Conspectus
NMR crystallography – the synergistic combination of X-ray diffraction, solid-state NMR spectroscopy, and computational chemistry – offers unprecedented insight into three-dimensional, chemically-detailed structure. From its initial role in refining diffraction data of organic and inorganic solids, NMR crystallography is now being developed for application to active sites in biomolecules, where it reveals chemically-rich detail concerning the interactions between enzyme site residues and the reacting substrate that is not achievable when X-ray, NMR, or computational methodologies are applied in isolation. For example, typical X-ray crystal structures (1.5 to 2.5 Å resolution) of enzyme-bound intermediates identify possible hydrogen-bonding interactions between site residues and substrate, but do not directly identify the protonation state of either. Solid-state NMR can provide chemical shifts for selected atoms of enzyme-substrate complexes, but without a larger structural framework in which to interpret them, only empirical correlations with local chemical structure are possible. Ab initio calculations and molecular mechanics can build models for enzymatic processes, but rely on chemical details that must be specified. Together, however, X-ray diffraction, solid-state NMR spectroscopy, and computational chemistry can provide consistent and testable models for structure and function of enzyme active sites: X-ray crystallography provides a coarse framework upon which models of the active site can be developed using computational chemistry; these models can be distinguished by comparison of their calculated NMR chemical shifts with the results of solid-state NMR spectroscopy experiments. Conceptually, each technique is a puzzle piece offering a generous view of the big picture. Only when correctly pieced together, however, can they reveal the big picture at highest resolution. In this Account, we detail our first steps in the development of NMR crystallography for application to enzyme catalysis. We begin with a brief introduction to NMR crystallography and then define the process that we have employed to probe the active site in the β-subunit of tryptophan synthase with unprecedented atomic-level resolution. This approach has resulted in a novel structural hypothesis for the protonation state of the quinonoid intermediate in tryptophan synthase and its surprising role in directing the next step in the catalysis of L-Trp formation.
Introduction
In the context of NMR spectroscopy, the term “crystallography” is used in a broad sense to mean the study of crystalline solids and the arrangement of atoms in crystals. Clearly, NMR has been engaged in crystallography since the earliest days of solid-state NMR. Still, the term “NMR crystallography” has only recently been adopted and is typically reserved for studies that are performed in conjunction with diffraction methods or where lattice parameters or crystal symmetry are explicitly determined.1–10 Several recent reviews provide excellent introductions to this subject and highlight the ways in which NMR spectroscopy and X-ray diffraction methods are complementary.11 In particular, X-ray diffraction methods are unmatched in their ability to determine framework structures for systems that have long-range crystalline order, while NMR spectroscopy is unmatched in its ability to determine local chemical structure. Together, the two can provide chemically-detailed, three-dimensional structures. The sophistication with which NMR and diffraction methods have been combined took a remarkable leap forward when they were paired with ab initio computational chemistry. In one of the first examples of this, Facelli and Grant1 demonstrated that structural models based upon the X-ray crystal structure framework could be distinguished by the comparison of experimental chemical shift tensor components with those predicted for competing structural models developed using ab initio computational chemistry. This work laid the foundation for the development of NMR crystallography in organic and inorganic molecular crystals, and it was suggested this approach might also be attractive for the study of active sites in biomolecules.1 Indeed, the atomic resolution of NMR crystallography allows changes in protonation and hybridization states during enzymatic transformations to be directly observed, and in this Account we detail our recent work on the development and application of NMR crystallography to a functioning enzyme active site.2
The field of enzymology has long held as its primary scientific objective the explanation of enzyme catalysis at a chemical and structural level. Over the past 40–50 years, great advances in the understanding of the relationship between protein structure and biological function have been achieved by a combination of bioorganic mechanistic studies, the determination of protein structures at near atomic resolution, and the modification of protein structure using the tools of molecular biology. What has become clear is that much of the chemistry of enzyme catalysis is based on simple organic chemical interactions consisting, for example, of Brønsted acid-base catalysis, Lewis acid catalysis, and nucleophilic and electrophilic catalysis. Yet it remains unclear how the enormous rate accelerations achieved by enzyme active sites occur within the context of transition-state kinetic theory.
To move our understanding to the next level, a more detailed knowledge of the chemical microenvironment of the catalytic site is needed. High-resolution X-ray crystal structures provide part of the answer by identifying the protein residues and cofactors that interact with the reacting substrate. However, the resolution of most protein structures (1.5 to 2.5 Å) generally is insufficient to determine the protonation states of the acid-base functional groups. Thus investigators usually infer protonation states from hydrogen-bonding patterns and intuition based on the aqueous solution pKa values of small molecule models. NMR spectroscopy can provide another part of the answer, as the interactions of chemical shift (both isotropic and anisotropic), dipolar coupling, and quadrupolar coupling are extremely sensitive probes of the chemical microenvironment and can distinguish, for example, direct protonation of an ionizable group or the change in hybridization of a reacting substrate.12 Yet, interpretations of NMR properties such as chemical shifts are often limited to the chemical state of the reporter atom, even though the shift is sensitive in the next significant figure to the surrounding chemical microenvironment. This makes enzyme active sites a compelling target for NMR crystallography; computational chemistry provides the high-resolution link between specific models of the chemical structure, built upon the coarse X-ray framework, and observable spectroscopic parameters.
In this Account, we highlight our first steps in the development of NMR crystallography for the determination of chemically-rich crystal structures in the active sites of enzymes. By chemically-rich, we mean that the three-dimensional location of all atoms, including protons, are specified. We develop a protocol for the interrogation of the active sites of enzymes by NMR crystallography, following as an example our recent determination of the chemically-rich crystal structure for the indoline quinonoid intermediate in the β-site reaction of S. typhimurium tryptophan synthase.2 We start by placing the X-ray crystal structure of the indoline quinonoid intermediate within the larger structural and mechanistic context of catalysis in tryptophan synthase. We discuss the strategy for obtaining 13C and 15N chemical shifts of key sites within the enzyme-bound intermediate under conditions of active catalysis and their implications for the chemical structure of the intermediate. Finally, we detail the strategy to determine the atomic-resolution, three-dimensional structure of the active site using computational models that are consistent with both the X-ray and NMR data and the novel structural and mechanistic hypothesis that results.
Framework Structure: X-Ray Crystallography
NMR crystallography requires a structural framework as a starting point for building, refining, and testing three-dimensional chemical models of the enzyme active site. Here we review the progression of crystal structures in tryptophan synthase and the pivotal role these advances have played in understanding mechanism and allostery, and how these advances motivate the use of even higher-resolution, mechanistic probes.
