 27 | 123 | ||||||||
| 19 | 123 | ||||||||
| T55 | M | Testis (teratoma) | 25 | 61 | 113 | 59 | 154 | 33 | 116 |
| T56 | M | Testis (seminoma) | 28 | 55 | 134 | 55 | 118 | 24 | 122 |
| 22 | 118 | ||||||||
| 20 | 125 | ||||||||
| 16 | 147 | ||||||||
| T57 | M | Testis (teratocarcinoma) | 30 | 58 | 165 | 57 | 129 | 24 | 151 |
| T58 | M | Testis (seminoma) | 25 | 52 | 152 | 59 | 137 | 22 | 144 |
| 15 | 185 | ||||||||
| Ratio of variance to mean (average value for each population) | 1.73 | 1.78 | 1.62 | ||||||
M = male; F = female.
Offspring born after cancer treatment of survivor in birth order.
Table III.
Characteristics of childhood and young adult cancer survivor, partner and offspring groups and radiation-induced G2 chromatid aberration frequencies
| Group | Number of individuals |
Male/female | Median age in years (range) |
Median aberration frequency per 100 cells (range) |
P1 | Mean aberration frequency ± SE per 100 cells |
Sensitive response |
||
|---|---|---|---|---|---|---|---|---|---|
| Number of individuals | % | P2 | |||||||
| Cancer survivors | 29 | 17/12 | 48 (32–68) |
133.0 (74–206) |
0.576 | 137.2 ± 2.9 | 7 | 24.1 | 0.297 |
| Partners | 29 | 12/17 | 48 (32–75) |
130.0 (75–204) |
- | 131.1 ± 2.8 | 3 | 10.3 | - |
| Offspring | 53 | 29/24 | 18 (1–41) |
128.0 (66–207) |
0.497 | 137.4 ± 2.1 | 11 | 20.8 | 0.358 |
Significance value obtained when using the non-parametric Mann-Whitney U-test to compare the median induced aberration frequency of the partner controls group with those of the cancer survivor and offspring groups, respectively.
Significance value obtained when using Fisher’s exact tests to compare the number of individuals displaying a radiosensitive response in the partner controls group with those of the cancer survivor and offspring groups, respectively.
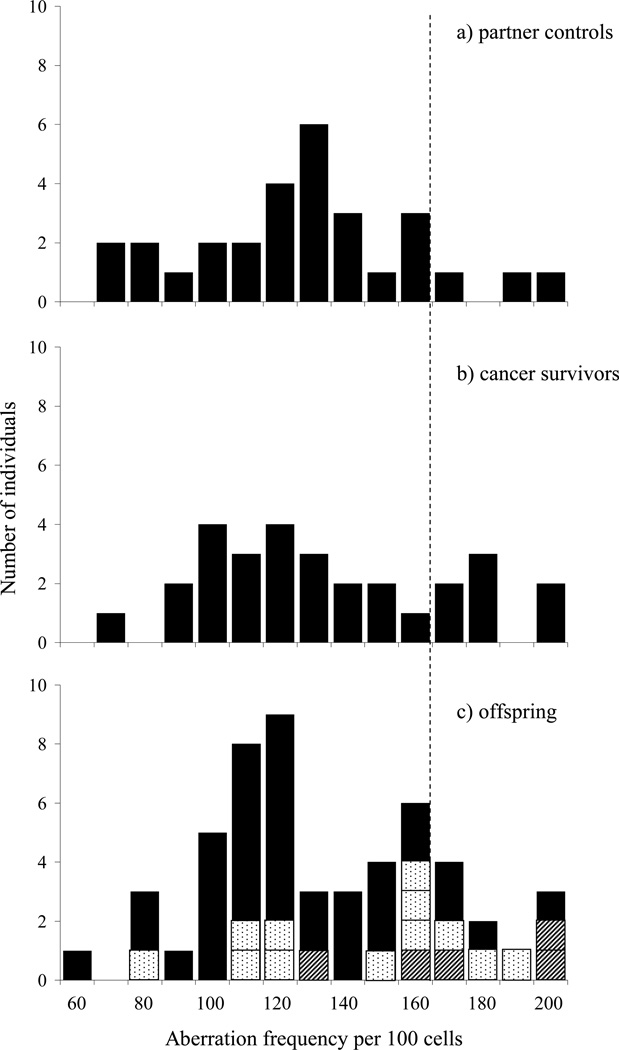
Distributions of the G2 aberration frequencies for the three groups are illustrated in Figure 1. The cut-off point for a radiosensitive/non-radiosensitive response, as determined by the 90th percentile for the partner control group, was 167.8 aberrations per 100 cells and resulted in 24.1, 10.3 and 20.8% of the cancer survivors, partners and offspring displaying an enhanced radiosensitive response, respectively (Table III). An illustration of parentage is provided with offspring being identified on the basis of 0, 1 or 2 parents who had radiosensitivity scores greater than the 90th percentile of the partner group (Figure 1). Whilst the proportion of individuals displaying elevated sensitivity in both the cancer survivor and offspring groups was greater than that of the partner control group, neither reached statistical significance when using the partner group for comparison (P = 0.297, P = 0.358, respectively).
