Abstract
Purpose
To identify metabolic pathways that are perturbed in pancreatic ductal adenocarcinoma (PDAC), we investigated gene-metabolite networks with integration of metabolomics and transcriptomics.
Experimental design
We have performed global metabolite profiling analysis on two independent cohorts of resected PDAC cases to identify critical metabolites alteration that may contribute to the progression of pancreatic cancer. We then searched for gene surrogates that were significantly correlated with the key metabolites by integrating metabolite and gene expression profiles.
Results
55 metabolites were consistently altered in tumors as compared with adjacent nontumor tissues in a test cohort (N=33) and an independent validation cohort (N=31). Weighted network analysis revealed a unique set of free fatty acids (FFAs) that were highly co-regulated and decreased in PDAC. Pathway analysis of 157 differentially expressed gene surrogates revealed a significantly altered lipid metabolism network, including key lipolytic enzymes PNLIP, CLPS, PNLIPRP1, and PNLIPRP2. Gene expressions of these lipases were significantly decreased in pancreatic tumors as compared with nontumor tissues, leading to reduced FFAs. More importantly, a lower gene expression of PNLIP in tumors was associated with poorer survival in two independent cohorts. We further demonstrated that two saturated FFAs, palmitate and stearate significantly induced TRAIL expression, triggered apoptosis, and inhibited proliferation in pancreatic cancer cells.
Conclusions
Our results suggest that impairment in a lipolytic pathway involving lipases and a unique set of FFAs, may play an important role in the development and progression of pancreatic cancer and provide potential targets for therapeutic intervention.
Keywords: Pancreatic ductal adenocarcinoma, Metabolic Profiling, Integrative Analysis, Free Fatty Acids, Pancreatic Lipase
Introduction
Pancreatic cancer is the fourth leading cause of cancer death in the United States with an estimated 44, 920 new cases and 37, 390 deaths in 2013 (1). Pancreatic cancer cases and deaths have been on the rise since 1998. The median survival of all PDAC cases is less than 6 months, and only 6% of patients survive 5 years after diagnosis. The mortality rate has not improved significantly, due to late diagnosis and resistance to available chemotherapy. Therefore, a better understanding of molecular mechanisms of disease progression and discovery of novel therapeutic targets are desperately needed to improve outcomes in patients with PDAC.
Genetic alterations have been extensively characterized in pancreatic cancer. We and others have previously identified gene markers of PDAC which have prognostic and therapeutic significance (2, 3). However the impact of those gene alterations on metabolism and gene-metabolite networks in PDAC has not been clearly defined. Several oncogenes and tumour suppressors such as P53 and c-Myc, control the activity of different metabolic pathways to support the metabolic transformation of a cancer cell (4). A growing body of evidence demonstrated the strong connection between cancer and metabolism, including the discovery that some key metabolic enzymes such as succinate dehydrogenase, fumarate hydratase, isocitrate dehydrogenase and phosphoglycerate dehydrogenase, if mutated, could lead to different forms of cancer (5). The development of pancreatic cancer has also been linked to abnormal glucose metabolism which is induced by long-term type 2 diabetes, a known risk factor for pancreatic cancer (6). Metabolites are central in intermediary metabolism, and they provide substrates for biological processes, and have an active role in regulating cell cycle, proliferation and apoptosis (7). Recently, there has been an increased interest in global analysis of metabolites for cancer biomarker discovery and identification of potential novel therapeutic targets. New technologies applying chromatography-mass spectrometry (MS) provides sensitive and reproducible detection of hundreds to thousands of metabolites in a single biofluid or tissue sample, and allows non-targeted high-throughput metabolic profiling analysis (8). Furthermore, integration of comprehensive gene expression profile with metabolic profiling has been shown to be an innovative way to reveal the complex regulatory networks involving genes and metabolic pathways in cancers (9, 10).
In the present study, we have performed global metabolite profiling analysis in two independent cohorts of PDAC cases to identify critical metabolite alterations that may contribute to the progression of pancreatic cancer. We then searched for gene surrogates that were significantly correlated with the key metabolites using transcriptomic profiling data of the same samples in the test cohort from our previous study (2). The integrative analysis of metabolomics and transcriptomic data revealed a lipid metabolism network involving 4 lipases and a unique set of FFAs that may play an important role in pancreatic tumor progression and could provide potential targets for therapeutic intervention.
Materials and Methods
Tissue Collection
Primary pancreatic tumor and adjacent nontumor tissues were collected from patients with PDAC at the University of Medicine, Göttingen, Germany, and from the University of Maryland Medical Center at Baltimore, Maryland through the NCI-UMD resource contract. Tissues were flash frozen immediately after surgery. Demographic and clinical information for each tissue donor, including age, sex, clinical staging, resection margin status, survival times from diagnosis, and receipt of adjuvant chemotherapy were collected. Tumor histopathology was classified according to the World Health Organization Classification of Tumor system(11). Use of these clinical specimens was reviewed and approved by the NCI-Office of the Human Subject Research (OHSR, Exempt # 4678) at the National Institutes of Health, Bethesda, MD.
Metabolic Profiling of PDAC
Metabolic profiling of PDAC samples (tumor and adjacent nontumor tissues) was carried out at Metabolon Inc. using the general protocol as outlined earlier (12, 13) (see more details in the supplementary file). Metabolon analytical platform incorporates two separate ultra-high performance liquid chromatography/tandem mass spectrometry (UHPLC/MS/MS2) injections and one gas chromatography GC/MS injection per sample. The UHPLC injections are optimized for basic species and acidic species. This integrated platform enabled the high-throughput collection and relative quantitative analysis of analytical data and identified a large number and broad spectrum of molecules with a high degree of confidence (12). A total of 469 known metabolites were measured.
