Abstract
Objective
Ribavirin improves treatment response to pegylated-interferon (PEG-IFN) in chronic hepatitis C but the mechanism remains controversial. We studied correlates of response and mechanism of action of ribavirin in treatment of hepatitis C.
Design
70 treatment-naïve patients were randomized to 4 weeks of ribavirin (1000–1200 mg/d) or none, followed by PEG-IFN alfa-2a and ribavirin at standard doses and durations. Patients were also randomized to a liver biopsy 24 hours before, or 6 hours after starting PEG-IFN. Hepatic gene expression was assessed by microarray and interferon-stimulated gene (ISG) expression quantified by nCounter platform. Temporal changes in ISG expression were assessed by qPCR in peripheral-blood mononuclear cells (PBMC) and by serum levels of IP-10.
Results
After four weeks of ribavirin monotherapy, HCV levels decreased by 0.5±0.5 log10 (p=0.009 vs. controls) and ALT by 33% (p<0.001). Ribavirin pretreatment, while modestly augmenting ISG induction by PEG-IFN, did not modify the virological response to subsequent PEG-IFN and ribavirin treatment. However, biochemical, but not virological response to ribavirin monotherapy predicted response to subsequent combination treatment (rapid virological response, 71% in biochemical responders vs. 22% non-responders, p=0.01; early virological response, 100% vs. 68%, p=0.03, sustained virological response 83% vs. 41%, p=0.053). Ribavirin monotherapy lowered serum IP-10 levels but had no effect on ISG expression in PBMC.
Conclusion
Ribavirin is a weak antiviral but its clinical effect seems to be mediated by a separate, indirect mechanism, which may act to reset interferon-responsiveness in HCV-infected liver.
Keywords: Interferon-stimulated genes, hepatic gene expression, ribavirin mechanism, hepatitis C virus, viral kinetics
INTRODUCTION
Despite recent advances and the approval of direct-acting antivirals[1] for chronic hepatitis C, pegylated-interferon (PEG-IFN) and ribavirin (RBV) remain the backbone of treatment. PEG-IFN is thought to exert its activity through induction of interferon-stimulated genes (ISGs), many of which have antiviral effects[2]. Combination therapy with RBV markedly improves the response to PEG-IFN[3] but RBV monotherapy is not sufficient to achieve viral clearance[4].
The mechanism of action of RBV is unclear. Several mechanisms have been suggested: direct inhibition of the viral RNA polymerase, viral mutagenesis leading to error catastrophe, GTP depletion through inhibition of IMPDH, or modulation of T-cell responses[5, 6, 7]. Recent in vitro[8] and in vivo[9] studies have indicated that RBV may act as a potentiator of IFN signaling by augmenting ISG induction. In this study, we aimed to determine the effects of RBV monotherapy and to identify potential mechanisms of action of RBV in treatment of hepatitis C.
METHODS
Patients And Study Design
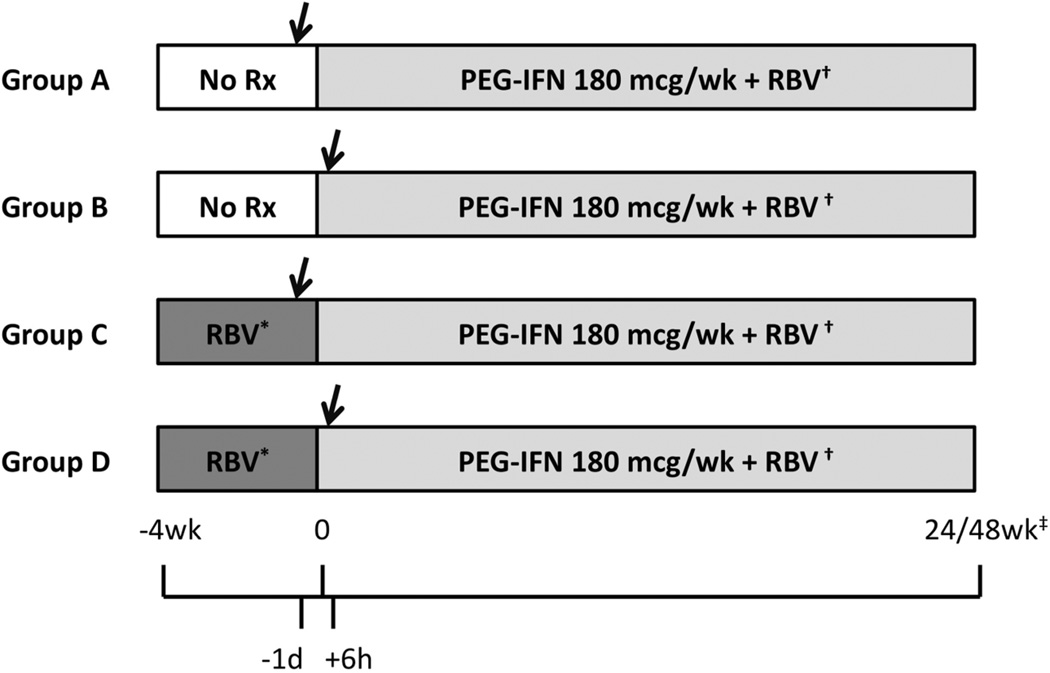
We enrolled 70 adult treatment-naïve patients with chronic hepatitis C into a prospective, randomized, 2X2, open-label controlled trial. Patients with serious comorbidities, other liver diseases, HIV coinfection, contraindications to liver biopsy or to antiviral therapy were excluded. Patients were randomized evenly to receive 4 weeks of ribavirin monotherapy (weight-based 1000–1200 mg/d for all genotypes) or no treatment, after which they received combination treatment with PEG-IFN and RBV with standard doses (180 µg/wk of PEG-IFN alfa-2a for all patients, 800 mg/d of RBV for genotypes 2/3, and 1000–1200 mg/d for all other genotypes) and duration (24 weeks for genotypes 2/3, 48 weeks for all other genotypes). Patients were also randomized to undergo a percutaneous liver biopsy 24 hours before, or 6 hours after the first PEG-IFN injection. This yielded 4 groups with different drug exposure at the time of the biopsy: group A - no treatment, group B – PEG-IFN alone (with the first dose of RBV given in the evening, 6 hours after the biopsy), group C – RBV alone, and group D – PEG-IFN and RBV. Time 0 was defined as the time of the first PEG-IFN injection (Figure 1). Randomization was stratified by genotype (2/3 vs. all others) and race (African-American vs. all others).
Figure 1.
Study design. Arrows note timing of liver biopsy. *Ribavirin dose 1000 mg/d for patients weighing less than 75 kg, 1200 mg/d for heavier patients, irrespective of viral genotype. †Ribavirin dose 800 mg/d for patients with genotype 2/3, weight-based 1000–1200 mg/d for all other genotypes. ‡Treatment duration 24 weeks for genotypes 2/3, 48 weeks for all others.
Patients were seen and blood was drawn on a weekly basis (groups C & D) or every other week (groups A & B) from week −4 to time 0; at 6, 24, 48 and 72 hours after the first injection; on a weekly basis from week 1 to week 4; and then every 4–8 weeks until 24 weeks after the end of treatment.
The study was approved by the NIDDK Institutional Review Board and all patients gave written informed consent (Clinicaltrials.gov registration NCT00718172). None of the authors has a competing interest regarding this study.
Sample Acquisition
Percutaneous liver biopsies were obtained under conscious sedation and local anesthesia using a 14G Klatskin needle. A 2–5 mm segment was immediately deposited in RNALater (Qiagen, Valencia, Ca), maintained at 4C for 24 hours and subsequently stored at −80C.