Figure 1.
Radiation-induced G2 chromatid aberration frequencies. The dotted vertical line represents the cut-off point for a radiosensitive/non-radiosensitive response (167.8 aberrations per 100 cells) as determined by the 90th percentile of the partner control group: a) partners, b) survivors of childhood and young adult cancer, c) offspring with 

 parents displaying elevated radiosensitivity.
parents displaying elevated radiosensitivity.
Heritability analysis
Analysis of the induced G2 aberration frequency data from the 29 families, performed using the SOLAR software v4.2.0, estimated heritability to be 0.780 ± 0.12 (P < 0.001). Table IV provides details of heritability of the radiosensitive phenotype as investigated by segregation analysis. There was borderline evidence to reject the non-genetic sporadic model in comparison with the general transmission model using a likelihood ratio test (P = 0.070), indicating the general model fitted the data significantly better than the sporadic model. The Mendelian model was not rejected when compared to the general transmission model (P = 0.436), indicating the general model did not show a significantly better fit to the data than the Mendelian model. In addition, the Mendelian model had the lowest Akaike information criterion (AIC) value compared to both the general and sporadic models. This suggested that the data were better fit by the Mendelian model, which should be accepted over both the general and sporadic models. Based on the AIC criterion, further comparison of the Mendelian model (AIC 1071.51) with an autosomal dominant model (AIC 1074.01) and an additive model (AIC 1073.77) also suggested the unrestricted Mendelian model provided the best fit for the data. The most parsimonious model was a Mendelian model with no gender covariate (AIC 1069.64). With genotype frequencies of 0.044, 0.332, 0.623 and genotype means of 131.00, 169.96 and 117.94 respectively (Table IV), the genetic variance attributable to the putative major locus under this model becomes 587.54, with a residual variance of 426.75, resulting in a heritability estimate of 587.54 / (587.54 + 426.75) = 0.579, slightly lower than the estimate provided by SOLAR.
Table IV.