Weighted Coexpression Network Analysis
Weighted coexpression network analysis (WGCNA) has been implemented in R, a free and open source statistical programming language (14). We followed the protocols of WGCNA to create metabolite networks. Briefly, for each metabolite profiling data set, Pearson correlation coefficients were calculated for all pairwise comparisons of metabolites across all tumor samples. The resulting Pearson correlation matrix was transformed into an adjacency matrix using a power function, which resulted in a weighted network (14). WGCNA defines modules as a group of densely interconnected molecules with high topological overlap in weighted network analysis. For each data set, we used average linkage hierarchical clustering with a dynamic tree-cutting algorithm to identify modules on the basis of the topological overlap dissimilarity measure (15).
Ingenuity Pathways Analysis
Canonical pathway analysis identified the pathways from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the data set. The association between the data set and the canonical pathway was measured in 2 ways: (i) The statistical significance: Fischer's exact test was used to calculate a p-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone; (ii) A ratio of the number of genes from the data set that map to the given pathway divided by the total number of genes in that canonical pathway (16).
RNA Isolation and Transcriptome Profiling
RNA from frozen tissue samples was extracted using standard TRIZOL (Invitrogen) protocol. RNA quality was confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies) before the microarray gene expression profiling. Tumors and paired non-tumor tissues from the Germany cohort were profiled using the Affymetrix GeneChip Human 1.0 ST arrays according to the manufacturer's protocol at the LMT microarray core facility at National Cancer Institute, Frederick, MD. All arrays were RMA normalized and gene expression summaries were created for each gene by averaging all probe sets for each gene using Partek Genomics Suite 6.5. All data analysis was performed on gene summarized data. The microarray gene expression data has been deposited in the National Center for Biotechnology Information's (NCBIs) Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) with accession number GSE28735.
Quantitative RT-PCR (qRT-PCR)
Total RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR reactions in 384 well plates were performed using Taqman Gene Expression Assays on an ABI Prism 7900HT Sequence Detection instrument from Applied Biosystems. Expression levels of GAPDH were used as the endogenous controls. All assays were performed in triplicate. For quality control, any sample with a gene cycle value greater than 36 was considered of poor quality and removed. All the primers for qRT-PCR in the present study were purchased from Applied Biosystems (Table S1).
Cell lines and culture conditions
Human pancreatic carcinoma cell lines hTERT-HPNE (CRL-4023™), MIApaca2 (CRL-1420™) and Panc1 (CRL-1469™) were obtained from American Type Culture Collection ATCC (Rockville, MD, USA). Cells were maintained in GIBCO® RPMI Media 1640 supplemented with GlutaMAX™-I (Invitrogen), penicillin-streptomycin (50 IU/ml and 50 mg/ml, respectively), and 10% (v/v) fetal calf serum (FCS). Fatty acids were purchased from Sigma (MO, USA). The stock solutions of fatty acids bound to BSA were prepared as described earlier (17). A 5% FFA-free BSA solution was prepared in H2O and dissolved for 30mins at 55°C in an a water bath. The appropriate amount of 50 mM FFA stock solution was added to BSA solution and incubated for 8 hours at 37 °C under nitrogen atmosphere to prevent oxidization. The FFA/BSA stock solution was then cooled to 25 °C, filter sterilized, and stored at −20 °C.
MTT assay
Cells were seeded in 96-well plates (3,000–5000 cells/well) and incubated for 2–10 days. Then, the MTT solution was added and incubated for 4 hours. The solution was aspirated and 100ul DMSO was added to each well. The absorbance was measured at 570nm and 650 nm.
Apoptosis Assay
Panc1 and MIApaca2 cells were seeded and incubated for 24 h in standard medium. After 12 h of serum starvation, cells were incubated in serum-free medium with BSA-bound fatty acids or BSA control for 12 and 24 hours. Caspase activity was measured by Apo-ONE™ Homogeneous Caspase-3/7 Assay (Promega, Madison, WI) and potency of caspase activation was calculated compared to control cells.
Quantification of TRAIL protein level by Enzyme-Linked Immunosorbent Assays (ELISA)
TRAIL protein level was directly quantified in pancreatic cancer cells treated with 0.1 μM FFA or BSA for 24 hours. Cells were lysed (5 × 105cells/ 100ul lysate buffer) and TRAIL production in cell lysates was measured using Quantikine Human TRAIL/TNFSF10 ELISA KIT (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Optical density of each well was then determined using a microplate reader set to 450 nm. TRAIL concentrations were calculated using a standard curve and linear regression analysis.
Statistical Analysis
A student's T-test was used to compare metabolite or gene expression between tumors and non-tumor tissues using Graphpad Prism 5.0 (Graphpad Software Inc, San Diego, California). Correlation analysis and Kaplan-Meier analysis were performed with Graphpad Prism 5.0. Fisher's exact test, correlation analysis and Cox Proportional-hazards regression analysis were performed using Stata 11 (StataCorp LP, College Station, Texas). Univariate Cox regression was performed on genes and clinical covariates to examine the influence of each on patient survival. For these analyses, resection margin status was dichotomized as positive (R1) vs negative (R0); TNM staging was dichotomized based on non-metastatic (I–IIA) vs metastatic (IIB–IV) disease; histological grade was dichotomized based on well and moderately differentiated (G1&2) vs poorly differentiated (G3&4). All Cox regression models were tested for proportional hazards assumptions based on Schoenfeld residuals, and no model violated these assumptions. The statistical significance was defined as P<0.05. All P values reported were 2- sided.