Acid-Citrate-Dextrose-anticoagulated blood samples were drawn at week −4, 0, 6 hr, 24 hr, week 1 and week 4 and PBMC were separated immediately on Ficoll–Histopaque (Mediatech, Manassas, VA) density gradients. Approximately 1.5X106 cells were placed in 500 µl of TRizol (Qiagen) and stored at −80C. Serum samples were drawn on every visit and stored at −80C.
Gene Expression
RNA was extracted from thawed liver samples using TRizol and Gene expression was measured using GeneChip Human Genome U133 Plus 2.0 microarray (Affymetrix, Santa Clara, CA). Microarray results were deposited in the GEO database (GSE386663).
Hepatic expression of ISGs was quantified using the nCounter Gene Expression assay[10] (NanoString, Seattle, WA). The custom codeset contained genes that were found to be significantly affected by PEG-IFN (group B) in the microarray data (> 3-fold change), genes that were shown to affect HCV in a culture model[2], and the genes for interferons α, β, γ and λ (Supplementary Table 1). 100 ng of total RNA from the liver biopsy samples were utilized. Results were normalized by the expression of 4 housekeeping genes.
RNA from frozen PBMCs was extracted using RNeasy Mini (Qiagen) and cDNA was synthesized using First-Strand cDNA (Origene, Rockville, MD). Expression of candidate genes was measured using quantitative PCR with FastStart TaqMan Probe Master mix (Roche, Indianapolis, IN) and primers (Integrated DNA Technologies, Coralville, IA).
Cytokine Measurements
Serum cytokine levels were measured on thawed serum samples using cytometric bead array (BD Biosciences, San Jose, CA).
Viral and Biochemical Kinetics
Serum HCV-RNA level was measured using a COBAS AmpliPrep/COBAS TaqMan HCV Test with a lower limit of detection of 10 IU/ml and lower limit of quantification of 25–43 IU/ml. Virological and biochemical responses to RBV monotheraphy were defined to capture responses that are significantly greater than the natural variation in viral levels in untreated patients. Using the 90% percentile of variations in viral levels and ALT in controls as a reference, we defined virological response to RBV as ≥0.5 log10 decline from baseline (week −4) to time 0, or ≥0.3 log10 decline at both week −1 and time 0. In the same fashion biochemical response to RBV monotherapy was defined as >33% decline in ALT activity from week −4 to week 0. The 1st phase decline after PEG-IFN was defined as the logarithmic decline in viral level from time 0 to the nadir during the first three days post injection. The second phase slope was the slope of the viral level decline from week 1 to week 4, or to the first non-quantifiable measurement if it occurred earlier. Non-quantifiable or non-detectable measurements were imputed conservatively as the lower limit of quantification or detection, respectively. Rapid virological response (RVR) was defined as a negative serum HCV-RNA on week 4 of combination therapy. Early virological response (EVR) was defined as ≥2 log10 decline from time 0 or undetectable virus at week 12. A sustained virological response (SVR) was defined as a negative HCV-RNA result at least 24 weeks after stopping treatment.
End Points and Statistical Analyses
The study was designed to compare the effects of RBV on viral levels and hepatic gene expression before and after the addition of PEG-IFN. We did not hypothesize that ribavirin pre-treatment would affect overall clinical response to treatment and therefore, achievement of SVR was not a primary end-point.
Microarray and nCounter data were analyzed using Partek Genomics Suite 6.5 (Partek Inc., St. Louis, MO) using a 2-way ANOVA for treatment-group assignment and presence of cirrhosis. Pathway analysis was performed using IPA (Ingenuity Systems, Redwood City, CA). Statistical analyses were performed using SPSS Statistics v. 19 (IBM), Prism v. 5.0a (GraphPad Software, La Jolla, CA) and Excel 2008 (Microsoft). Non-parametric tests were used when appropriate. Significance level for all analyses was set at α<0.05. The false detection rate (FDR) method[11] was used to correct for multiple comparisons in microarray and the nCounter assay with q<0.05.
RESULTS
Seventy patients were enrolled between September 2008 and September 2011 (Table 1). The groups were well balanced for most parameters, apart from cirrhosis, which was more frequent in group D.
Table 1.
patient characteristics
| Group A (n=17) |
Group B (n=18) |
Group C (n=18) |
Group D (n=17) |
p-value | |
|---|---|---|---|---|---|
| Age | 52.2±9.5 | 52.6±7.8 | 48.3±12.6 | 53±8.5 | 0.461 |
| Female Gender (n [%]) | 9 (53%) | 6 (33%) | 6 (33%) | 11 (65%) | 0.172 |
| Race/Ethnicity (n [%]) | 0.652 | ||||
| Non-Hispanic White | 9 (53%) | 9 (50%) | 10 (56%) | 10 (59%) | |
| Asian | 3 (18%) | 3 (17%) | 4 (22%) | 4 (24%) | |
| African-American | 3 (18%) | 6 (33%) | 3 (17%) | 2 (12%) | |
| Hispanic | 2 (12%) | 0 (0%) | 1 (6%) | 0 (0%) | |
| Mixed | 0 (0%) | 0 (0%) | 0 (0%) | 1 (6%) | |
| BMI (kg, mean±SE) | 27±5 | 32±8 | 29±5 | 30±5 | 0.181 |
| HCV-RNA (Log10IU/ml, mean±SE) | 6.5±0.4 | 6.1±0.7 | 6.1±0.76 | 6.2±0.6 | 0.421 |
| Viral Genotype | 0.842 | ||||
| 1 | 10 (59%) | 9 (50%) | 9 (50%) | 10 (59%) | |
| 2/3 | 6 (35%) | 6 (33%) | 6 (33%) | 5 (29%) | |
| 4 | 1 (6%) | 3 (17%) | 1 (6%) | 1 (6%) | |
| 6 | 0 (0%) | 0 (0%) | 2 (11%) | 1 (6%) | |
| ALT (U/L) | 66±33 | 116±71 | 112±85 | 78±46 | 0.071 |
| Cirrhosis | 1 (6%) | 2 (11%) | 3 (17%) | 8 (47%) | 0.0262 |
ANOVA
Chi-square
Virological and Biochemical Response to Ribavirin
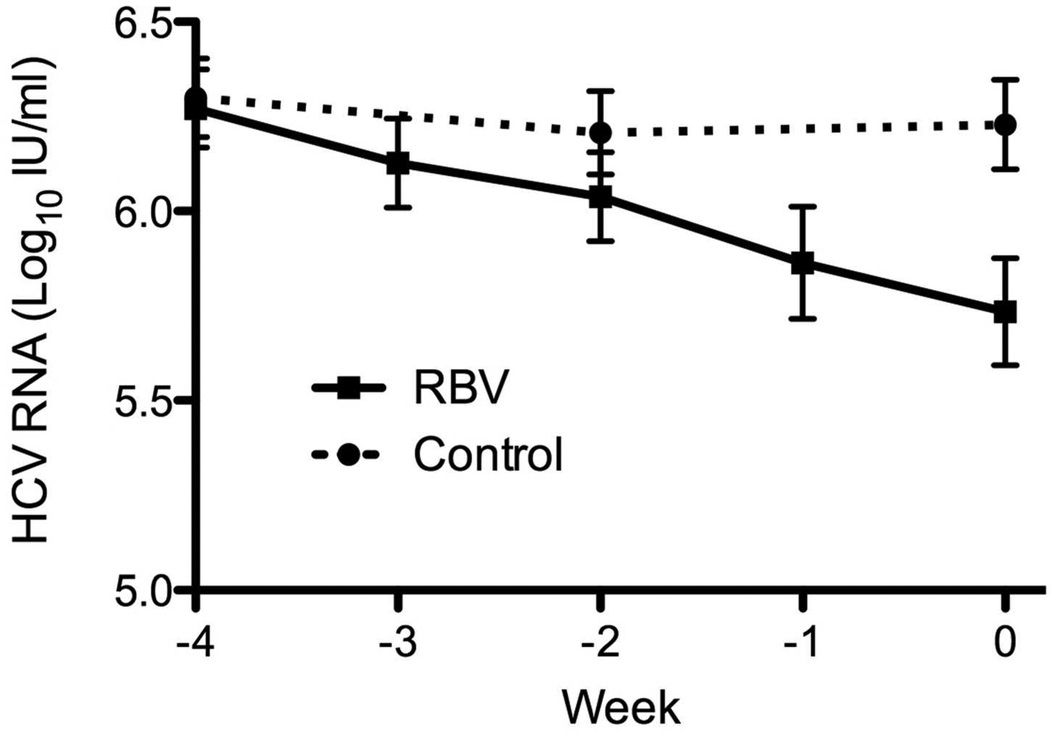
During the 4 weeks leading to combination PEG-IFN and RBV, serum HCV-RNA levels in control patients (groups A and B) did not change (6.3±0.6 log10IU/ml at week −4 vs. 6.3±0.7 at time 0, p=0.51, paired T test). In contrast, patients who were pre-treated with RBV monotherapy (groups C and D) displayed a decline in viral levels over the 4 weeks of pre-treatment at a rate of 0.13±0.13 log10/week (p<0.0001, repeated-measures ANOVA) (Figure 2a). At time 0, the viral level for patients treated with RBV was 0.55±0.53 log10 lower than their level at baseline (p<0.001, paired T-test), and was also lower than that of untreated patients at time 0 (p=0.009, T-test) (Figure 2a). Nineteen treated patients (58%) met the criterion for virological response to RBV with an average virological decline of 0.83±0.53 log10 over 4 weeks of RBV therapy; there was large between-patient variability in temporal pattern and magnitude of response, including patients demonstrating early and late response, primary non-response and a pattern of rapid response followed by a virological rebound (Supplementary Figure 1). In contrast, only 2 (6%) of untreated patients met the criterion for virological response, both of whom had marked fluctuations of HCV-RNA levels at baseline.