Model parameters from segregation analysis of G2 chromosomal radiosensitivity in 29 Danish families of survivors of childhood and young adult cancer
| Parameter | Models analysed |
|||
|---|---|---|---|---|
| General | Equal τ (sporadic) |
Mendelian | Mendelian (no gender effect) |
|
| QA | 0.499 ± 0.083 | 0.217 ± 0.208 | 0.210 ± 0.055 | 0.210 ± 0.055 |
| ψAA | 0.249 ± 0.082 | 0.047 ± 0.090 | 0.044 ± 0.023 | 0.044 ± 0.023 |
| ψAB | 0.499 ± 0.000 | 0.340 ± 0.234 | 0.332 ± 0.064 | 0.332 ± 0.064 |
| ψBB | 0.250 ± 0.083 | 0.611 ± 0.325 | 0.623 ± 0.087 | 0.623 ± 0.087 |
| τAA | 0.852 ± 0.120 | QA | 1.000 | 1.000 |
| τAB | 0.000 | QA | 0.500 | 0.500 |
| τBB | 1.000 | QA | 0.000 | 0.000 |
| βAA | 167.29 ± 7.18 | 151.99 ± 53.89 | 131.19 ± 12.46 | 131.00 ± 12.53 |
| βAB | 118.35 ± 5.53 | 151.99 ± 19.10 | 169.87 ± 6.03 | 169.96 ± 6.00 |
| βBB | 124.26 ± 8.26 | 120.17 ± 9.12 | 117.89 ± 4.05 | 117.94 ± 4.07 |
| Residual variance | 575.12 ± 123.21 | 750.20 ± 220.17 | 425.72 ± 96.65 | 426.75 ± 97.32 |
| λ (Box-Cox parameter) | 0.90 ± 0.36 | 0.54 ± 0.49 | 1.12 ± 0.35 | 1.13 ± 0.36 |
| Residual ρpo=ρss | 0.539 ± 0.053 | 0.510 ± 0.068 | 0.386 ± 0.104 | 0.388 ± 0.104 |
| Gender covariance | 2.54 ± 4.58 | −1.96 ± 5.34 | −1.54 ± 4.36 | - |
| −2 ln L | 1053.89 | 1060.96 | 1055.51 | 1055.64 |
| AIC | 1071.89 | 1076.96 | 1071.51 | 1069.64 |
| P – value | - | 0.0701 | 0.4361 | - |
A = dominant allele; B = recessive allele; QA = frequency of dominant allele; ψ = genotype frequencies; τ = transmission probabilities; β = genotype dependent mean (induced aberration frequency per 100 cells); ρpo = correlation between parent of offspring pairs, ρss = correlation between siblings; AIC = Akaike information criterion
likelihood ratio test compared with the general model
Discussion
The G2 chromosomal radiosensitivity assay requires stringent technical conditions to produce reproducible results. Problems associated with blood sample storage and transport conditions, in particular transit over long distances, have been reported (Scott et al. 1996, Scott et al. 1999, Bryant et al. 2002). In contrast, our laboratory and others have found blood storage conditions to have no effect on aberration yield (Scott et al. 1996, Scott et al. 1999, Bryant et al. 2002, Smart et al. 2003). Additionally, transportation over short distances has not been found to have an effect on reproducibility (Roberts et al. 1999, Riches et al. 2001, Smart et al. 2003). High standards of intrinsic assay reproducibility have previously been demonstrated in our laboratory (Smart et al. 2003) as well as by others using the technique (Vral et al. 2002).
The same internal control individual was used in this and the previous study (Curwen et al. 2005). However, whilst in the previous study repeated sampling indicated no significant intra-individual variability and the CV was 13.6%, repeated sampling of the same healthy adult donor throughout the present study period revealed significant variation in aberration yield and a CV of 20.7%. Nevertheless, the mean aberration yield of 113.6 ± 3.4 per 100 cells was similar to that of 108.0 ± 2.4 per 100 cells derived from the previous study data using the revised scoring criteria. Samples collected from two donors to monitor the transportation system showed even greater intra-individual variation with CV of 31.2% and 29.9%, respectively. Moreover, the mean aberration frequency for transport control 1 of 131.3 ± 5.9 per 100 cells was significantly greater than that of 88.9 ± 2.4 per 100 cells for the same individual recalculated from the previous study (P = 0.036, two-tailed student’s t-test). Intra-individual variability was also reported for this transport control in the previous study, although it was speculated that this could be attributable to hormonal influences associated with pregnancy (Curwen et al. 2005). It is of interest that the mean aberration frequencies of all three controls were less than the radiosensitive cut-off point of 167.8 per 100 cells determined from the partner data, although one of the seven samples from transport control 1 did have a higher value. The samples were sent from Denmark at ambient temperature, by road and by air and in the possession of a personal courier at all times, to ensure that they were not subjected to significant temperature fluctuations and thus minimise any potential transportation effect. The three repeatedly sampled controls showed no common pattern of high or low G2 scores associated with specific sampling times, nor with high or low scores determined for cancer family samples taken and processed at the same time (data not shown), suggesting that factors not intrinsic to the G2 assay, such as varying characteristics of the individual, are more likely than assay variability to be responsible for the intra-individual variation. Changes in hormone levels have been reported to influence in vitro radiosensitivity (Roberts et al. 1997, Ricoul et al. 1998), although this has been disputed in more recent studies (Baeyens et al. 2005). Nevertheless, since all three controls were female the influence of fluctuating hormonal levels cannot be ruled out.