Results
Metabolomics of PDAC
The characteristics of the patients with PDAC in a test cohort (N=33) and a validation cohort (N=31) are shown in Table S1. The two cohorts were similar in TNM staging, resection margin status, grade and cancer-specific mortality (P=0.76, Kaplan-Meier log rank) with 1-year survival rate of 50.8% for the test cohort and 51% for the validation cohort.
Metabolite profiling was performed using liquid and gas chromatography coupled with mass spectrometry to identify and statistically compare the relative metabolite expression levels between tumor and nun-tumor tissues from PDAC cases. We identified 55 metabolite that were differentially expressed in tumors as compared to nontumor tissues (P<0.01) in both test and validation cohorts (Table S2).
Weighted network analysis identified a unique set of fatty acids that are co-regulated in PDAC
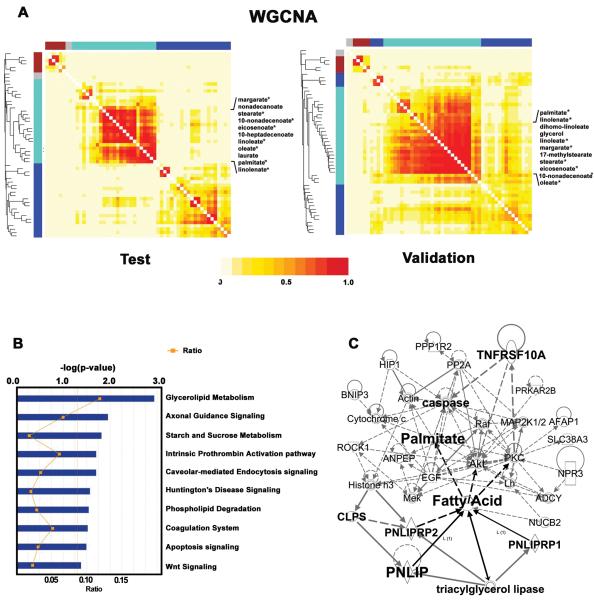
We constructed coexpression networks using the 55 metabolite profiling data in 2 independent cohorts. According to recently described methodology (14), the connectivity (k) was determined for all metabolites in the network by taking the sum of their connection strengths (coexpression similarity) with all other nodes in the network. To identify modules of highly co-regulated metabolites, we used average linkage hierarchical clustering to group metabolites based on the topological overlap of their connectivity (see Materials and Methods for details). WGCNA identified 3 modules of highly connected metabolites. Each module was assigned a unique color identifier (Figure 1A), with the remaining, poorly connected metabolites colored gray. In a topological overlap matrix (TOM) plot, the increasing color intensity indicates higher connectivity among metabolites in the network (Figure 1A). Metabolites with the greatest connectivity index represent network “hubs” and are localized in the center of individual modules. Highly connected and correlated hub metabolites are often sharing common pathways and tightly co-regulated within the same metabolism networks (18).
Figure 1. Integration of metabolomics and transcriptomics revealed altered lipid metabolism pathways in PDAC.
(A) Weighted network analysis showed that the turquoise-colored module was highly conserved in both test and validation cohort. 8 fatty acids (indicated by stars) with high connectivity (IMconn > 6, Table 1) in both cohorts represent the main hubs in the metabolite network that were chosen for integration analysis. (B) Canonical pathways analysis identified 10 pathways from the Ingenuity Pathways Analysis library of canonical pathways that were most significant (P<0.05) to the data set of 157 surrogate genes for the unique set of 8 fatty acids in PDAC. Glycerolipid metabolism was the most significantly enriched pathway. (C) IPA analysis revealed a lipid metabolism network involving 4 lipases and fatty acids highlighted here, that may potentially regulate and interact with apoptosis signaling pathways. Lipolytic genes PNLIP, CLPS, PNLIRP1 and PNLIPRP2 from the surrogate gene data set, play central roles in lipolysis.
Because of the indicated importance of hubs in the network, we ranked metabolites within turquoise module (Table S3) based on their intramodular connectivity to identify module hubs (Table 1). Eight metabolites with high connectivity (IMconn > 6) in the turquoise module represented the main hubs in both test and validation cohorts. Interestingly, this set of highly co-regulated hub metabolites are all free fatty acids, suggesting that fatty acid metabolism may be significantly altered in PDAC.
Table 1.
A set of co-regulated fatty acids are identified using WGCNA on two independent cohorts.
| Test Cohort | Validation Cohort | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Metabolite ID | P-value* | Ratio† | IMconn† | P-value* | Ratio† | IMconn‡ |
| linolenate (18:3n3 or 6) | 2.3E-04 | 0.53 | 6.91 | 1.9E-06 | 0.42 | 9.83 |
| palmitate (16:0) | 4.2E-04 | 0.53 | 6.00 | 1.1E-04 | 0.48 | 6.60 |
| margarate (17:0) | 1.0E-03 | 0.56 | 6.79 | 5.5E-07 | 0.40 | 13.24 |
| stearate (18:0) | 1.5E-03 | 0.58 | 6.85 | 1.3E-06 | 0.41 | 13.50 |
| linoleate (18:2n6) | 1.9E-03 | 0.58 | 6.79 | 4.6E-06 | 0.43 | 12.20 |
| oleate (18:1n9) | 3.1E-03 | 0.59 | 7.86 | 4.3E-06 | 0.43 | 13.62 |
| eicosenoate (20:1n9 or 11) | 3.9E-03 | 0.59 | 6.43 | 6.7E-05 | 0.47 | 12.22 |
| 10-nonadecenoate (19:1n9) | 4.1E-03 | 0.59 | 7.50 | 2.6E-05 | 0.45 | 12.12 |
P-value calculated using t-test in each cohort.