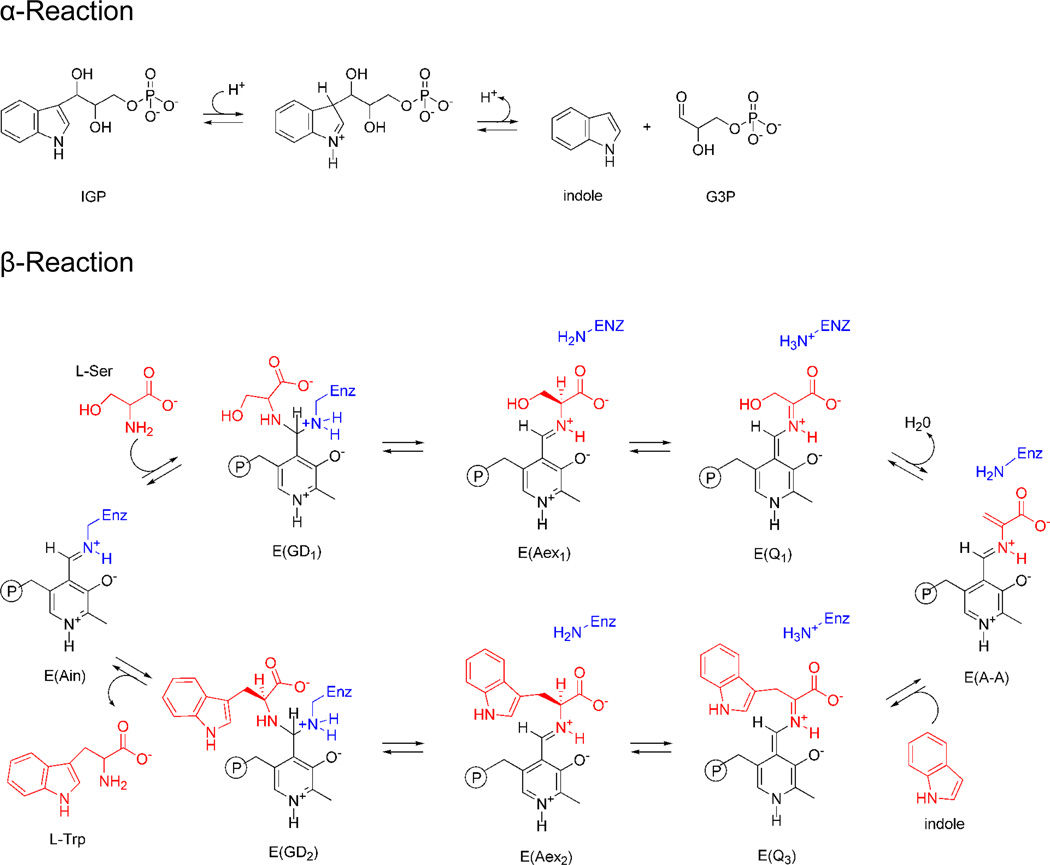
Tryptophan synthase catalyzes the last two steps in the biosynthesis of L-tryptophan (L-Trp) (Scheme I). Early work established that the substrates for the bienzyme complex are 3-indole-D-glycerol-3′-phosphate (IGP) and L-serine (L-Ser), the bienzyme complex has the subunit composition α2β2, the β-subunits require pyridoxal 5′-phosphate (PLP) for catalytic activity, and that indole is a channeled intermediate.13 Many details important to the catalytic mechanism of the α- and β-subunits were revealed by the first two X-ray structures.14 Prior to the determination of these structures, there was no agreed upon mechanism for the channeling of the intermediate, indole, between the α- and β-sites, many protein residues were suspect candidates for involvement in catalysis,15 the protein folds and the αββα linear arrangement of the subunits were not known, and there was no clear explanation for the dependence of the activities of the subunits on the state of aggregation. Many of these and other details were revealed by subsequent X-ray structures determined over the next 20 years.
Scheme I.
Mechanistic pathway for the biosynthesis of L-Trp in tryptophan synthase. In the α-site, 3-indole-D-glycerol-3′-phosphate (IGP) is cleaved to D-glyceraldehyde-3-phosphate (G3P) and indole. In stage I of the β-reaction, L-Ser reacts with the internal aldimine, E(Ain), giving in sequence gem-diamine, E(GD1), L-Ser external aldimine, E(Aex1), quinonoid, E(Q1), and aminoacrylate Schiff base, E(A-A), species and a water molecule. In stage II, indole, channeled from the α-site, makes a nucleophilic attack on E(A-A) giving E(Q3), E(Aex2), E(GD2) intermediates and finally the product, L-Trp.
However, all of the 30+ X-ray structures published between 1988 and 2007 for the wild-type enzyme showed the same global conformation of the β-subunit, designated the open conformation. As it turns out, the open β-subunit conformation has either very low, or no, catalytic activity. Nevertheless, during this time period, catalytic mechanism studies provided indirect evidence that the switching of the β-subunit between open and closed conformational states is essential to the catalysis and allosteric regulation of channeling.16–21
When the structure of the wild-type enzyme with the β-subunits in the closed conformation was solved in 2007,22 it became obvious which residues play key chemical roles in the catalytic activity of the β-site and how the conformational transition between open and closed states both prevents the escape of indole and couples the activities of the α- and β-sites so that the reactions catalyzed by the two subunits become synchronized during the overall αβ-catalytic cycle.20,22–24 The closed subunit structures also provided a structural context for the α- and β-site transformations shown in Scheme I. The formation of the closed conformation of the α-subunit brings catalytic residues αGlu49 and αAsp60 into the correct bonding positions to catalyze the cleavage of IGP to indole. It is postulated that αAsp60 provides coulombic charge-charge interactions at the indolyl ring N-1 that stabilize development of partial positive charge on the ring as bond scission at the ring C-3 position occurs, and that αGlu49 plays an acid-base catalytic role in facilitating the C-C bond scission step. The switch from the open to the closed conformation in the β-subunit rearranges the interactions within the β-site so that the hydrogen bonding between the β-hydroxyl of the reacting substrate L-serine and the carboxylate of βAsp305, which stabilizes the external aldimine, E(Aex1), is replaced by interactions that facilitate the conversion of E(Aex1) to E(Q1) and then E(Q1) to E(A-A) and release of a water molecule (Stage I of the β-reaction, Scheme I). The rearrangement of the βAsp305 side chain carboxylate allows formation of a salt bridge between βArg141 and βAsp305 which locks the β-subunit into the closed conformation and blocks access from solution into the β-site by providing a steric/electrostatic cap over the opening. Within this closed conformation, the active site acid base catalytic groups, βLys87 and βGlu109, are correctly positioned to facilitate the chemical transformations of the reacting substrate.