Whilst some studies of G2 chromosomal radiosensitivity have reported no significant intra-individual variability with CV in the range of 7.0 – 10.3% (Roberts et al. 1999, Scott et al. 1999, Baria et al. 2001, Papworth et al. 2001), a number of studies have experienced problems with high intra-individual variability, and some laboratories have reported levels of variation for some individuals not significantly different to inter-individual variability (Baria et al. 2002, Vral et al. 2002, Smart et al. 2003, Vral et al. 2004, Howe et al. 2005). This has led to suggestions that the G2 assay is unlikely to detect any real differences in chromosomal radiosensitivity between individuals within a normal population (Vral et al. 2002, Smart et al. 2003, Vral et al. 2004, Howe et al. 2005).
Analysis of the three family groups revealed a greater proportion of radiosensitive individuals in the cancer survivor (24.1%) and offspring (20.8%) groups than in the partner control group (10.3%) but no comparisons reached statistical significance. Similarly, no statistically significant differences in median or mean aberration yields were found. In our previous study of a group of 23 childhood cancer survivors, their partners and their 38 offspring, no association between G2 chromosomal radiosensitivity and predisposition to cancer was observed when using the 90th percentile value of the partner control group as the cut-off point for a radiosensitive/non-radiosensitive response. However, comparisons with an in-house control group did suggest heightened radiosensitivity profiles in the cancer survivor and offspring groups respectively (Curwen et al. 2005). A re-analysis of the previous data using the revised scoring criteria (i.e. induced score without subtraction of the spontaneous score) gave a slightly different radiosensitive cut-off point for the partners of 162.4 aberrations per 100 cells, and this is similar to that of 167.8 aberrations per 100 cells obtained in the current study. However, the current study does show a distinction in the radiosensitivity profiles between the three groups, with both cancer survivor and offspring groups demonstrating higher proportions of radiosensitive individuals than the partner control group, although these increases do not reach statistical significance.
In addition to sample size limitations, the failure to find a clear-cut relationship between G2 chromosomal radiosensitivity and cancer predisposition may reflect, in part, the nature of the malignancies selected for study. Whilst early-onset cancer is often associated with inherited susceptibility, in this study there is a preponderance of haematological disorders, which may have a stronger environmental than heritable aetiology (Hemminki and Bermejo 2005, Hemminki et al. 2005). Nevertheless, a hereditary component to non-Hodgkin and Hodgkin lymphoma has been suggested within twin and family studies (Schottenfeld and Fraumeni 2006). Although this familial aggregation may relate to immune system functionality, a role for deoxyribonucleic acid (DNA) repair mechanisms seems possible at least for Hodgkin lymphoma (Schottenfeld and Fraumeni 2006) and it is noted that six of the 22 cases of haematological cancers (27.3%) did exhibit enhanced radiosensitivity. Specifically, in the current study four out of 14 survivors of Hodgkin lymphoma exhibited enhanced radiosensitivity compared to four out of ten in the previous study (Curwen et al. 2005) and three out of six reported by Baria et al. (2002). Only one out of seven cases of non-haematological cancers in the current study exhibited elevated radiosensitivity.