Ratio of tumor vs. non-tumor.
Intramodular connectivity represents the strength of coexpression for each metabolite in network analysis
Integration of metabolomics and transcriptomics revealed altered lipid metabolism pathway in PDAC
In order to define genetic alterations and the molecular pathways associated with 8 fatty acids identified from WGCNA, we first searched for gene surrogates that were significantly correlated with these 8 fatty acids within the same 33 samples in test cohort. Using transcriptomic profiling data from our previous study (2), we performed Pearson correlation analysis between gene expression and metabolite profile data of 8 fatty acids. These analyses identified 157 gene surrogates that were highly correlated with the set of 8 fatty acids (Pearson correlation P<0.01) and differentially expressed between tumors and nontumors (T-test, FDR-corrected P<0.01, |fold change|>1.5, Table S4).
Furthermore, Ingenuity Pathways Analysis demonstrated that 157 gene surrogates of 8 fatty acids were highly enriched for glycerolipid metabolism, axonal guidance signaling, starch and sucrose metabolism, intrinsic prothrombin activation pathway and other pathways. The glycerolipid metabolism pathway is the top hit with Fisher's exact test p-value of 0.001 representing the most significant pathway that is associated with our dataset (Figure 1B). Significant enrichments were also observed for surrogate genes associated with apoptosis and Wnt signaling, which are known to play critical roles in tumorigenesis. IPA's network analysis further identified interacting modules involved in lipid metabolism and apoptosis signaling networks, which include fatty acids and their surrogate genes PNLIP (pancreatic lipase), CLPS (colipase, pancreatic), PNLIPRP1 (pancreatic lipase-related protein 1) and PNLIPRP2 (pancreatic lipase-related protein 2) (Figure1C).
Lypolytic enzymes PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 are significantly decreased in PDAC
It should be noted that surrogate genes PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 which encode key lipolytic enzymes playing central roles in glycerolipid metabolism, were decreased in tumors as compared with adjacent nontumor tissues in the test cohort (Figure 2A), which is consistent with the down-regulation of FFAs in pancreatic tumors (Table 1).
Figure 2. Gene expressions of PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 in two independent cohorts of PDAC.
(A) PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 are decreased in tumors as compared with adjacent non-tumor tissues in test and validation cohorts. Dot plots represent gene expression level with relative intensity (log2) of microarray data in the test cohort or relative threshold cycle value (Ct) normalized with endogenous control gene GAPDH using qRT-PCR data in validation cohort. Bars indicate median value. Student t-tests P value and tumor: non-tumor ratios (T:N) are indicated in the graphs. (B) Kaplan Meier analysis of PNLIP in test and validation cohorts. Gene expression of PNLIP is dichotomized into high and low groups using a median cutoff. Log-rank P value is indicated in the graphs.
To validate the association of these lipases with the set of 8 fatty acids identified in metabolic profiling, we then used the validation cohort of PDAC cases (N= 31) to examine the gene expression of PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 by qRT-PCR. Our data confirmed that the expression of all 4 lipases were decreased in tumors, as compared with surrounding nontumor tissues in two independent cohorts (Figure 2), and consistently all 8 fatty acids were also decreased in tumors from both cohorts (Table 2). Particularly, qRT-PCR data in the validation cohort showed ~100- to 1000-fold decrease in the gene expression of these lipases in pancreatic tumors. Our data also showed that gene expression of PNLIP, CLPS, PNLIPRP1 and PNLIPR2 are lower in PDAC cell lines Panc1 and MIApaca2, as compared to non-tumorigenic hTERT-HPNE cells (Figure S1). These data are consistent with other publically available transcriptional profiling data in Oncomine database (Table S5), suggesting that the gene expression of these lipases are potential diagnostic markers for PDAC.
Table 2.
Univariate Cox regression analysis on test and validation cohorts.
| Test cohort | Validation cohort | |||
|---|---|---|---|---|
|
|
||||
| Variables (comparison/referent) | HR (95% CI) | P | HR (95% CI) | P |
| PNLIP (high/low)* | 0.36 (0.15–0.88) | 0.023 | 0.36 (0.15–0.87) | 0.020 |
| CLPS (high/low)* | 0.34 (0.14–0.84) | 0.027 | 0.46 (0.20–1.09) | 0.077 |
| PNLIPRP1 (high/low)* | 0.37 (0.15–0.89) | 0.028 | 0.56 (0.24–1.31) | 0.182 |
| PNLIPRP2 (high/low)* | 0.35 (0.14–0.85) | 0.020 | 0.50 (0.20–1.21) | 0.122 |
| Grading (G3&4/1&2) | 1.94 (0.89–4.26) | 0.097 | 1.11 (0.47–2.62) | 0.819 |
| Resection Margin (R1/R0) | 1.18 (0.54–2.58) | 0.677 | 0.80 (0.32–1.96) | 0.622 |
| Tumor stage (IIB-IV/I-IIA) | 1.07 (0.54–2.14) | 0.844 | 0.95 (0.39–2.32) | 0.914 |
Gene expression value was dichotomized into high and low groups using median. P-value was calculated using univariate cox regression analysis.