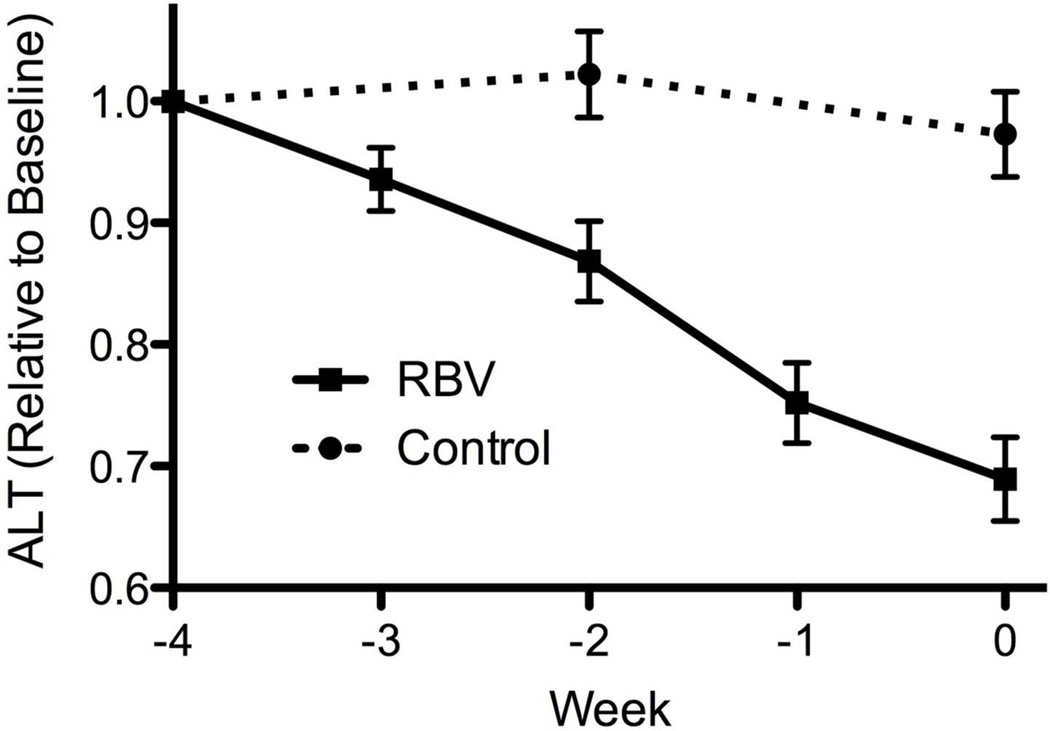
Figure 2.
Effect of ribavirin monotherapy for 4 weeks on serum levels of HCV-RNA (A) and ALT activity (B). Solid line – ribavirin treatment. Dotted line – untreated controls. Mean ± Standard Error (SE).
No baseline factor was associated with the virological decline in response to RBV, including age, gender, race, BMI, baseline HCV-RNA, ALT and fibrosis. The average decline from week −4 to time 0 was numerically greater in patients with genotype 2/3 infection, compared to genotype 1, but was not statistically significant (−0.76±0.76 vs. −0.49±0.37 log10, p=0.32, T-test).
RBV monotherapy was also associated with a linear decline in serum ALT activity with a slope of 10 U/L per week (p<0.0001, repeated-measures ANOVA). During the 4 weeks, ALT levels decreased by 31±20% in RBV pre-treated patients (mean decreasing from 98 to 60 U/L) but did not change in controls (3±21%, p<0.001, T-test, mean decreasing from 90 to 87 U/L)(Figure 2b). A biochemical response was achieved in 14 (40%) of the patients pre-treated with RBV. The relative decline in ALT during RBV pre-treatment was greater for patients with lower baseline viral level (r=0.58, p<0.001) and higher baseline ALT activity (r=−0.54, p=0.001), but was not associated with any other baseline factor, including viral genotype (p=0.21). There was no correlation between biochemical and virological responses (Supplementary Figure 2a).
Hemoglobin levels decreased after 4 weeks of RBV by 2.3±1.2 g/dl. There was no need for dose reduction during this phase. Hemoglobin decrease was not associated with the decrease in HCV-RNA but was positively, albeit weakly, correlated with the decrease in ALT (Spearman’s rho=0.36, p=0.046, Supplementary Figure 2b).