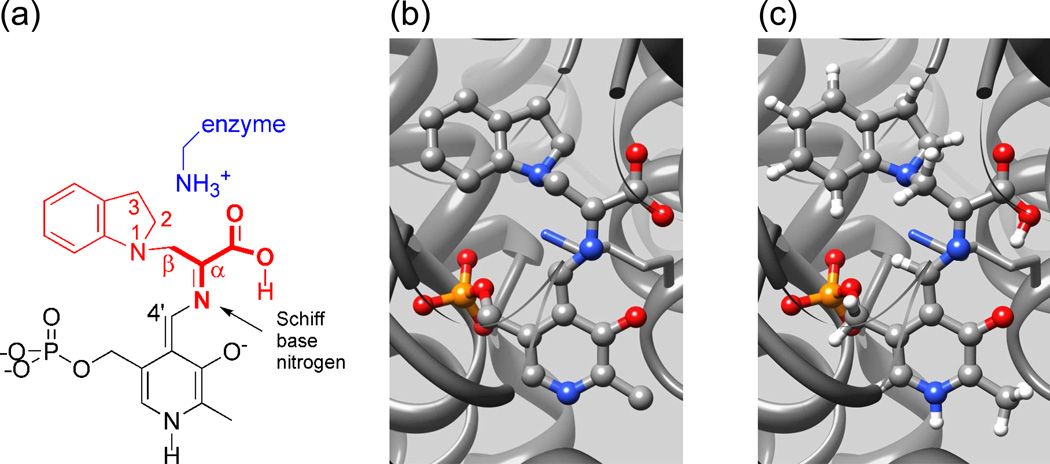
Despite these successes, significant questions regarding the chemical mechanism for the substrates' transformation remain because the resolution of the X-ray structures does not allow for protonation states to be established on the reacting substrate, PLP coenzyme, or catalytic residues. The chemical transformations at the α- and β-catalytic sites involve Brønsted acid-base catalyzed proton transfers between catalytic side-chain sites (Glu49 and Asp60 at the α-site; Lys87 and Glu109 at the β-site) and the reacting substrates. The catalytic activity of the PLP-requiring β-site is also dependent upon the protonation states of the ionizable groups on the PLP moiety (the PLP phenolic hydroxyl, pyridine ring nitrogen, and phosphoryl group) and on the reacting substrate (the Schiff base nitrogen linked to the PLP C4′ carbon, and the carboxylate of the reacting substrate), which are highlighted in Figure 1(a). Therefore, to fully understand the mechanism requires chemically-detailed structural models of the intermediates in the enzyme active site.
Figure 1.
Three views of the indoline-quinonoid intermediate in tryptophan synthase: (a) Line drawing highlighting the components of the intermediate derived from the PLP co-enzyme (black), and the substrates indoline (red) and serine (bold, red). Also indicated are the catalytically-important βlysine-87 side chain (blue) and the Schiff base nitrogen. (b) The X-ray framework structure. (c) The chemically-rich crystal structure from NMR crystallography.
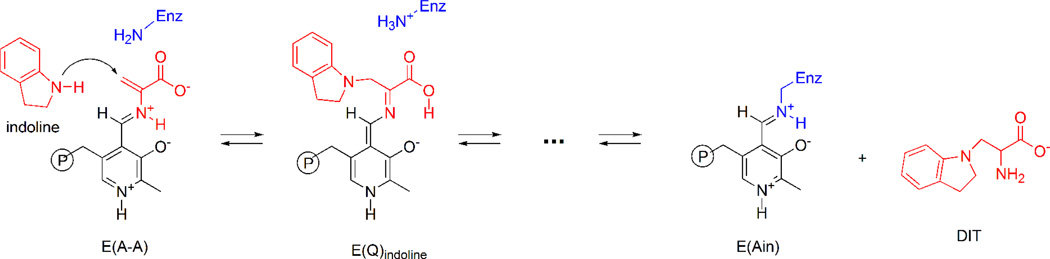
The challenge to NMR crystallography is to provide this atomic-resolution structural characterization. In the remainder of this Account, we detail how this goal can be accomplished, taking as an example our determination of the chemically-rich crystal structure for the quinonoid intermediate formed by the reaction of the indole analogue, indoline, with E(A-A) in the β-site of tryptophan synthase (Scheme II).2 Quinonoid intermediates are ubiquitous to almost all PLP-requiring enzymes and thus play central roles in the catalytic mechanisms of this class of enzymes.25 The rapid formation of E(Q)indoline and its slow conversion to dihydroiso-L-tryptophan (DIT) make this species an ideal subject for NMR crystallography. The kinetic behavior of indoline as a substrate has been well-characterized,23 and two X-ray structures of E(Q)indoline have been determined.2,24 Figure 1(b) shows the starting point: the framework structure of the enzyme-intermediate complex from X-ray crystallography.
Scheme II.
The indole analogue, indoline, reacts rapidly with E(A-A) to give the indoline quinonoid species, E(Q)indoline, which slowly converts to dihydroiso-L-tryptophan (DIT).
Chemical Structure: NMR Spectroscopy
Our work has focused on the use of isotropic chemical shifts for NMR crystallography in tryptophan synthase. We note that, when available, anisotropic NMR interactions26 can provide significant additional restraints for refining models.1,4–6,27 NMR crystallography uses isotropic chemical shifts in two specific ways. First, the chemical shift is used in its analytical role to directly report on the chemical state of a probe atom, often answering chemical questions regarding the direct protonation or hybridization state at that site. For example, McDowell et al. showed that when labeled L-[3-13C]Ser was supplied as a substrate to tryptophan synthase, solid-state 13C NMR spectroscopy could identify the change in hybridization at the beta carbon as water is lost to form the E(A-A) intermediate.28 Second, the chemical shift is used in NMR crystallography to report on the chemical and structural environment surrounding a probe nucleus. This is the manner in which chemical shifts are used in the structural refinement of organic and inorganic molecular crystals using NMR crystallography;2,3,5,7–9 the chemical state of the probe atom is known, but its shift depends not only on its chemical structure, but also its three-dimensional molecular conformation and the precise location and chemical state of nearby atoms in the surrounding crystal lattice. Interpreting the chemical shift of a probe nucleus in terms of the immediate chemical and structural environment is challenging, but, as illustrated below, when chemical shifts for multiple probe nuclei are measured and analyzed collectively within the context of NMR crystallography, atomic-resolution structural models can be uniquely determined.
Solid-state NMR spectra can be acquired on microcrystalline protein samples under essentially the same conditions used to determine X-ray crystal structures, making solid-state NMR the method of choice for NMR crystallography. Acquisition of NMR spectra in the solid-state does not necessarily decrease sensitivity relative to solution-state NMR; it is our experience that in larger systems, such as tryptophan synthase, 13C and 15N sensitivities are better under cross-polarization magic-angle-spinning (CP MAS) conditions in solid-state NMR (using 20 mg of microcrystalline protein in an 80 µl, 4 mm MAS rotor) than in solution-state NMR (the same 20 mg of protein in a 350 µl, 5 mm restricted-volume tube). Acquisition in the solid-state also does not decrease biological relevance; microcrystalline samples of tryptophan synthase,29 as well as other enzymes,30 are able to retain catalytic activity.