Pancreatic lipases directly hydrolyse triglycerolipids into fatty acid and glycerol, therefore regulating fatty acid turnover and signaling (19). In both test and validation cohorts, gene expressions of these lipases are significantly correlated with the set of 8 FFAs (Table S6), indicating that markedly decreased expression of PNLIP, CLPS, PNLIPRP1 and PNLIPRP2 may have a role in reduced fatty acid level in pancreatic tumors.
PNLIP is a predictor of cancer-specific mortality in PDAC
There's no significant association between 8 fatty acids and cancer-specific mortality in both cohorts of PDAC cases (data not shown). We then tested whether the 4 lipolytic genes were associated with patient outcome. The association of gene expression with cancer-specific mortality was evaluated in both test and validation cohort using Cox regression analysis, in which we dichotomized high and low gene expression as values above and below the median. In our study, univariate Cox regression analysis (Table 2) for all cases showed that only PNLIP, was associated with prognosis in both test cohort (hazard ratio (HR), 0.36; 95% CI, 0.15–0.88; P=0.023) and validation cohort (HR, 0.36; 95% CI, 0.15–0.87; P=0.02). Therefore, surrogate gene PNLIP maybe a prognostic marker for pancreatic cancer.
Palmitate and stearate inhibits cell growth in vitro
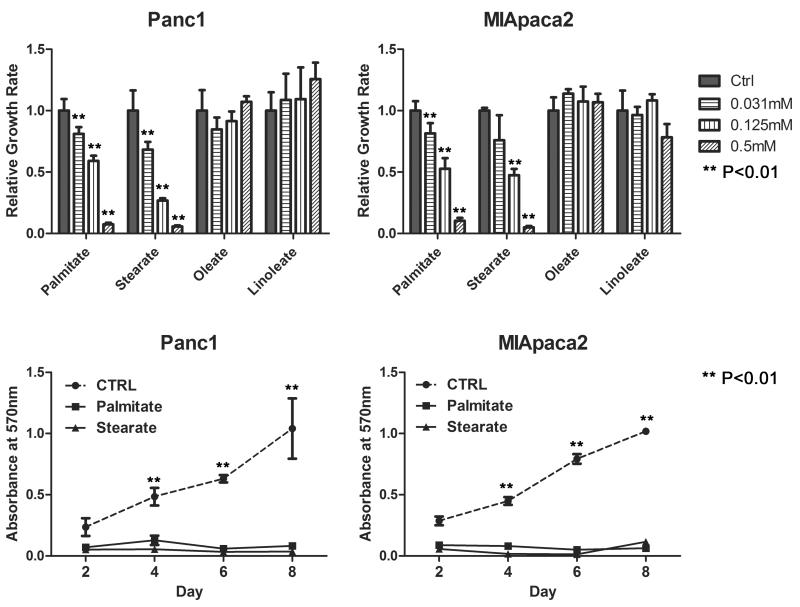
IPA Canonical Pathway analysis identified an enriched set of interactions between the lipid metabolism and the apoptosis signaling pathway (Figure1B & C), leading to the hypothesis that fatty acids may regulate apoptosis and cell growth of pancreatic tumors. Therefore, we studied the effects of this unique set of FFAs on the proliferation of human pancreatic cancer cell lines Panc1 and MIApaca2. Palmitate, stearate, linoleate and oleate were chosen because they are the most abundant fatty acids in animals (17). MTT cell proliferation assays showed that palmitate and stearate significantly inhibit Panc1 and MIApaca2 cell growth in a dose-dependent manner. In contrast, linoleate and oleate had little effect on cell proliferation in both cells (Figure 3A). Cell counting and BrdU assays in addition to MTT assay, also showed significant growth inhibitory effect of palmitate and stearate on Panc1 and MIApaca2 cells (Figure S2). To better assess the growth-inhibitory effect of palmitate and stearate, cell growth curve was generated by incubating Panc1 and MIApaca2 cells in the media containing either BSA alone (as a control) or 0.25mM FFAs over a 8-day period (Figure 3B). Our data showed that palmitate and stearate substantially inhibit the growth of pancreatic cancer cells.
Figure 3. Palmitate and Stearate inhibit pancreatic cancer cell proliferation.
(A) Dose dependence of palmitate and stearate on inhibition of cell proliferation in Panc1 and MIApaca2 cells. After 16 hours of starvation, cells are treated with BSA control or BSA-bound FFAs for 72 hours. Saturated fatty acids palmitate and stearate show significant inhibition of cell proliferation at the concentration of 0.031–0.5mM. Unsaturated fatty acids linolate and oleate have little or no effect on cell proliferation. (B) There are significant decreases in cell growth of Panc1 and MIApaca2 treated with 0.25mM palmitate and stearate as compared with control cells over a 8-day period. Data are presented as means ± S.D. from 3 independent experiments. ** t-test P <0.01, ANOVA P<0.01.
Palmitate and stearate induce TRAIL expression and promote apoptosis in pancreatic cancer cells
To determine whether palmitate and stearate could affect apoptosis, we examined the caspase-3 activity in Panc1 and MIApaca2 cells following treatment of FFAs for 24 hours. Our data showed that palmitate and stearate (0.25mM) significantly increased caspase-3/7 activity by 2- to 3- fold as compared to controls in both cell lines (P<0.01, Figure 4A). To elucidate the potential underlying mechanisms of apoptosis regulation by palmitate and stearate, we then analyzed the gene expression of apoptosis related genes in response to FFA treatment using qRT-PCR and found that palmitate and stearate significantly induced the expression of the pro-apoptotic gene TRAIL by 2- to 3- fold in pancreatic cancer cell lines as compared to controls (P<0.01, Figure 4B). Consistent with the gene expression, the protein level of TRAIL was also increased by about 3- to 4- fold following 24 hour incubation with 0.25mM palmitate or stearate. Taken together, these results show that palmitate and stearate induce TRAIL expression and trigger apoptosis in pancreatic cancer cells.