Effect of RBV Pre-Treatment on Subsequent Response to PEG-IFN and RBV
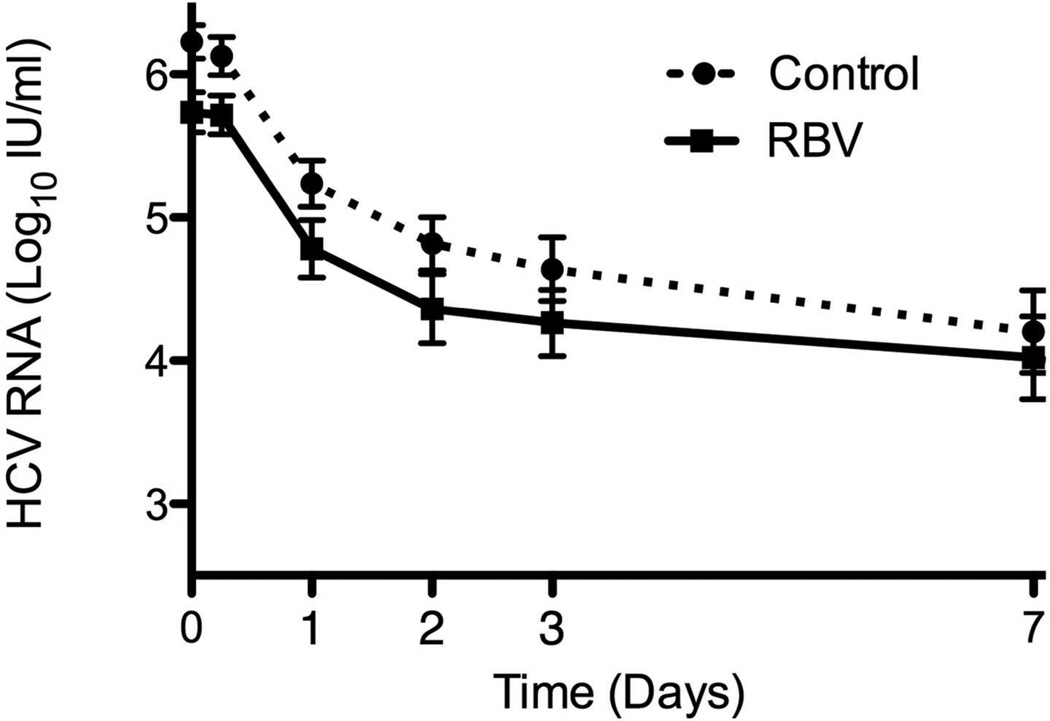
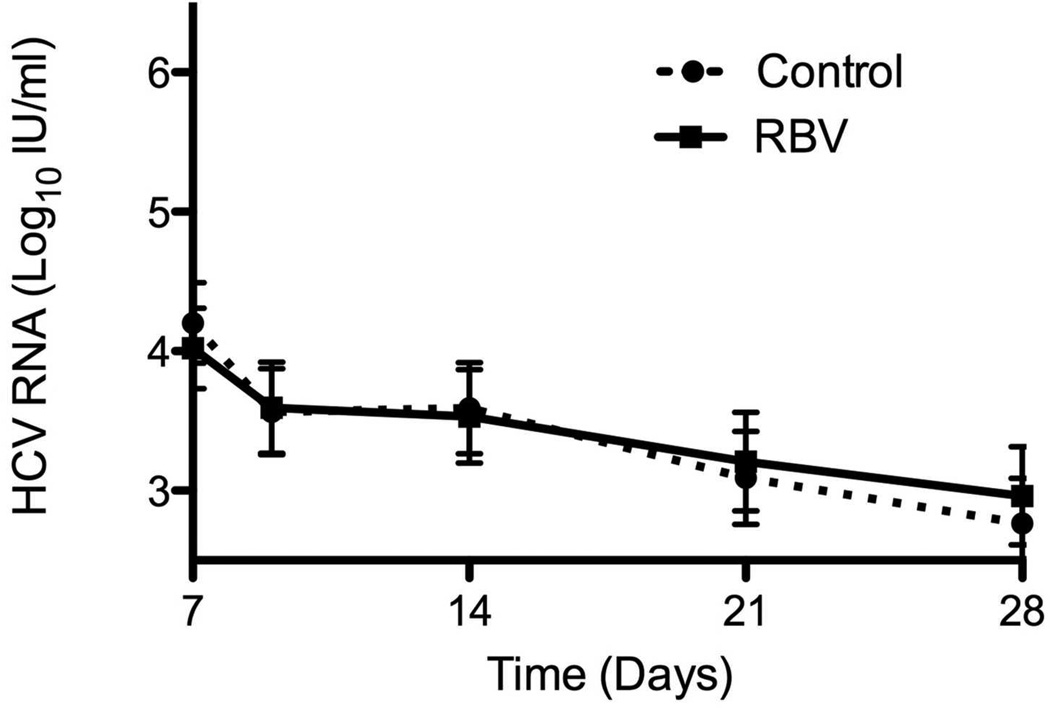
Virological response to PEG-IFN and RBV combination was not affected by RBV pretreatment (Table 2). At the start of combination treatment, viral levels were lower by 0.5 log10 in patients who received RBV, as a result of prior monotherapy. This difference persisted until day 7 of treatment, after which no further difference was seen (Figure 3a-b). RBV pre-treatment had no effect on rates of RVR, EVR or SVR (Table 2). The results were similar when analysis was limited to patients with genotype 1 infection (Table 2) and when the analysis was controlled for the presence of cirrhosis (data not shown).
Table 2.
Parameters of early viral kinetics by PEG-IFN treatment and treatment response
| All Patients | Virological Response to RBV Pre- treatment5 |
Biochemical Response to RBV Pre treatment 5 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | RBV Pre- treatment |
p | Non- Responder6 |
Responder6 | p | Non- Responder7 |
Responder7 | p | |
| 1st phase decline (log10) | 1.7±0.9 | 1.5±0.9 | 0.441 | 1.2±0.9 | 1.8±0.9 | 0.081 | 1.3±0.8 | 1.9±0.9 | 0.061 |
| 2nd phase slope (log10/week) | −0.52±0.3 | −0.52±0.5 | 0.981 | −0.57±0.64 | −0.47±0.32 | 0.621 | −0.48±0.57 | −0.62±0.28 | 0.561 |
| Rapid Virological Response (n, %)3 | 16 (46%) | 14 (44%) | 12 | 5 (39%) | 9 (47%) | 0.732 | 4 (22%) | 10 (71%) | 0.012 |
| Early Virological Response (n, %)3 | 30 (88%) | 27 (82%) | 0.512 | 10 (71%) | 17 (90%) | 0.362 | 13 (68%) | 14 (100%) | 0.032 |
| Sustained Virological Response (n, %)3,4 | 16 (52%) | 17 (59%) | 0.612 | 6 (50%) | 11 (65%) | 0.472 | 7 (41%) | 10 (83%) | 0.0532 |
| Genotype 1 Only | Virological Response to RBV Pre- treatment5 |
Biochemical Response to RBV Pre- treatment5 |
|||||||
| Control | RBV Pre- treatment |
p | Non- Responder6 |
Responder6 | p | Non- Responder7 |
Responder7 | p | |
| 1st phase decline (log10) | 1.4±0.9 | 1.1±0.6 | 0.211 | 0.7±0.3 | 1.4±0.7 | 0.021 | 1.0±0.7 | 1.1±0.5 | 0.951 |
| 2nd phase slope (log10/week) | −0.45±0.3 | −0.39±0.3 | 0.601 | −0.36±0.33 | −0.43±0.37 | 0.671 | −0.32±0.37 | −0.55±0.22 | 0.191 |
| Rapid Virological Response (n, %)3 | 5 (26%) | 3 (17%) | 0.692 | 1 (11%) | 2 (22%) | 0.532 | 1 (8%) | 2 (33%) | 0.182 |
| Early Virological Response (n, %)3 | 15 (79%) | 13 (68%) | 0.712 | 6 (60%) | 7 (78%) | 0.412 | 7 (54%) | 6 (100%) | 0.0442 |
| Sustained Virological Response (n, %)3,4 | 7 (41%) | 7 (41%) | 1.02 | 3 (38%) | 4 (44%) | 0.772 | 3 (25%) | 4 (80%) | 0.0362 |
Student’s t-test
Fisher’s exact test
Modified intention-to-treat limited to patients who received at least one dose of PEG-IFN
Limited to patients who completed at least 48 (genotypes 2/3) or 72 weeks (other genotypes) of follow-up (n=60) at time of writing.
Limited to patients pre-treated with RBV
Virological response defined as HCV-RNA decline > 0.5 log10 from week −4 to week 0, or decline greater than >0.3 log10 on both weeks −1 and 0
Biochemical response defined as ALT decline > 33% from week −4 to week 0
Figure 3.