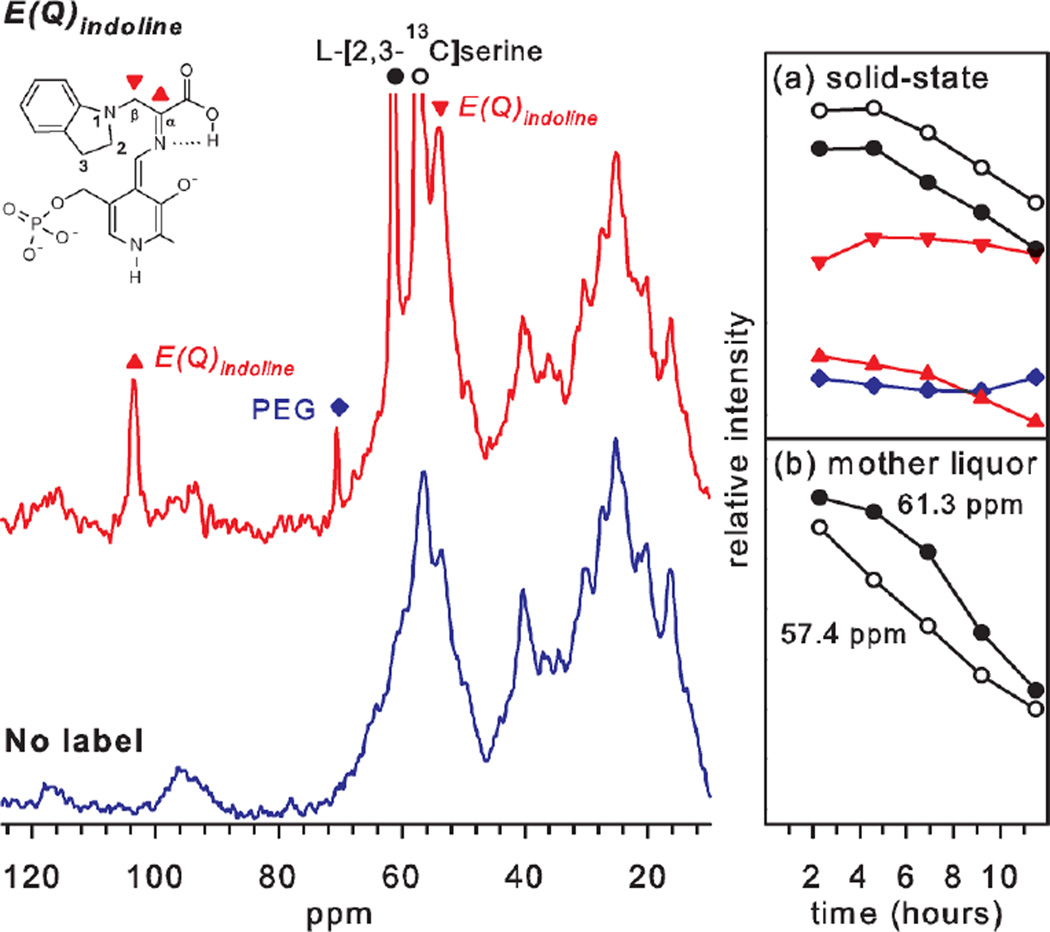
While every system will present unique challenges to the acquisition and assignment of solid-state NMR spectra,13C and 15N chemical shifts for the E(Q)indoline intermediate in tryptophan synthase can be obtained by supplying isotopically-labeled L-serine and indoline substrates to microcrystals of the tryptophan synthase bienzyme complex (Figure 2). The crystal lattice is greater than 50% by mass water, and the substrates diffuse freely within lattice channels to establish steady-state concentrations of the intermediates in the catalytically-active enzyme. As the reaction progresses, it is possible to follow with NMR the depletion of the labeled substrate in the mother liquor (as well as the formation of the product, DIT) by using direct 13C excitation with low-power 1H decoupling. Bound substrates are selected using CP MAS with high-power 1H decoupling, allowing the assignment of the 13C and 15N resonances for the fragment of the intermediate derived from serine and indoline (Table 1). The serine-derived Cα and Cβ chemical shifts and the serine- and indoline-derived nitrogen chemical shifts of the bound intermediate can be resolved in 1D CP-MAS experiments. The carboxylate carbon and the indoline carbon resonances overlap those for non-specifically bound (and/or solubility-limited) substrate, but can be resolved via double difference experiments.2
Figure 2.
13C- CPMAS solid-state NMR spectra of the indoline quinonoid intermediate in tryptophan synthase formed with L-[2,3-13C]Ser (red spectrum) or unlabeled L-Ser (blue spectrum). (Inset) Time courses for the disappearance and appearance of the enriched 13C signals in the (a) bound (solid) state and (b) mother liquor. The resonances at 103.6 ppm (red triangles) and 54.1 ppm (inverted red triangles) are assigned to the sp2-hybridized 2-13C (Cα) and sp3-hybridized 3-13C (Cβ) of E(Q)indoline, respectively, while the peaks at 61.3 ppm and 57.4 ppm (black and open circles) are assigned to L-[2,3-13C]-Ser nonspecifically bound to the microcrystals. The time courses on the right show the consumption of the labeled serine as the reactions progresses, while the quinonoid species persists until all substrate is consumed. Reprinted with permission from J. Am. Chem. Soc. 2011 133, 4–7. Copyright 2011 American Chemical Society.
Table 1.
Experimental and theoretical (ab initio) chemical shifts for two forms of the E(Q)indoline intermediate and their fast-exchange equilibrium. (Site labels are shown in Figures 1 and 2).
| Exp. /ppm |
Calculated PSB Form /ppm |
Calculated Acid Form /ppm |
Two-Site Equilib. /ppm |
|
|---|---|---|---|---|
| Cα | 103.6 | 106.0 | 101.3 | 102.9 |
| Cβ | 54.1 | 54.0 | 49.6 | 51.1 |
| C' | 173.0 | 172.3 | 169.3 | 170.3 |
| N | 296.5 | 215.1 | 337.2 | 295.7 |
| C2 | 50.5 | 50.1 | 51.5 | 51.1 |
| C3 | 28.5 | 31.9 | 32.5 | 32.3 |
| N1 | 83.5 | 85.0 | 91.1 | 89.0 |
An immediate goal of NMR crystallography is to determine the protonation states of ionizable groups in the active site. Fig. 3(b) shows possible sites of protonation on or near the intermediate complex, including the phosphoryl group, pyridoxal phenolic oxygen, Schiff base nitrogen, both carboxylate oxygens (derived from L-Ser), and the neighboring Nε of βLys87. The Schiff base nitrogen is the only ionizable group with a directly measured chemical shift. Previous measurement of PLP-Schiff base complexes in model compounds31,32 and within the inhibited PLP-dependent alanine racemase33 place protonated Schiff bases near 190 ppm, while neutral Schiff base linkages fall above 315 ppm (referenced to liq-NH3). At 296.5 ppm, the experimental chemical shift of this intermediate's Schiff base nitrogen is outside (and in between) the range expected for either a fully protonated or deprotonated Schiff base, suggesting that the observed shift is due to a fast exchange equilibrium between the two forms. The partner structures, including the protonation states of the other ionizable groups, cannot readily be established from the empirical correlation of shift with structure. But in the following section we show that when combined with ab initio computational chemistry and X-ray crystallography, the chemical shifts can be identified with high certainty to an equilibrium between two specific tautomeric forms.