Figure 4. Palmitate and stearate induce TRAIL expression and promote apoptosis.
(A) There are significant increases in caspase-3/7 activity in Panc1 and MIApaca2 cells with palmitate and stearate treatment. Relative Caspase3/7 activity represents the effect of FFAs on apoptosis compared to control cells after 24h incubation. (B) Palmitate and stearate upregulate TRAIL expression. Cell lysates were collected after 24h of incubation with 0.25mM FFAs or BSA control. Real-time PCR was performed to determine TRAIL mRNA levels. TRAIL protein level in cell lysates was measured using an ELISA kit (R&D Systems) according to the manufacturer's instructions. Data represent means ± S.D. from 3 independent experiments. ** t-test P <0.01, ANOVA P<0.01.
Discussion
Altered metabolism is considered as one of the hallmarks of cancer (20). Genetic alterations enable cancer cells to reprogram metabolism to meet increased energy demands for cell proliferation and to survive in hypoxic and nutrient-deprived tumor microenvironment (21). In this regard, a better understanding of metabolic dysregulation in pancreatic cancer could lead to the discovery of novel therapeutic targets (22). Integrative post-genomic studies and systems biology approaches have emerged with the aim of developing a more comprehensive understanding of cellular physiology and metabolism (23, 24). To the best of our knowledge, here we report for the first time the implementation of a systems biology approach to investigate gene-metabolite networks and metabolic dysregulation in pancreatic cancer, with integration of metabolomics and transcriptomics.
Metabolomics allow for global assessment of a cellular state within the context of the immediate environment, taking into account genetic regulation, altered kinetic activity of enzymes, and changes in metabolic reactions (25). Thus, compared with transcriptomics or proteomics, metabolomics reflects changes in phenotype and therefore cellular function (26). Metabolomics strategies have been applied to tissues, serum and other body fluid, to develop novel early diagnostic biomarkers in human cancers (27–30).
In this study, we identified 55 differentially expressed metabolites using metabolic profiling in two independent cohorts of PDAC cases. Next, we applied WGCNA to analyze metabolic networks in PDAC. WGCNA is a systems biology–based network analysis that has been demonstrated to be an important alternative and more meaningful tool for discovery of molecular interaction networks and candidate biomarkers (31–35). Using WGCNA, we have identified 8 highly connected fatty acid hubs in a conserved lipid module, which are decreased in tumors as compared to adjacent non-tumor tissues in two independent cohorts of PDAC. Further data mining revealed significant involvement of these fatty acids in cell proliferation and tumorigenesis (36). Free fatty acids play an important role in numerous biological functions. They serve as a source of energy and as precursors of many signaling and cellular components. The effect of different types of FFAs on cell proliferation and apoptotic activity in pancreatic cancer remains unclear. In this study, we showed for the first time that two major saturated FFAs, palmitate and stearate, exert a strong growth inhibitory effect in pancreatic cancer cells. Our data also showed that palmitate and stearate significantly induce apoptosis in pancreatic cancer cells. These findings are consistent with previous reports on induction of apoptosis by palmitate in other cell types, including breast cancer cells (37), hematopoietic cells (38), pancreatic β-cells (39), and cardiomyocytes (40). In contrast, many studies reported contradictory findings with respect to the role of unsaturated fatty acids in tumor growth, particularly for oleic acid, in breast cancer cells and tumor xenograft models (41). Our functional investigation in different pancreatic cancer cell lines showed that unsaturated FFAs linoleate and oleate have no significant effect on the proliferation of pancreatic cancer cells.
It has been proposed that excess palmitate could induce cell death through increased intracellular concentration of ceramide (38, 39), a metabolite exclusively produced from saturated FFAs. However, other studies suggested that apoptosis induced by palmitate could occur through the generation of reactive oxygen species (ROS) rather than ceramide synthesis (40). FFA-induced production of mitochondrial ROS is linked to the activation of protein kinase C (PKC) and the redox sensitive transcription factor NF-κB (42), which might be involved in the regulation of apoptosis (43). However, the biochemical pathways by which fatty acids influence pancreatic cancer cell growth and death have not been adequately defined. Our data show that palmitate and stearate can upregulate TRAIL expression in pancreatic cancer cells, which may contribute to the apoptosis induced by these two fatty acids.
To further define the genetic alterations and molecular pathways associated with this unique set of fatty acids identified by metabolic profiling, we integrated transcriptomics and metabolomics, and identified 157 gene surrogates for the fatty acid set that is associated with PDAC. Pathway and network analysis revealed that the expected lipid metabolism, particularly in lipolytic pathway involving gene surrogates PNLIP, CLPS, PNLIPRP1 and PNLIPRP2, is significantly altered in PDAC (Figure1B and 1C). Pancreatic lipase, also known as pancreatic triacylglycerol lipase, is encoded by PNLIP, and secreted by the pancreas, and is the primary lipase that hydrolyzes lipids, converting triglyceride substrates to monoglycerides and free fatty acids (44). Unlike some pancreatic enzymes that are activated by proteolytic cleavage, pancreatic lipase is secreted in its final form. Colipase encoded by the CLPS gene, is a protein co-enzyme required for optimal enzyme activity of pancreatic lipase (45). PNLIPRP1 and PNLIPRP2 code for two novel human pancreatic lipase-related proteins in pancreatic juice, referred to as PNLIP-related proteins 1 and 2, each showing an amino acid sequence identity of 68% to PNLIP. PNLIPRP2 shows a lipolytic activity that is only marginally dependent on the presence of colipase, whereas the function of PNLIPRP1 remains unclear (44). Overall, these functionally related lipases play key roles in direct regulation of fatty acid turnover and signaling. Therefore, decreased expressions of lipases eventually lead to reduced levels of free fatty acids. Consistent with the function of lipases, our data showed positive correlations between FFA levels and gene expression of these lipases in tumor samples, indicating that a profound dysregulation of the lipolytic network exists in PDAC and may play an important role in tumor growth (Figure 1C).