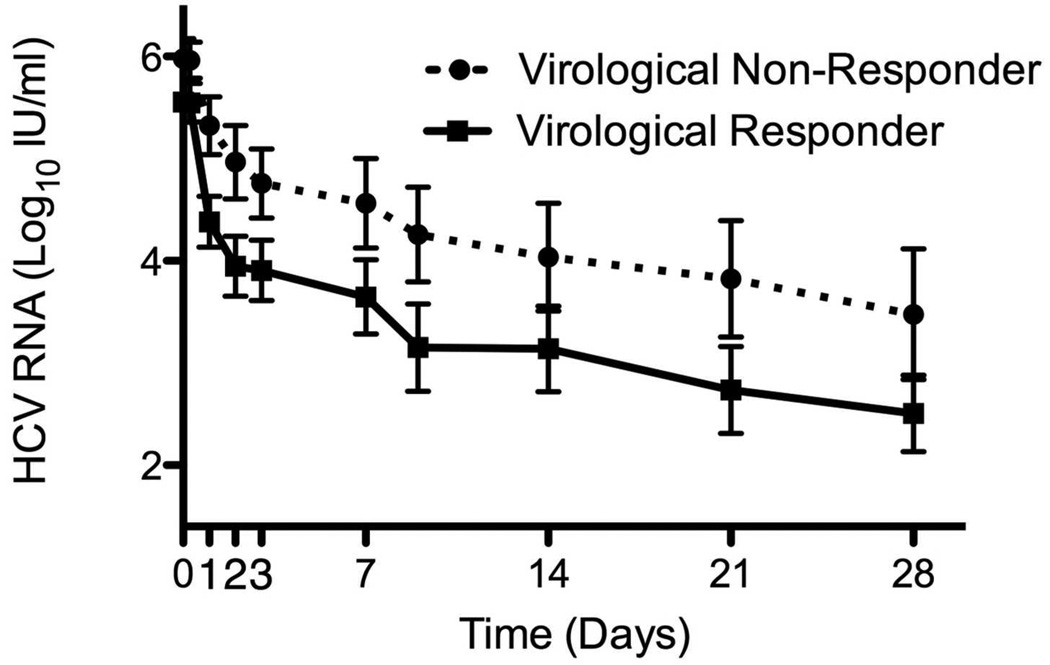
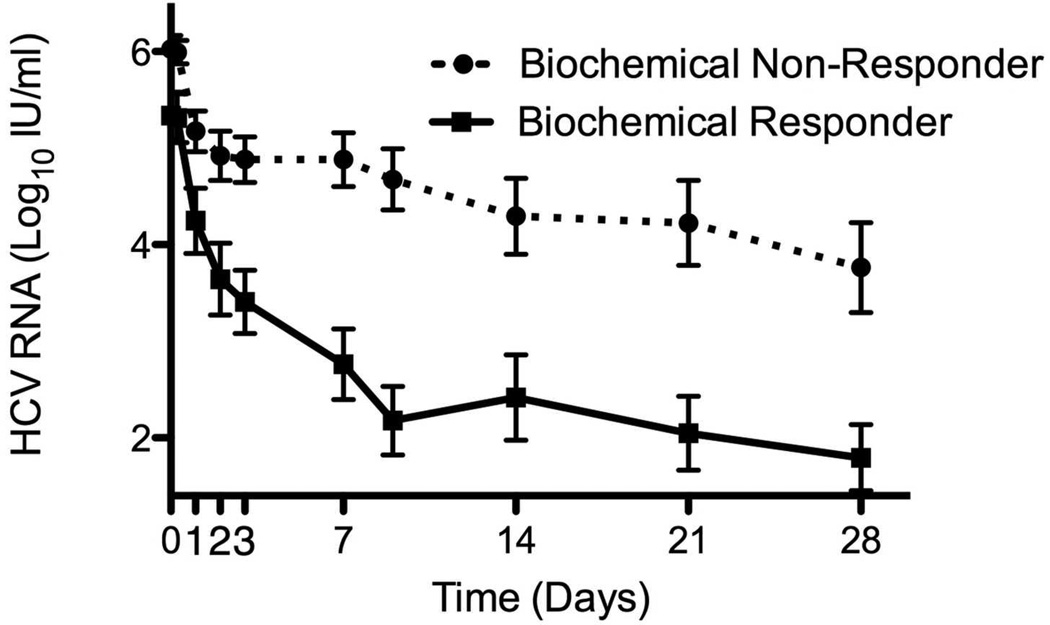
Virological response to PEG-IFN and RBV of patients pre-treated with 4 weeks of RBV monotherapy and controls during the first 7 days (A) and weeks 1 to 4 (B) of combination treatment. Virological response to Peg-IFN and RBV combination therapy stratified by virological (C) and biochemical (D) response to previous RBV monotherapy. Means ± SE.
Although RBV pre-treatment itself did not affect response to subsequent combination therapy, among the patients who were pre-treated with RBV, virological response to pre-treatment was associated with slightly better viral kinetics after the addition of PEG-IFN, compared to virological non-responders, but the difference was not significant, except for 1st phase decline in patients with genotype 1 (Table 2, Figure 3c). Biochemical response to RBV, on the other hand, was significantly associated with improved virological response to subsequent combination treatment (Table 2, Figure 3d). Patients whose ALT decreased by >33% on RBV monotherapy had higher rates of RVR, EVR and SVR and multivariable logistic regression showed biochemical, but not virological response to RBV pretreatment to be associated with RVR (Odds ratio=9.4, 95% CI 1.8–48.5, p=0.008 for biochemical and OR=1.8 [0.3–9.8], p=0.49 for virological response) and with SVR (OR=7.6 [1.2–48.1], p=0.031 for biochemical and OR=2.1 [0.4–11.6], p=0.38 for virological response). Similar findings were seen when the analysis of SVR predictors was limited to genotype 1 patients (OR=2.6 [0.2–32.1], p=0.46 for virological response to RBV pretreatment, OR=16 [0.97–268], p=0.053 for biochemical response).
Effect of RBV on Hepatic Gene Expression
Hepatic gene expression was assessed by microarray in 52 patients (15 in group A, 14 in B, 11 in C and 12 in group D). Samples were not available from 18 patients; in 4 (2 each from groups C and D) the biopsy was cancelled because of RBV-induced anemia as pre-specified by the protocol, 2 patients (C and D) refused biopsy, 7 had insufficient samples for analysis (1, 2, 2 and 2 subjects from treatment group A, B, C and D, respectively) and 5 were enrolled after the microarray analysis was done. As expected, we found a significant effect of PEG-IFN on hepatic gene expression; 314 probes, reflecting 243 unique transcripts, were significantly different between group B and group A, with 213 transcripts up- and 30 down-regulated (Supplementary Table 2). In contrast, no significant effect on hepatic gene expression was detected for RBV alone compared to controls (group C vs. group A) nor for adding RBV to PEG-IFN compared to PEG-IFN alone (group D vs. group B).
To determine whether the magnitude of treatment-response to RBV was related to gene expression profiles, we analyzed hepatic gene response after 4 weeks of RBV pre-treatment (group C only) and compared patients who had a virological response to RBV pre-treatment (n=8) to those who did not (n=3). No genes were significantly different in expression using the strict significance FDR criteria. Using a less stringent criterion (p<0.001), only 8 of 54,613 microarray targets were different between the two groups, and these were not able to correctly cluster patients according to treatment assignment (Supplementary Figure 3a-b). In contrast, when comparing biochemical responders (n=4) within group C to non-responders (n=7), 48 transcripts (Supplementary Table 3) were differentially expressed using the less stringent criterion and these genes clustered patients into two distinct groups, within which patients were clearly separated according to treatment assignment (Supplementary Figure 3c-d). Pathway analysis did not identify a specific pathway associated with these transcripts.