Direct support for the suggestion that chromosomal radiosensitivity has a heritable component has come from studies on breast cancer patients and their families (Roberts et al. 1999, Scott 2000). First-degree relatives of radiosensitive breast cancer patients were found to be more radiosensitive compared with first-degree relatives of breast cancer patients with normal responses to radiation. An indication that the trait has a heritable component can be seen in the distribution of offspring radiosensitivity scores in relation to the radiosensitivity status of their parents, using the 90th percentile of the partner distribution to distinguish those with an elevated response (Figure 1). However, although the cut-off point of the 90th percentile is an easy visual way to illustrate and test the differences in radiosensitivity between different groups, it has no real bearing on the heritability analysis which examines different models for goodness of fit to the data using the G2 assay (aberration frequency per 100 cells) as a quantitative trait outcome. Segregation analysis suggested that a model with a single major gene, with two alleles combining in an additive manner to give three phenotypes, could account for 82.0% of the variability in G2 sensitivity. Heritability of the phenotype was also demonstrated in our previous study of childhood and adolescent cancer survivors and their families (Curwen et al. 2005). In that study, 67.3% of the variance of G2 chromosomal radiosensitivity could be attributed to a major gene locus with dominant effect. This was confirmed in the SOLAR analysis which indicated a value of 60.7%.
Heritability analysis in the current study performed using both the SOLAR software and segregation analysis indicated that the G2 chromosomal radiosensitive phenotype was a heritable trait, with SOLAR suggesting strong heritability. SOLAR analysis suggested an estimate of heritability of 78.0% and segregation analysis 57.9%. However, in the current study, segregation analysis indicated that the unrestricted Mendelian model (with no gender effect) was the most parsimonious, unlike in the previous study (Curwen et al. 2005) for which the most parsimonious model was an autosomal dominant model with no residual correlation (and no gender effect). Given the complexity of segregation analysis and the large number of parameters that are estimated (using a relatively small number of individuals with relatively few different relationship types), one should beware of over-interpreting these results. However, the results are certainly consistent with an underlying genetic basis for this phenotype.
Conclusion
In conclusion, this study has provided suggestive, albeit not conclusive, evidence of an association between chromosomal radiosensitivity and cancer predisposition. The failure to demonstrate a statistically significant difference in radiosensitive profiles between cancer survivors and their partners may reflect in part the types of malignancy studied and/or sample size limitations. The finding of significant intra-individual variation in the three repeatedly sampled controls is a concern and indicates that reliance cannot be placed on a single individual result. Nevertheless, the study provides further evidence for heritability of the G2 chromosomal radiosensitive phenotype and suggests that future studies should focus on specific cancer types which are most likely to have a fault in DNA damage recognition and repair mechanisms.