Excessive production of pancreatic lipase may indicate the presence of certain disorders, most notably inflammation of the pancreas, or pancreatitis (46). Elevated levels of pancreatic lipases also occur in bowel obstruction or kidney disease (47, 48). On the other hand, individuals with Crohn's disease, cystic fibrosis, and celiac disease suffer from lipase deficiency, in which the cells of the pancreas responsible for producing this enzyme may be irreversibly damaged (49). The most common symptoms associated with lipase deficiency are, muscle spasms, acne, arthritis, gallbladder stress and formation of gallstones, bladder problems and cystitis. Therefore, pancreatic lipase supplements are used to treat PNLIP deficiency diseases (49). However, the role of PNLIP in pancreatic cancer remains unknown. In this study, we have shown striking decreases (>100 fold) in the gene expression of all four lipases including PNLIP in pancreatic tumors, and consistently immunohistochemical staining on paraffin sections also showed that PNLIP protein level is lower in pancreatic ductal carcinomas as compare to normal ducts (Figure S3). In addition, our study also provided the first evidence that a lower expression of PNLIP is associated with poor outcome in PDAC.
In summary, we have used a systems biology approach and identified an altered lipolytic network involving lipase genes and a unique set of fatty acids that are associated with pancreatic cancer. This approach moves beyond single gene investigation to provide a systems level perspective on the potential relationships among members of a biological network in pancreatic cancer. Our results suggest that the impaired lipolytic pathway may contribute to the development and progression of PDAC. In conclusion, our study showed significant decrease in fatty acids and their surrogate lipase genes in PDAC, and demonstrated tumor inhibitory roles of palmitate and stearate in pancreatic cancer. Furthermore, to the best of our knowledge, this is the first report indicating the potential prognostic significance of PNLIP in pancreatic cancer. Further studies are needed to determine the mechanism underlying the altered expression of these lipase genes, and to explore their potential clinical application in PDAC. These studies will lead to a better understanding of the aggressiveness of pancreatic cancer and may facilitate therapeutic target discovery.
Supplementary Material
Translational Relevance.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies worldwide. Altered metabolism is considered as one of the hallmarks of cancer. Therefore, a better understanding of metabolic dysregulation in pancreatic cancer could lead to the discovery of novel therapeutic targets. In the present study, we investigated metabolic dysregulation and gene-metabolite networks in pancreatic cancer, with integration of metabolomics and transcriptomics. Our results suggest that the impaired lipolytic network involving 4 lipases and a unique set of fatty acids may play an important role in the development and progression of pancreatic cancer. Furthermore, our study demonstrated that integration of omics data provide a systems level perspective of pancreatic cancer that could facilitate the development of novel treatments for this disease.
Acknowledgements
We would like to thank Dr. Xinwei Wang and Dr. Stefan Ambs (National Cancer Institute) for helpful discussions. We also thank Dr. Matthias Gaida for histopathological evaluation of pancreatic tissue samples. Handling of clinical samples and maintenance of the frozen tissue database by Ms. Elise Bowman is greatly appreciated. We thank Drs.Jeff Pfohl and Ryan Michalek (Metabolon) for helpful discussions. We would also like to thank the personnel at UMMS for their contribution to the collection of clinical samples under NCI-UMD contract.
Grant support: Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH.
Footnotes
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Zhang G, Schetter A, He P, Funamizu N, Gaedcke J, Ghadimi BM, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e31507. doi: 10.1371/journal.pone.0031507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeh JJ. Prognostic signature for pancreatic cancer: are we close? Future Oncol. 2009;5:313–21. doi: 10.2217/fon.09.12. [DOI] [PubMed] [Google Scholar]
- 4.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–61. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 5.Dang CV. Links between metabolism and cancer. Genes Dev. 26:877–90. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F, Herrington M, Larsson J, Permert J. The relationship between diabetes and pancreatic cancer. Mol Cancer. 2003;2:4. doi: 10.1186/1476-4598-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 6:1060–83. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 9.Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:10205–10. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, Wenner BR, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton SR, Aaltonen LA. Pathology and genetics of tumours of the digestive system. In: Hamilton SR, Aaltonen LA, editors. WHO Classification of tumors. Vol. 204. IARC Press; Lyon, France: 2000. [Google Scholar]
- 12.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 13.Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 144:1066–75. e1. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4(Article17) doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–20. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 16.Connor SC, Hansen MK, Corner A, Smith RF, Ryan TE. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Molecular bioSystems. 2010;6:909–21. doi: 10.1039/b914182k. [DOI] [PubMed] [Google Scholar]