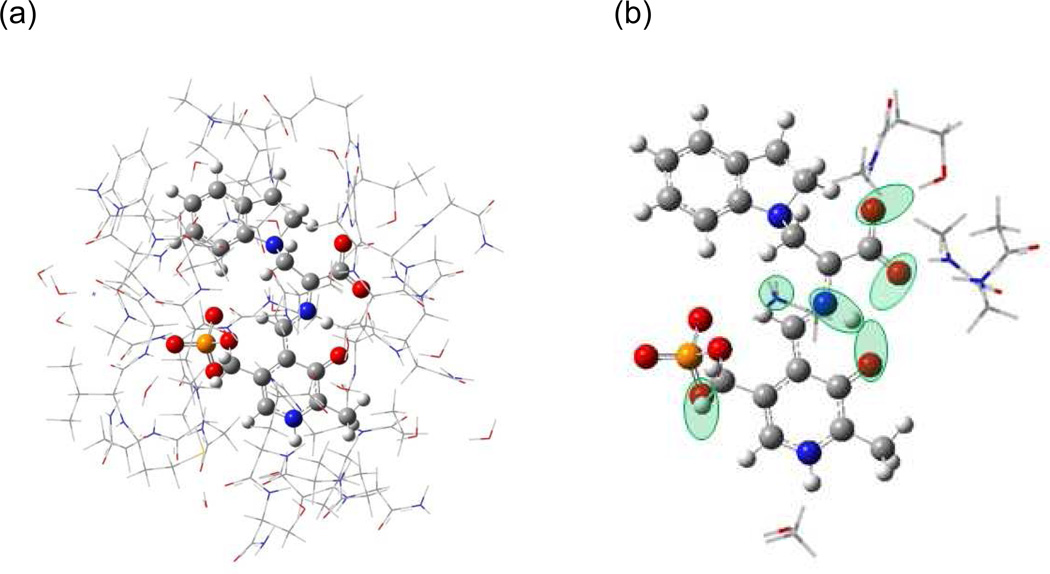
Figure 3.
(a) Model of the tryptophan synthase β-subunit enzyme active site showing (wireframe) the side chains and (ball and stick) the PLP-ligand complex. (b) Substructure used for calculating NMR chemical shifts. Green ovals indicate possible sites of protonation. Reprinted with permission from J. Am. Chem. Soc. 2011 133, 4–7. Copyright 2011 American Chemical Society.
Chemically-Rich Structure: Adding Ab Initio Computational Chemistry
Computational chemistry plays two important roles in NMR crystallography. First, it allows chemically-detailed, three-dimensional models to be built upon the coarse X-ray framework. Second, it provides first-principles predictions of spectroscopic properties, such as NMR chemical shifts, that allow competing structural models to be judged and ranked based on their agreement with experiment. Enabling the latter has been the development of highly-accurate, ab initio (first-principles) methods for the calculation of NMR parameters in chemical and biomolecular systems.34–36
In NMR crystallography, the X-ray crystal structure provides the scaffolding upon which chemically-rich structures are constructed. In building models of enzyme active sites, we have adopted a strategy in which we fix the (non-hydrogen) backbone and side chain atoms of the enzyme at their crystallographically-determined coordinates and use computational chemistry to geometry optimize the non-peptide components such as the coenzyme and substrates. The motivation to refine the latter derives from the larger relative uncertainty expected in their structural coordinates: in solving X-ray crystal structures, the backbone and side chain geometries are refined using restraints derived from residue-specific distributions of bond lengths, angles, and torsions found within protein databases of high-resolution structures;37 substrates and coenzymes are modeled directly to the experimentally-determined electron density, making their structural coordinates potentially less accurate. Still, only relatively minor changes to bond lengths (<0.2 Å) and angles (~5°) for the intermediates are expected during geometry optimization, and weaker constraints can be implemented to maintain gross structural features, such as the relative orientation of ring planes that are defined by multiple atoms, to compensate for physical interactions (e.g., dispersion forces) that are potentially poorly modeled computationally.
Protein systems are currently too large to be treated fully using ab initio computational methods, but combined classical, semi-empirical, and first-principles quantum-mechanical techniques have been developed that enable computational modeling of enzyme active sites and entire protein systems. Mixed techniques, such as ONIOM,38 treat selected regions of the system at higher levels of theory, allowing the substrates, coenzyme, and catalytic residues in the active site to be modeled at a quantum-mechanical level, while the surrounding active site residues are treated at a less computationally costly, semi-empirical or classical (molecular mechanics) level. ONIOM methods calculate the total energy as the sum of the full system energy calculated at the low level of theory plus the difference in the energy of the isolated high-layer region calculated at high and low levels of theory. In order to use molecular mechanics for the low layer, both the peptide and non-peptide components must be accurately parameterized.