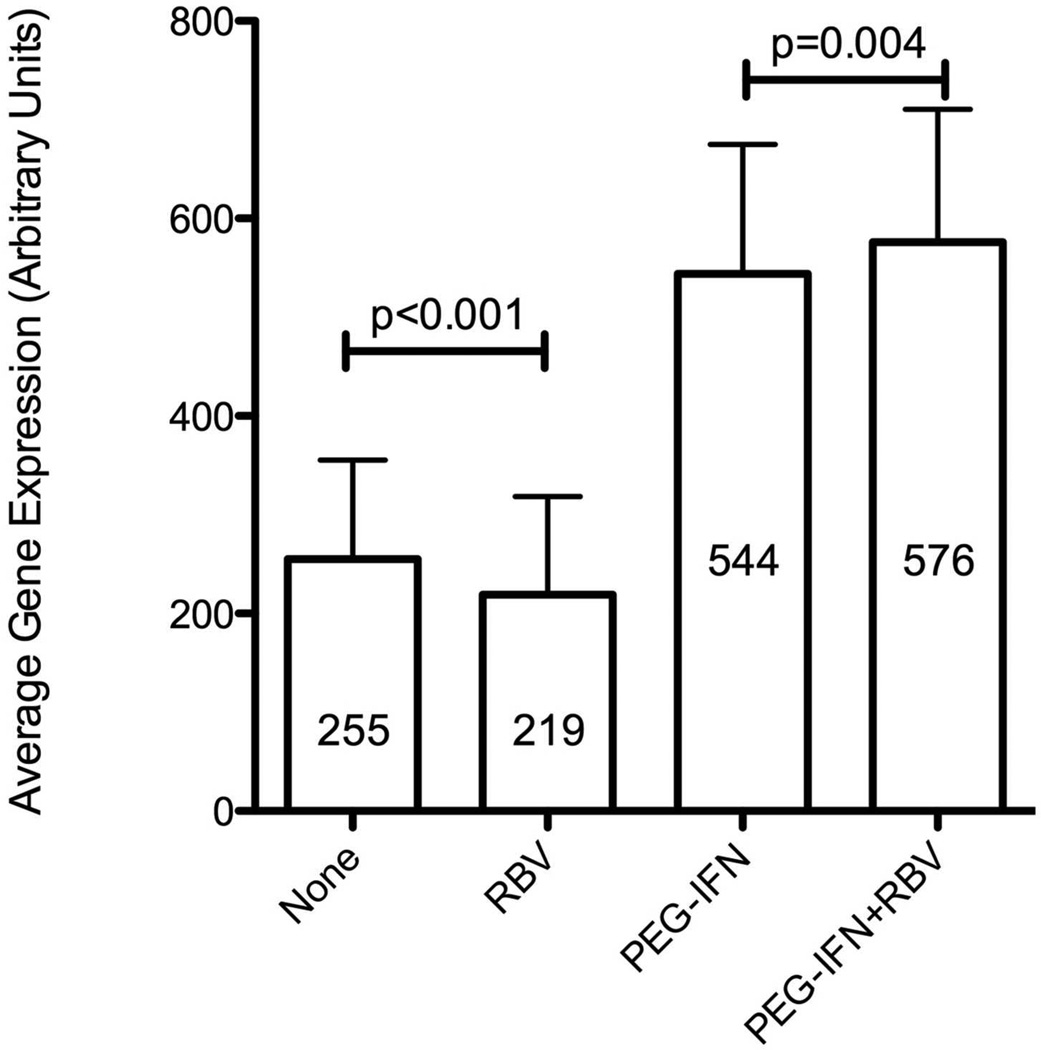
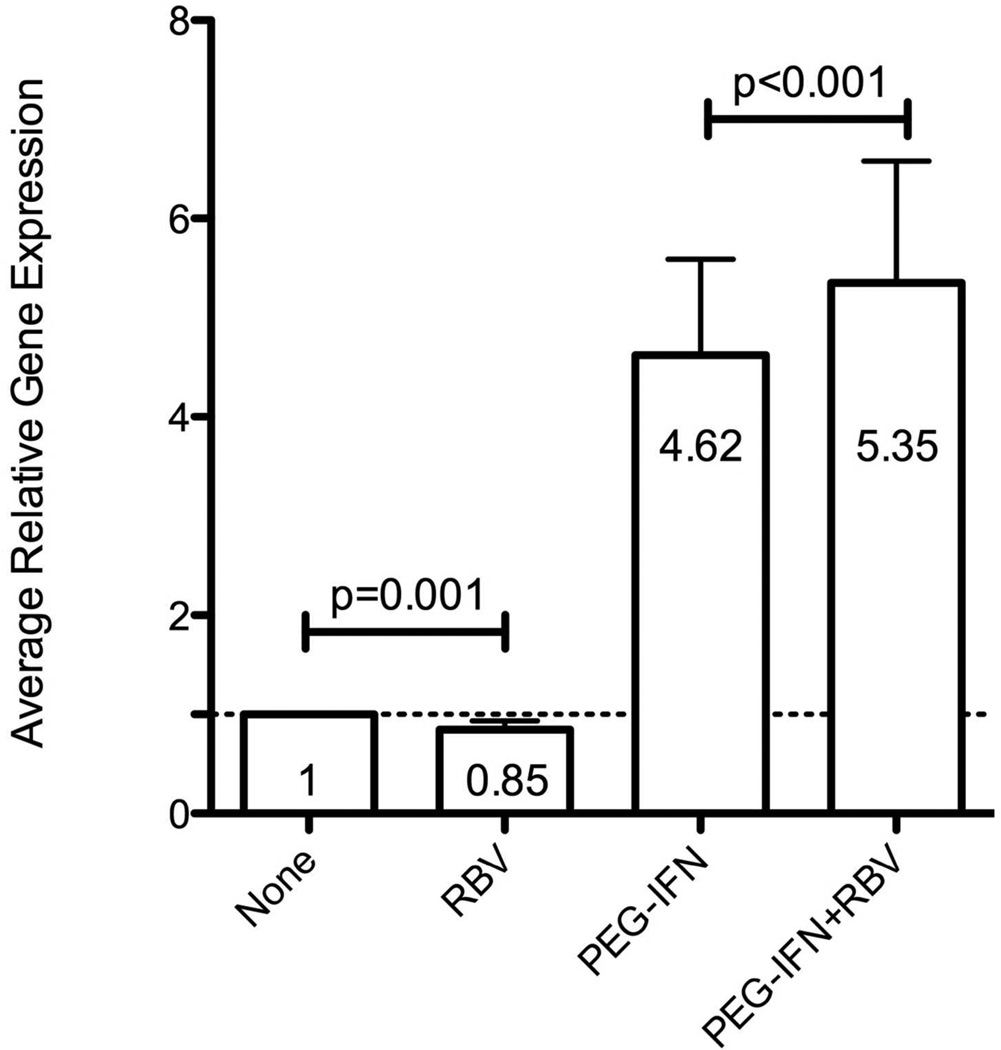
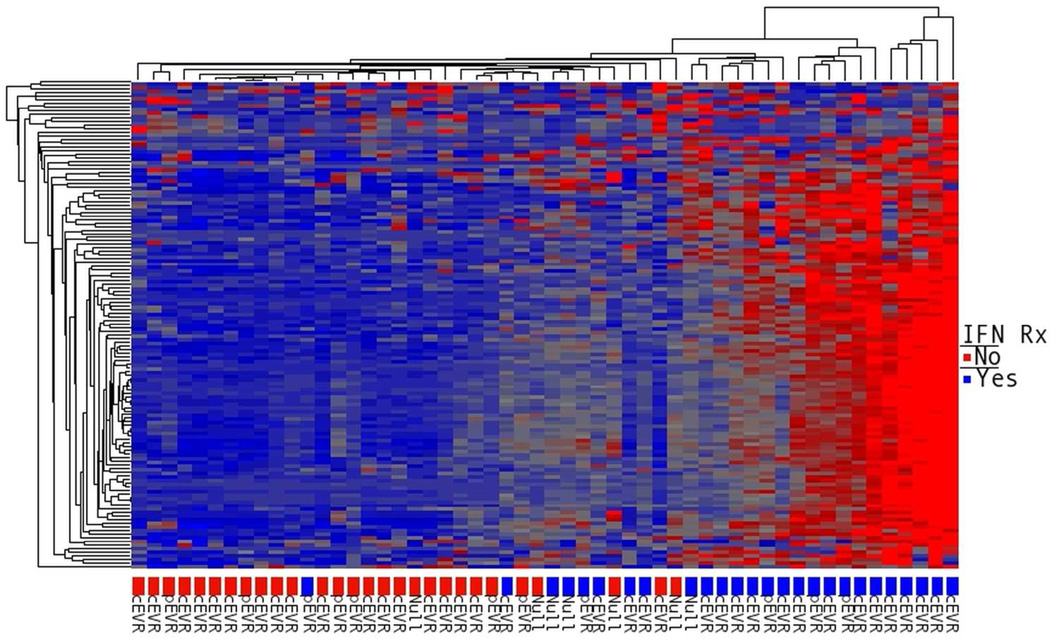
Although RBV did not affect overall hepatic gene expression, we tested whether it had a specific effect on hepatic ISG expression. We utilized the nCounter Gene Expression assay to analyze the expression of 135 ISGs in liver biopsy samples (Supplementary Table 4) from 54 patients (16 from group A, 14 group B, 12 group C and 12 from group D), including 51 that were analyzed by microarray (one excluded because of insufficient RNA) and 3 additional patients who enrolled after the microarray analysis. When comparing PEG-IFN in combination with RBV to PEG-IFN alone (group D vs. B), the overall hepatic ISG expression, obtained by averaging the individual ISG expression levels, was higher for the combination group (Figure 4, p<0.001). In contrast, the average expression of ISGs in group C (RBV alone) was lower than in group A (control, p<0.001). No specific ISG could be identified to show a significant induction in these analyses (examples in supplementary Figure 4), either when analyzing data from all of the patients or when limiting it to the 30 patients with genotype 1. The average ISG expression in group D (PEG-IFN/RBV) was 6.6-fold higher than that in group C (RBV alone), while the ratio of average ISG expression between group B (PEG-IFN alone) to group A (controls) was 4.5 (p=0.008, Mann-Whitney), suggesting a stronger induction of ISGs by PEG-IFN in the presence of RBV. Among patients pretreated with RBV (group C only), average and individual ISG expression did not differ between patients with good virological response to RBV and those without it, nor did it differ between biochemical responders and non-responders. Consistent with previous observations[9, 12], expression of ISGs was higher in patients treated with interferon, and patients who subsequently did not respond to treatment had higher ISG expression prior to interferon (Figure 5).
Figure 4.