- 17.Hardy S, Langelier Y, Prentki M. Oleate activates phosphatidylinositol 3-kinase and promotes proliferation and reduces apoptosis of MDA-MB-231 breast cancer cells, whereas palmitate has opposite effects. Cancer Res. 2000;60:6353–8. [PubMed] [Google Scholar]
- 18.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 19.Mead JF. Lipid Metabolism. Annual review of biochemistry. 1963;32:241–68. doi: 10.1146/annurev.bi.32.070163.001325. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Le A, Rajeshkumar NV, Maitra A, Dang CV. Conceptual framework for cutting the pancreatic cancer fuel supply. Clin Cancer Res. 2012;18:4285–90. doi: 10.1158/1078-0432.CCR-12-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson RU, Shaw RJ. Cancer metabolism: Tumour friend or foe. Nature. 2012;485:590–1. doi: 10.1038/485590a. [DOI] [PubMed] [Google Scholar]
- 23.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 24.Tomita M, Kami K. Cancer. Systems biology, metabolomics, and cancer metabolism. Science. 2012;336:990–1. doi: 10.1126/science.1223066. [DOI] [PubMed] [Google Scholar]
- 25.Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40. doi: 10.1158/1078-0432.CCR-08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan TW, Lane AN, Higashi RM. The promise of metabolomics in cancer molecular therapeutics. Current opinion in molecular therapeutics. 2004;6:584–92. [PubMed] [Google Scholar]
- 27.Claudino WM, Quattrone A, Biganzoli L, Pestrin M, Bertini I, Di Leo A. Metabolomics: available results, current research projects in breast cancer, and future applications. J Clin Oncol. 2007;25:2840–6. doi: 10.1200/JCO.2006.09.7550. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Tso VK, Slupsky CM, Fedorak RN. Metabolomics and detection of colorectal cancer in humans: a systematic review. Future Oncol. 2010;6:1395–406. doi: 10.2217/fon.10.107. [DOI] [PubMed] [Google Scholar]
- 29.Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, et al. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev. 2010;20:140–7. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]
- 30.Clyne M. Kidney cancer: Metabolomics for targeted therapy. Nature reviews Urology. 2012;9:355. doi: 10.1038/nrurol.2012.127. [DOI] [PubMed] [Google Scholar]
- 31.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–4. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–7. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci U S A. 2006;103:12741–6. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritz V, Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene. 2010;29:4369–77. doi: 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy S, El-Assaad W, Przybytkowski E, Joly E, Prentki M, Langelier Y. Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin. J Biol Chem. 2003;278:31861–70. doi: 10.1074/jbc.M300190200. [DOI] [PubMed] [Google Scholar]
- 38.Paumen MB, Ishida Y, Muramatsu M, Yamamoto M, Honjo T. Inhibition of carnitine palmitoyltransferase I augments sphingolipid synthesis and palmitate-induced apoptosis. J Biol Chem. 1997;272:3324–9. doi: 10.1074/jbc.272.6.3324. [DOI] [PubMed] [Google Scholar]
- 39.Shimabukuro M, Zhou YT, Levi M, Unger RH. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc Natl Acad Sci U S A. 1998;95:2498–502. doi: 10.1073/pnas.95.5.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Listenberger LL, Ory DS, Schaffer JE. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J Biol Chem. 2001;276:14890–5. doi: 10.1074/jbc.M010286200. [DOI] [PubMed] [Google Scholar]
- 41.Fay MP, Freedman LS, Clifford CK, Midthune DN. Effect of different types and amounts of fat on the development of mammary tumors in rodents: a review. Cancer Res. 1997;57:3979–88. [PubMed] [Google Scholar]
- 42.Evans JL, Goldfine ID, Maddux BA, Grodsky GM. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes. 2003;52:1–8. doi: 10.2337/diabetes.52.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Romeo G, Liu WH, Asnaghi V, Kern TS, Lorenzi M. Activation of nuclear factor-kappaB induced by diabetes and high glucose regulates a proapoptotic program in retinal pericytes. Diabetes. 2002;51:2241–8. doi: 10.2337/diabetes.51.7.2241. [DOI] [PubMed] [Google Scholar]
- 44.Reboul E, Berton A, Moussa M, Kreuzer C, Crenon I, Borel P. Pancreatic lipase and pancreatic lipase-related protein 2, but not pancreatic lipase-related protein 1, hydrolyze retinyl palmitate in physiological conditions. Biochim Biophys Acta. 2006;1761:4–10. doi: 10.1016/j.bbalip.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 45.van Tilbeurgh H, Bezzine S, Cambillau C, Verger R, Carriere F. Colipase: structure and interaction with pancreatic lipase. Biochim Biophys Acta. 1999;1441:173–84. doi: 10.1016/s1388-1981(99)00149-3. [DOI] [PubMed] [Google Scholar]
- 46.Yadav D, Ng B, Saul M, Kennard ED. Relationship of serum pancreatic enzyme testing trends with the diagnosis of acute pancreatitis. Pancreas. 2011;40:383–9. doi: 10.1097/MPA.0b013e3182062970. [DOI] [PubMed] [Google Scholar]
- 47.Brooks S, Phelan MP, Chand B, Hatem S. Markedly elevated lipase as a clue to diagnosis of small bowel obstruction after gastric bypass. The American journal of emergency medicine. 2009;27:1167, e5–7. doi: 10.1016/j.ajem.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Ozkok A, Elcioglu OC, Cukadar T, Bakan A, Sasak G, Atilgan KG, et al. Low serum pancreatic enzyme levels predict mortality and are associated with malnutrition-inflammation-atherosclerosis syndrome in patients with chronic kidney disease. Int Urol Nephrol. 2012 doi: 10.1007/s11255-012-0237-6. [DOI] [PubMed] [Google Scholar]
- 49.Dominguez-Munoz JE. Pancreatic enzyme therapy for pancreatic exocrine insufficiency. Curr Gastroenterol Rep. 2007;9:116–22. doi: 10.1007/s11894-007-0005-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.