Acknowledgements
We thank the Danish families for agreeing to participate in this study and volunteering blood samples, Brian Møllgren, Rigshospitalet, for collection of blood samples from the families and the transport controls, Patricia Jonas, Newcastle NHS Trust, for collection of blood samples from the in-house assay control. Permissions were granted from the Danish Data Protection Agency (2001-41-1113) and the Danish Scientific Ethical Committee ([KF] 01 – 150/01, [KF] 11 – 129/02 and [H-KF] 01 – 150/01). This work was funded by a grant from the National Institutes of Health, USA. (Grant Number 1 RO1 CA 104666) through Vanderbilt University Medical Center, USA. We are grateful for the support provided by the Danish Cancer Society. Some of the results of this paper were obtained by using the program package S.A.G.E., which is supported by a US Public Health Service Resource Grant (RR03655) from the National Center for Research Resources.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American Journal of Human Genetics. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens A, Thierens H, Claes K, Poppe B, Messiaen L, De Ridder L, Vral A. Chromosomal radiosensitivity in breast cancer patients with a known or putative genetic predisposition. British Journal of Cancer. 2002;87:1379–1385. doi: 10.1038/sj.bjc.6600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens A, Vandersickel V, Thierens H, Ridder LD, Vral A. Effects of estradiol and progesterone on the variability of the micronucleus assay. Mutation Research. 2005;578:308–316. doi: 10.1016/j.mrfmmm.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Baria K, Warren C, Roberts SA, West CM, Scott D. Chromosomal radiosensitivity as a marker of predisposition to common cancers? British Journal of Cancer. 2001;84:892–896. doi: 10.1054/bjoc.2000.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baria K, Warren C, Eden OB, Roberts SA, West CM, Scott D. Chromosomal radiosensitivity in young cancer patients: possible evidence of genetic predisposition. International Journal of Radiation Biology. 2002;78:341–346. doi: 10.1080/09553000110117359. [DOI] [PubMed] [Google Scholar]
- Boice JD, Jr, Tawn EJ, Winther JF, Donaldson SS, Green DM, Mertens AC, Mulvihill JJ, Olsen JH, Robison LL, Stovall M. Genetic effects of radiotherapy for childhood cancer. Health Physics. 2003;85:65–80. doi: 10.1097/00004032-200307000-00013. [DOI] [PubMed] [Google Scholar]
- Bryant PE, Gray L, Riches AC, Steel CM, Finnon P, Howe O, Kesterton I, Vral A, Curwen GB, Smart V, Tawn EJ, Whitehouse CA. The G2 chromosomal radiosensitivity assay. International Journal of Radiation Biology. 2002;78:863–866. doi: 10.1080/09553000210144484. [DOI] [PubMed] [Google Scholar]
- Curwen GB, Winther JF, Tawn EJ, Smart V, Whitehouse CA, Rees GS, Olsen JH, Guldberg P, Rechnitzer C, Schroder H, Bryant PE, Sheng X, Lee HS, Chakraborty R, Boice JD. G2 chromosomal radiosensitivity in Danish survivors of childhood and adolescent cancer and their offspring. British Journal of Cancer. 2005;93:1038–1045. doi: 10.1038/sj.bjc.6602807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ruyck K, de Gelder V, Van Eijkeren M, Boterberg T, De Neve W, Vral A, Thierens H. Chromosomal radiosensitivity in head and neck cancer patients: evidence for genetic predisposition? British Journal of Cancer. 2008;98:1723–1738. doi: 10.1038/sj.bjc.6604345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K, Bermejo JL. Relationships between familial risks of cancer and the effects of heritable genes and their SNP variants. Mutation Research. 2005;592:6–17. doi: 10.1016/j.mrfmmm.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Forsti A, Lorenzo Bermejo J. Single nucleotide polymorphisms (SNPs) are inherited from parents and they measure heritable events. Journal of Carcinogenesis. 2005;4:2. doi: 10.1186/1477-3163-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe O, Daly P, Seymour C, Ormiston W, Nolan C, Mothersill C. Elevated G2 chromosomal radiosensitivity in Irish breast cancer patients: a comparison with other studies. International Journal of Radiation Biology. 2005;81:373–378. doi: 10.1080/09553000500147642. [DOI] [PubMed] [Google Scholar]