Average hepatic expression level of 135 ISGs according to treatment at the time of liver biopsy, expressed in arbitrary units (A, mean ± SE) or as fold induction relative to the average expression in controls (B, geometric mean ± 95% confidence intervals, CI). Significance tested using Wilcoxon’s signed-ranks test.
Figure 5.
Hierarchical clustering of patients according to the hepatic expression of 135 ISGs (nCounter assay). The color labels at the bottom mark whether tissue was obtained before the initial PEG-IFN injection (red: groups A & C), or 6 hours after the first injection (blue: groups B & D). Subsequent responses after 12 weeks of combination treatment are labeled beneath color code as cEVR (complete EVR, non-detectable virus at week 12, pEVR (partial EVR, > 2 log10 decline but still detectable HCV-RNA) or Null responder (< 2 log10 decline).
Among the genes for human interferons, PEG-IFN treatment significantly increased the relative hepatic expression of IFNA4 (p=0.01, Mann-Whitney U), IL28A/B (p<0.001) and IL28RA, (p=0.042). Ribavirin numerically decreased the expression of interferon γ with borderline significance (p=0.078) but it had no effect on expression of the other interferon genes, nor did it have any effect on their expression when added to PEG-IFN (Supplementary Figure 5).
ISG Expression in PBMC
Analysis of serial ISG expression in PBMC was performed in 39 patients (20 with RBV pre-treatment and 19 controls). We analyzed 5 genes of interest, based on the microarray results (Supplementary Table 2) and previous literature[8, 12]: CD274, ISG15, IRF7, CXCL10 and RSAD2. At baseline, there were no statistically significant differences between the groups in PBMC gene expression. Following 4 weeks of ribavirin monotherapy, PBMC expression of the 5 genes did not differ between treated patients and controls. After initiating combination treatment with PEG-IFN, the peak levels of the 5 genes were numerically lower in the RBV pre-treated group, but with no statistical significance, and by week 1 essentially became identical (Supplementary Figure 6a-e).