In our application of NMR crystallography to the indoline quinonoid intermediate in tryptophan synthase, we first constructed a truncated model of the active site consisting of residues located within 7 Å of the intermediate (PLP + reacting substrates) (Figure 3(a)). Peptide chains at the boundary of this region were terminated by replacing backbone N-terminal nitrogens with hydrogens and C-terminal carbonyls with carboxamides. This provided a framework for computationally optimizing the structure of the intermediate in the presence of local charges, hydrogen bonding, and steric interactions with the active site side chains. We adopted an ONIOM38 method (implemented in Gaussian03) which the atoms of the PLP-ligand complex defined the high layer (treated at the density-functional level of theory, B3LYP/6-31G(d,p)), while the amino acid residues making up the catalytic pocket were included in the low layer (treated at the semi-empirical, PM3 level of theory). Second, candidate structures for the intermediate in the active site were systematically generated with varying protonation states for the six ionizable sites on or near the PLP-ligand complex shown in Fig. 3(b). Models with more than a single proton placed at either the pyridoxal oxygen, the Schiff base nitrogen, or the closest carboxylate oxygen were not considered, nor were structures with a doubly-protonated carboxylate. This gave 28 model structures that were each fully geometry optimized. Third, chemical shieldings were calculated (B3LYP/ 6-311++G(d,p)) for each refined intermediate model complex using substructures such as the one shown in Figure 3(b), which included fragments of residues that were potentially charged or hydrogen bonded to the intermediate. Calculated chemical shieldings were converted to chemical shifts referenced to TMS for 13C using implicitly (polarizable continuum model)39 solvated benzene as a secondary shift reference and liquid ammonia for 15N using explicitly hydrated urea as a secondary shift reference.2 Finally, the agreement between the calculated chemical shifts for the model structures and the experimental shifts was quantified and ranked using the reduced χ2 statistic (the weighted deviation of the model and experimental shifts) with weightings corresponding to root-mean-square deviations of 2 ppm for 13C and 4 ppm for 15N.40,41
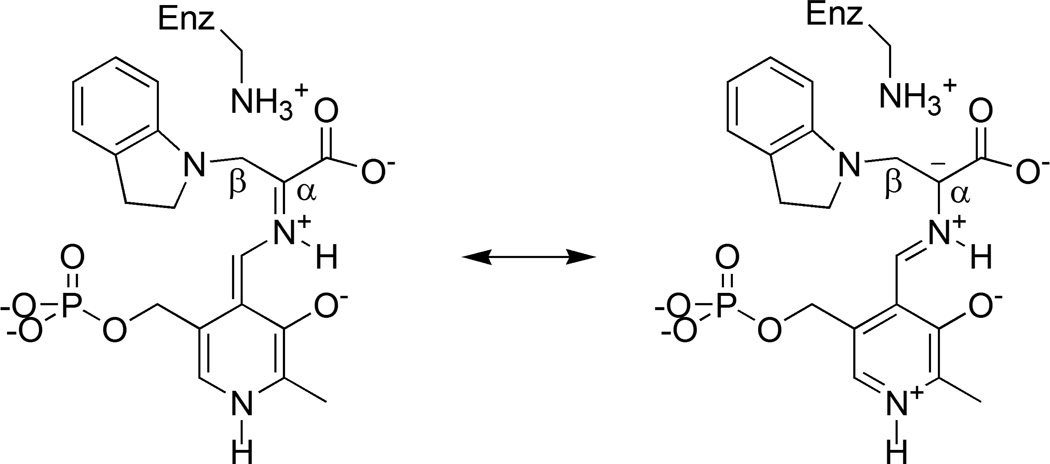
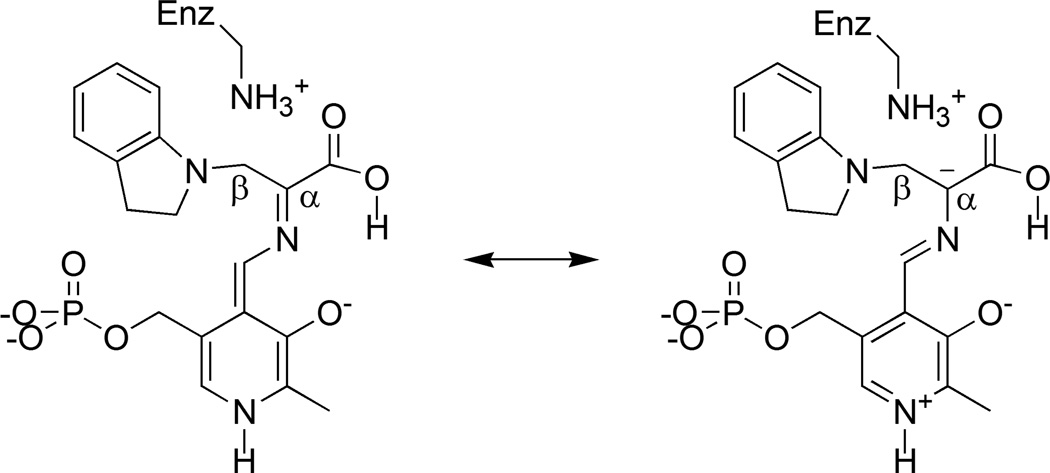
For the indoline quinonoid intermediate, no single static structural model was found to reproduce all of the experimental chemical shift values (lowest reduced χ2 of 4.9) due, in part, to a large discrepancy at the Schiff base nitrogen. To account for the chemical shift at this site, it was necessary to invoke a fast-exchange equilibrium model, in which exchange partners that differed by the position of a single proton were systematically paired and the relative populations of the tautomers optimized for best agreement with the experimental chemical shifts. These models were ranked and the best-fit equilibrium, with a reduced χ2 of 1.39, was found to be a 34:66 ratio between the canonical protonated Schiff-base (PSB) form (Scheme III) and the carboxylic acid form in which the proton moves from the Schiff base nitrogen to the nearest carboxylate oxygen (Scheme IV). In both partners, the side chain of βK87 is positively charged and the phosphoryl is dianionic; the latter is in agreement with the interpretation of the phosphoryl 31P chemical shift in solution.42 The solution-state 31P chemical shift can be used to rule out many of the competing equilibrium models including the second-best population-optimized model (reduced χ2 of 1.48), which was found to be a similar PSB-acid equilibrium, but with the phosphoryl groups of both structures protonated (monoanionic). All other population-optimized equilibrium models with dianionic phosphoryl groups were found to have reduced χ2 > 3, making them unlikely candidates. The calculated chemical shifts for the PSB and acid structures, in addition to their fast-exchange average, are summarized in Table 1.
Scheme III.
Canonical protonated Schiff base form of the indoline quinonoid intermediate and a resonance structure that builds up negative charge at Cα.
Scheme IV.
Acid form of the indoline quinonoid intermediate and a resonance structure that builds up negative charge at Cα.
Figure 1(c) shows the refined, chemically-rich crystal structure of the acid form of the indoline quinonoid intermediate. This form is hinted at in the X-ray crystal structure, which shows a distance of 2.57 Å from the Schiff base nitrogen to the nearest carboxylate oxygen, and in the 15N NMR spectrum, which reports a mostly deprotonated Schiff base nitrogen. But it is only when multiple chemical shifts for the intermediate are analyzed within the context of NMR crystallography that this tautomer can be confidently identified. The acid tautomer had not previously been proposed to play a role in PLP catalysis, but its identification has fundamental implications for the mechanism in tryptophan synthase catalysis, which at the next step involves protonation at the Cα site. Structures that build up negative charge at Cα are anticipated to promote this transformation by helping to direct the proton to the target site and by charge stabilizing the transition state. Schemes III and IV each show an additional resonance structure that builds up electron density at the Cα site. Repulsion between negative charges causes this resonance structure to be less important for the PSB (and all other carboxylate-based structures) than for the acid form of the intermediate, making it a smaller contribution to the ground-state electronic structure. Natural bond orbital analysis43 calculations of atomic charges are consistent with this model: Cα is essentially neutral (-0.014 atomic units) for the PSB form and is -0.097 atomic units for the acid form. In addition to the buildup of negative charge at Cα, there is a concomitant buildup of positive charge at C4′ of the PLP, biasing the catalysis away from a competing transamination pathway.