- Lisowska H, Lankoff A, Wieczorek A, Florek A, Kuszewski T, Gozdz S, Wojcik A. Enhanced chromosomal radiosensitivity in peripheral blood lymphocytes of larynx cancer patients. International Journal of Radiation Oncology Biology Physics. 2006;66:1245–1252. doi: 10.1016/j.ijrobp.2006.07.1370. [DOI] [PubMed] [Google Scholar]
- Papworth R, Slevin N, Roberts SA, Scott D. Sensitivity to radiation-induced chromosome damage may be a marker of genetic predisposition in young head and neck cancer patients. British Journal of Cancer. 2001;84:776–782. doi: 10.1054/bjoc.2000.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches AC, Bryant PE, Steel CM, Gleig A, Robertson AJ, Preece PE, Thompson AM. Chromosomal radiosensitivity in G2-phase lymphocytes identifies breast cancer patients with distinctive tumour characteristics. British Journal of Cancer. 2001;85:1157–1161. doi: 10.1054/bjoc.2001.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricoul M, Lebeau J, Sabatier L, Dutrillaux B. Increased radiation-induced chromosome breakage after progesterone addition at the G1/S-phase transition. Mutation Research. 1998;403:177–183. doi: 10.1016/s0027-5107(98)00078-5. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Morgan GR, Danford N. Effect of hormones on the variation of radiosensitivity in females as measured by induction of chromosomal aberrations. Environmental Health Perspectives. 1997;105:1467–1471. doi: 10.1289/ehp.97105s61467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Spreadborough AR, Bulman B, Barber JB, Evans DG, Scott D. Heritability of cellular radiosensitivity: a marker of low-penetrance predisposition genes in breast cancer? American Journal of Human Genetics. 1999;65:784–794. doi: 10.1086/302544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S.A.G.E. Statistical Analysis for Genetic Epidemiology. 2009 Release 6.0.1: http://darwin.cwru.edu/
- Savage JRK. Sites of radiation induced chromosome exchanges. Current Topics in Radiation Research. 1970;6:129–194. [Google Scholar]
- Schottenfeld D, Fraumeni JF., Jr. Cancer epidemiology and prevention. 3rd ed. UK: Oxford University Press; 2006. [Google Scholar]
- Scott D, Spreadborough A, Levine E, Roberts SA. Genetic predisposition in breast cancer. Lancet. 1994;344:1444. doi: 10.1016/s0140-6736(94)90615-7. [DOI] [PubMed] [Google Scholar]
- Scott D, Spreadborough AR, Jones LA, Roberts SA, Moore CJ. Chromosomal radiosensitivity in G2-phase lymphocytes as an indicator of cancer predisposition. Radiation Research. 1996;145:3–16. [PubMed] [Google Scholar]
- Scott D, Barber JB, Spreadborough AR, Burrill W, Roberts SA. Increased chromosomal radiosensitivity in breast cancer patients: a comparison of two assays. International Journal of Radiation Biology. 1999;75:1–10. doi: 10.1080/095530099140744. [DOI] [PubMed] [Google Scholar]
- Scott D. Chromosomal radiosensitivity, cancer predisposition and response to radiotherapy. Strahlentherapie Onkologie. 2000;176:229–234. doi: 10.1007/s000660050005. [DOI] [PubMed] [Google Scholar]
- Scott D. Chromosomal radiosensitivity and low penetrance predisposition to cancer. Cytogenetic and Genome Research. 2004;104:365–370. doi: 10.1159/000077517. [DOI] [PubMed] [Google Scholar]
- Smart V, Curwen GB, Whitehouse CA, Edwards A, Tawn EJ. Chromosomal radiosensitivity: a study of the chromosomal G2 assay in human blood lymphocytes indicating significant inter-individual variability. Mutation Research. 2003;528:105–110. doi: 10.1016/s0027-5107(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Vral A, Thierens H, Baeyens A, De Ridder L. The micronucleus and G2-phase assays for human blood lymphocytes as biomarkers of individual sensitivity to ionizing radiation: limitations imposed by intraindividual variability. Radiation Research. 2002;157:472–477. doi: 10.1667/0033-7587(2002)157[0472:tmagpa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Vral A, Thierens H, Baeyens A, De Ridder L. Chromosomal aberrations and in vitro radiosensitivity: intra-individual versus inter-individual variability. Toxicology Letters. 2004;149:345–352. doi: 10.1016/j.toxlet.2003.12.044. [DOI] [PubMed] [Google Scholar]