Conclusion
Enzymes have evolved to achieve remarkably efficient and specific chemical transformations. Yet atomic-level details of enzyme mechanisms remain elusive: the intermediates are transient and the chemistry that drives the transformation, such as changes in hybridization and protonation states, is difficult to characterize in functioning enzyme systems. NMR crystallography is poised to make a significant contribution to this understanding by the synergistic combination of solid-state NMR spectroscopy, X-ray crystallography, and computational chemistry. This fusion allows specific models of the chemical structure to be built upon the coarse X-ray framework and then tested by comparison of predicted and assigned chemical shifts. The result is a unique and chemically-rich view into functioning enzyme catalysis, which for the case of tryptophan synthase leads to a new acid-form hypothesis for the indoline quinonoid intermediate.
Acknowledgements
This work has been supported by the National Institutes of Health Grant R01GM097569.
Biographies
Leonard J. Mueller is Professor of Chemistry at the University of California, Riverside. He received his B.S. in Chemistry from the University of Rochester (1988), C.P.G.S. in Natural Science (Chemistry) from the University of Cambridge (1989), and Ph.D. in Chemistry from the California Institute of Technology (1997). From 1996–1998 he was an American Cancer Society Postdoctoral Fellow at the Massachusetts Institute of Technology. Len’s research interests include nuclear magnetic resonance spectroscopy as a probe of molecular and biological structure and dynamics.
Michael F. Dunn is Professor Emeritus of Biochemistry at the University of California, Riverside. He received his B.S. in Chemical Engineering from the Colorado School of Mining Engineering (1961) and his Ph.D. in Chemistry from the Georgia Institute of Technology (1966). He was a National Institutes of Health Postdoctoral Fellow at the Institute of Molecular Biology, University of Oregon (1966–1969), and a NATO Postdoctoral Fellow at Denmark’s Technical University (1969–1970). Michael’s research interests include the investigation of the mechanisms of enzyme catalyzed reactions, and structure-function relationships in protein hormones. He utilizes optical spectroscopy and nuclear magnetic resonance spectroscopy, organic synthesis and rapid kinetics in his research.
References
- 1.Facelli JC, Grant DM. Determination of Molecular Symmetry in Crystalline Naphthalene Using Solid-State NMR. Nature. 1993;365:325–327. doi: 10.1038/365325a0. [DOI] [PubMed] [Google Scholar]
- 2.Lai JF, Niks D, Wang YC, Domratcheva T, Barends TRM, Schwarz F, Olsen RA, Elliott DW, Fatmi MQ, Chang CEA, Schlichting I, Dunn MF, Mueller LJ. X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase. J Am Chem Soc. 2011;133:4–7. doi: 10.1021/ja106555c. [DOI] [PubMed] [Google Scholar]
- 3.Olsen RA, Struppe J, Elliott DW, Thomas RJ, Mueller LJ. Through-bond C-13-C-13 correlation at the natural abundance level: Refining dynamic regions in the crystal structure of vitamin-D-3 with solid-state NMR. J Am Chem Soc. 2003;125:11784–11785. doi: 10.1021/ja036655s. [DOI] [PubMed] [Google Scholar]
- 4.Brouwer DH, Darton RJ, Morris RE, Levitt MH. A solid-state NMR method for solution of zeolite crystal structures. J Am Chem Soc. 2005;127:10365–10370. doi: 10.1021/ja052306h. [DOI] [PubMed] [Google Scholar]
- 5.Harper JK, Grant DM, Zhang YG, Lee PL, Von Dreele R. Characterizing challenging microcrystalline solids with solid-state NMR shift tensor and synchrotron X-ray powder diffraction data: Structural analysis of ambuic acid. J Am Chem Soc. 2006;128:1547–1552. doi: 10.1021/ja055570j. [DOI] [PubMed] [Google Scholar]
- 6.Harris RK, Joyce SA, Pickard CJ, Cadars S, Emsley L. Assigning carbon-13 NMR spectra to crystal structures by the INADEQUATE pulse sequence and first principles computation: a case study of two forms of testosterone. Phys Chem Chem Phys. 2006;8:137–143. doi: 10.1039/b513392k. [DOI] [PubMed] [Google Scholar]
- 7.Rajeswaran M, Blanton TN, Zumbulyadis N, Giesen DJ, Conesa-Moratilla C, Misture ST, Stephens PW, Huq A. Three-dimensional structure determination of N-(p-tolyl)-dodecylsulfonamide from powder diffraction data and validation of structure using solid-state NMR spectroscopy. J Am Chem Soc. 2002;124:14450–14459. doi: 10.1021/ja027978b. [DOI] [PubMed] [Google Scholar]
- 8.Salager E, Day GM, Stein RS, Pickard CJ, Elena B, Emsley L. Powder Crystallography by Combined Crystal Structure Prediction and High-Resolution (1)H Solid-State NMR Spectroscopy. J Am Chem Soc. 2010;132:2564–2566. doi: 10.1021/ja909449k. [DOI] [PubMed] [Google Scholar]
- 9.Webber AL, Emsley L, Claramunt RM, Brown SP. NMR Crystallography of Campho[2,3-c]pyrazole (Z '=6): Combining High-Resolution H-1-C-13 Solid-State MAS NMR Spectroscopy and GIPAW Chemical-Shift Calculations. Journal of Physical Chemistry A. 2010;114:10435–10442. doi: 10.1021/jp104901j. [DOI] [PubMed] [Google Scholar]
- 10.Luchinat C, Parigi G, Ravera E, Rinaldelli M. Solid-State NMR Crystallography through Paramagnetic Restraints. J Am Chem Soc. 2012;134:5006–5009. doi: 10.1021/ja210079n. [DOI] [PubMed] [Google Scholar]
- 11.Harris RK, Wasylishen RE, Duer MJ, editors. NMR Crystallography. West Sussex, U.K.: John Wiley & Sons Ltd; 2009. [Google Scholar]
- 12.McDermott A, Polenova T. Solid state NMR: new tools for insight into enzyme function. Curr Opin Struc Biol. 2007;17:617–622. doi: 10.1016/j.sbi.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Yanofsky C, Crawford IP. In: The Enzymes. Boyer PD, editor. New York: Academic Press; 1972. pp. 1–31. [Google Scholar]
- 14.Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. 3-Dimensional Structure of the Tryptophan Synthase Alpha-2-Beta-2 Multienzyme Complex from Salmonella-Typhimurium. Journal of Biological Chemistry. 1988;263:17857–17871. [PubMed] [Google Scholar]
- 15.Miles EW. Tryptophan synthase: structure, function, and subunit interaction. Advances in Enzymology and Related Areas of Molecular Biology. 1979;49:127–186. doi: 10.1002/9780470122945.ch4. [DOI] [PubMed] [Google Scholar]
- 16.Dunn MF, Aguilar V, Brzovic P, Drewe WF, Houben KF, Leja CA, Roy M. The Tryptophan Synthase Bienzyme Complex Transfers Indole between the Alpha-Sites and Beta-Sites Via a 25–30 a Long Tunnel. Biochemistry-Us. 1990;29:8598–8607. doi: 10.1021/bi00489a015. [DOI] [PubMed] [Google Scholar]
- 17.Brzovic PS, Ngo K, Dunn MF. Allosteric Interactions Coordinate Catalytic Activity between Successive Metabolic Enzymes in the Tryptophan Synthase Bienzyme Complex. Biochemistry-Us. 1992;31:3831–3839. doi: 10.1021/bi00130a014. [DOI] [PubMed] [Google Scholar]
- 18.Brzovic PS, Sawa Y, Hyde CC, Miles EW, Dunn MF. Evidence That Mutations in a Loop Region of the Alpha-Subunit Inhibit the Transition from an Open to a Closed Conformation in the Tryptophan Synthase Bienzyme Complex. Journal of Biological Chemistry. 1992;267:13028–13038. [PubMed] [Google Scholar]
- 19.Harris RM, Dunn MF. Intermediate trapping via a conformational switch in the Na+-activated tryptophan synthase bienzyme complex. Biochemistry-Us. 2002;41:9982–9990. doi: 10.1021/bi0255672. [DOI] [PubMed] [Google Scholar]
- 20.Dunn MF. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Archives of Biochemistry and Biophysics. 2012;519:154–166. doi: 10.1016/j.abb.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan P, Woehl E, Dunn MF. Protein architecture, dynamics and allostery in tryptophan synthase channeling. Trends in Biochemical Sciences. 1997;22:22–27. doi: 10.1016/s0968-0004(96)10066-9. [DOI] [PubMed] [Google Scholar]
- 22.Ngo H, Kimmich N, Harris R, Niks D, Blumenstein L, Kulik V, Barends TR, Schlichting I, Dunn MF. Allosteric regulation of substrate channeling in tryptophan synthase: Modulation of the L-Serine reaction in stage I of the ss-reaction by alpha-site ligands. Biochemistry-Us. 2007;46:7740–7753. doi: 10.1021/bi7003872. [DOI] [PubMed] [Google Scholar]
- 23.Dunn MF, Niks D, Ngo H, Barends TRM, Schlichting I. Tryptophan synthase: the workings of a channeling nanomachine. Trends in Biochemical Sciences. 2008;33:254–264. doi: 10.1016/j.tibs.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Barends TRM, Domratcheva T, Kulik V, Blumenstein L, Niks D, Dunn MF, Schlichting I. Structure and mechanistic implications of a tryptophan synthase quinonold intermediate. Chembiochem. 2008;9:1024–1028. doi: 10.1002/cbic.200700703. [DOI] [PubMed] [Google Scholar]
- 25.Dunathan HC. Conformation and Reaction Specificity in Pyridoxal Phosphate Enzymes. P Natl Acad Sci USA. 1966;55:712–716. doi: 10.1073/pnas.55.4.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller LJ. Tensors and Rotations in NMR. Concepts in Magnetic Resonance Part A. 2011;38A:221–235. [Google Scholar]
- 27.Wylie BJ, Sperling LJ, Nieuwkoop AJ, Franks WT, Oldfield E, Rienstra CM. Ultrahigh resolution protein structures using NMR chemical shift tensors. P Natl Acad Sci USA. 2011;108:16974–16979. doi: 10.1073/pnas.1103728108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell LM, Lee MS, Schaefer J, Anderson KS. Observation of an Aminoacrylate Enzyme Intermediate in the Tryptophan Synthase Reaction by Solid-State Nmr. J Am Chem Soc. 1995;117:12352–12353. [Google Scholar]
- 29.Mozzarelli A, Peracchi A, Rossi GL, Ahmed SA, Miles EW. Microspectrophotometric Studies on Single-Crystals of the Tryptophan Synthase Alpha-2-Beta-2-Complex Demonstrate Formation of Enzyme-Substrate Intermediates. Journal of Biological Chemistry. 1989;264:15774–15780. [PubMed] [Google Scholar]
- 30.Rozovsky S, McDermott AE. Substrate product equilibrium on a reversible enzyme, triosephosphate isomerase. P Natl Acad Sci USA. 2007;104:2080–2085. doi: 10.1073/pnas.0608876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharif S, Huot MC, Tolstoy PM, Toney MD, Jonsson KHM, Limbach HH. N-15 nuclear magnetic resonance studies of acid-base properties of pyridoxal-5 '-phosphate aldimines in aqueous solution. J Phys Chem B. 2007;111:3869–3876. doi: 10.1021/jp067334g. [DOI] [PubMed] [Google Scholar]
- 32.Sharif S, Schagen D, Toney MD, Limbach HH. Coupling of functional hydrogen bonds in pyridoxal-5 '-phosphate-enzyme model systems observed by solid-state NMR spectroscopy. J Am Chem Soc. 2007;129:4440–4455. doi: 10.1021/ja066240h. [DOI] [PubMed] [Google Scholar]
- 33.Copie V, Faraci WS, Walsh CT, Griffin RG. Inhibition of Alanine Racemase by Alanine Phosphonate - Detection of an Imine Linkage to Pyridoxal 5'-Phosphate in the Enzyme-Inhibitor Complex by Solid-State N-15 Nuclear Magnetic-Resonance. Biochemistry-Us. 1988;27:4966–4970. doi: 10.1021/bi00414a002. [DOI] [PubMed] [Google Scholar]
- 34.Dedios AC, Pearson JG, Oldfield E. Secondary and Tertiary Structural Effects on Protein Nmr Chemical-Shifts - an Abinitio Approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 35.deDios AC. Ab initio calculations of the NMR chemical shift. Prog Nucl Mag Res Sp. 1996;29:229–278. [Google Scholar]
- 36.Oldfield E. Chemical shifts in amino acids, peptides, and proteins: From quantum chemistry to drug design. Annual Review of Physical Chemistry. 2002;53:349–378. doi: 10.1146/annurev.physchem.53.082201.124235. [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt GJ, Jones TA. Databases in protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 38.Vreven T, Morokuma K. On the application of the IMOMO (integrated molecular orbital plus molecular orbital) method. Journal of Computational Chemistry. 2000;21:1419–1432. [Google Scholar]
- 39.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chemical Reviews. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 40.Gryff-Keller A. Theoretical modeling of C-13 NMR chemical shifts: How to use the calculation results. Concepts in Magnetic Resonance Part A. 2011;38A:289–307. [Google Scholar]
- 41.Cai L, Kosov DS, Fushman D. Density functional calculations of backbone N-15 shielding tensors in beta-sheet and turn residues of protein G. J Biomol Nmr. 2011;50:19–33. doi: 10.1007/s10858-011-9474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnackerz KD, Andi B, Cook PF. P-31 NMR spectroscopy senses the microenvironment of the 5 '-phosphate group of enzyme-bound pyridoxal 5 '-phosphate. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2011;1814:1447–1458. doi: 10.1016/j.bbapap.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Foster JP, Weinhold F. Natural Hybrid Orbitals. J Am Chem Soc. 1980;102:7211–7218. [Google Scholar]