Serum IP-10 Levels
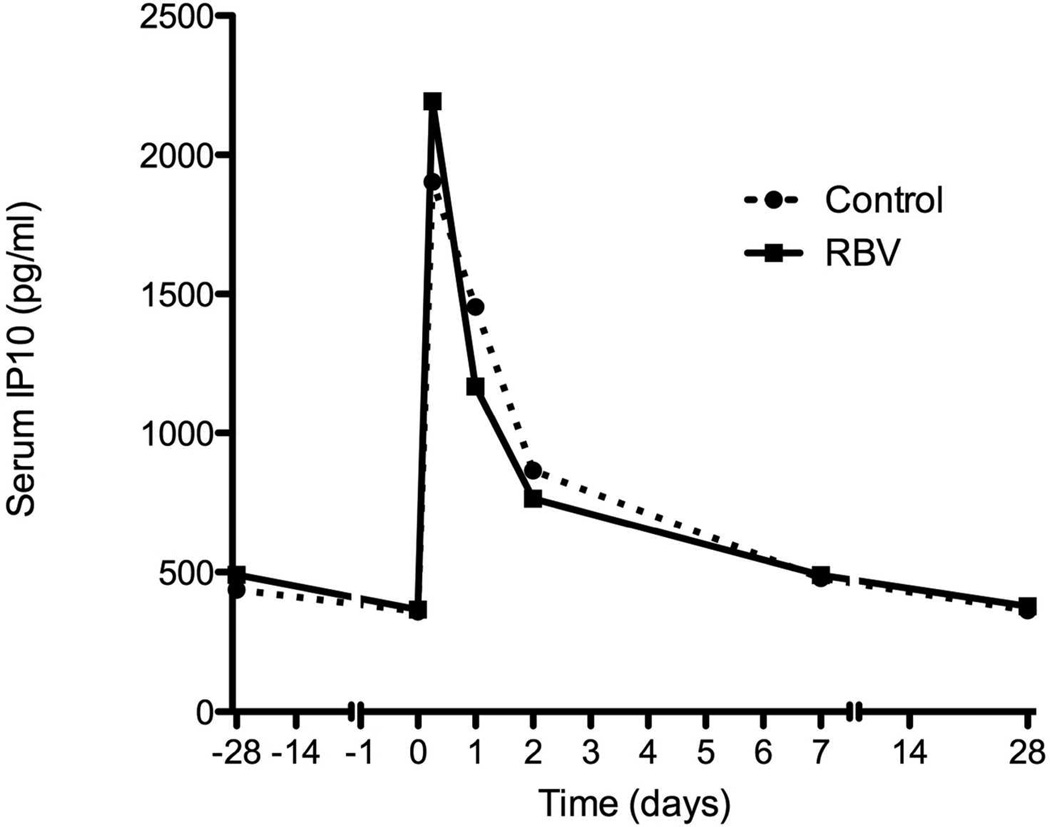
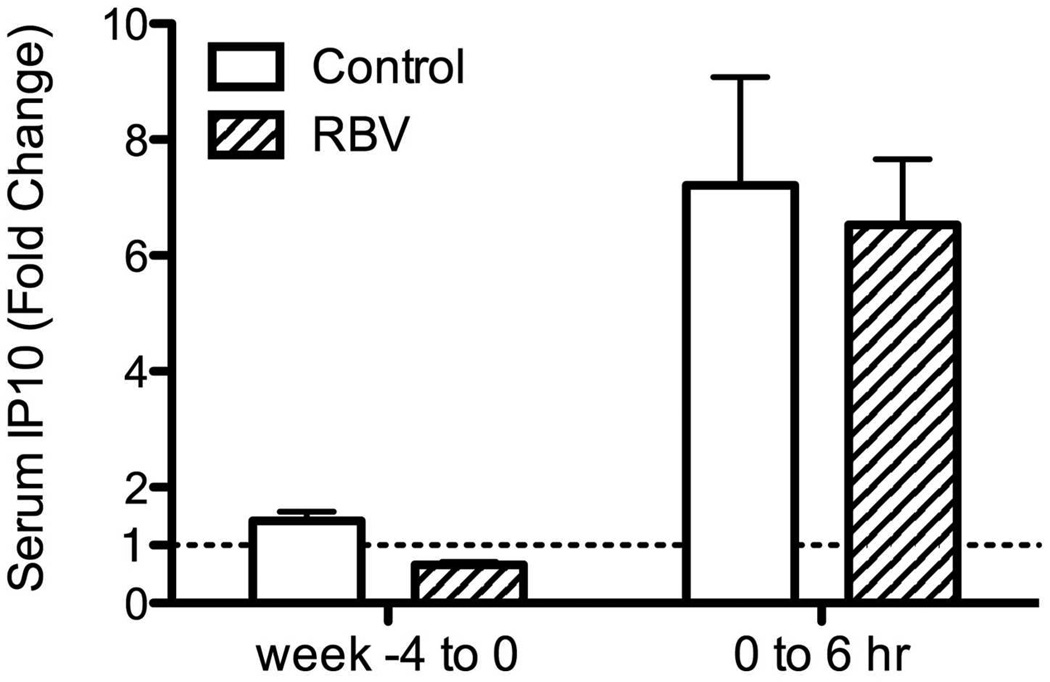
Serum levels of IP-10 were measured in 20 patients treated with RBV and 20 controls. As expected, baseline IP-10 levels were higher in patients who subsequently did not respond to the combination treatment (382±225 pg/ml in patients who achieved RVR vs. 707±439 pg/ml in those who did not, p=0.004, Mann-Whitney). Four weeks of RBV pre-treatment were associated with a decrease in serum IP-10 levels (0.66 fold-change vs. 1.42 in controls, p<0.001, Figure 6a-b). The decrease in IP-10 levels correlated with the decline in HCV-RNA (r=0.74, p=0.006) but not with ALT decline (r=−0.06, p=0.86, Supplementary Figure 7a-b). After starting PEG-IFN treatment, there was a rapid rise in IP-10 levels at 6 hours in both groups (6.5 fold in RBV pre-treated vs. 7.2 fold in controls, p= 0.79, Mann Whitney U), declining back to baseline levels by week 1 (Figure 6a). IP-10 serum levels were closely correlated with hepatic CXCL10 gene expression at the same time (Group A and C at time 0, Spearman rho=0.68, p=0.005; Groups B and D at 6 hours after peginterferon injection, rho=0.72, p=0.001, Supplementary Figure 7c-d). There was no correlation between serum IP-10 levels and PBMC CXCL10 expression at any time point.
Figure 6.
Serial measurements of serum IP10 levels during treatment (A). Solid line – patients treated with RBV monotherapy on weeks −4 to 0. Dotted line – controls with no pre-treatment. (B) Fold change in serum IP10 levels after 4 weeks of RBV or control and 6 hours after injecting interferon.
Effect of Host Genetic Variations
Genotype frequencies of the IL28B single nucleotide polymorphism (SNP) rs12979860 are shown in Supplementary Table 5. IL28B genotype did not modulate the effect of RBV pre-treatment on HCV levels (p=0.61, Supplementary Figure 8). In contrast, patients with rs12979860 CC had a greater decline in ALT after 4 weeks of RBV monotherapy compared to genotypes CT/TT (38±20% vs. 23±17%, p=0.035 Mann Whitney). IL28B genotype did not modify the association between biochemical response to RBV pre-treament and subsequent response to combination therapy. The inosine triphosphatase (ITPA) SNPs rs7270101 and rs1127354, which have been shown to be highly associated with RBV-induced anemia[13], were genotyped but the frequencies of the minor alleles were too low to allow meaningful analysis.
DISCUSSION
Ribavirin is a crucial component of anti-HCV therapy and may remain so even with the addition of direct-acting antivirials[14, 15]. In this study, we sought to determine its mechanism of action and specifically, whether RBV augments ISG induction by interferon. We show that RBV has a modest antiviral effect against HCV in 58% of subjects, with a mean 0.8 log10 decline from baseline in RBV responders. Clearly this effect is minimal and explains why RBV monotherapy is not an effective strategy towards HCV eradication. Consistent with what Pawlotsky et al.[16] described, we observed several patterns of response to RBV monotherapy including continuing response, non-response and response follow by breakthrough (Supplementary Figure 2). However the sampling frequency in our study was not sufficient to detect the transient early virological responses (within week 1 of therapy) reported in that paper.
We also demonstrate a marked decrease in serum ALT during RBV monotherapy, which was independent of the virological effect of RBV. This is consistent with previous reports showing that ribavirin monotherapy decreases ALT in the absence of a virological effect, and that the biochemical changes are correlated with histological improvement[17, 18]. The dissociation between the virological and biochemical response to RBV suggests that RBV may act through more than one mechanism in patients with chronic hepatitis C.
Although the weak effect of RBV on viral levels was completely offset by addition of PEG-IFN, the biochemical response to RBV was associated with subsequent response to interferon-based therapy. Biochemical responders to RBV showed a faster viral decline after adding PEG-IFN compared to non-responders and had higher rates of RVR, EVR and SVR. Furthermore, RBV-induced decrease in hemoglobin, which was previously shown to be associated with treatment response[19, 20], was also correlated with biochemical but not virological response to RBV. Thus, the contribution of RBV to treatment seems to be independent of its direct antiviral activity and may be a result of an indirect effect on liver injury and inflammation.
We were not able to show a significant effect of RBV on overall hepatic gene expression, either alone or in combination with PEG-IFN. However, RBV monotherapy was associated with a lower global expression of ISGs, while addition of RBV to PEG-IFN caused a modest but significant increase in ISG expression. The average fold-increase in ISG expression in the PEG-IFN/RBV group compared to RBV alone was significantly greater than that of PEG-IFN compared to controls. This observation raises the possibility that RBV may act by lowering the baseline ISG expression and resetting the IFN responsiveness in the liver, thereby rendering the liver more permissive to the action of exogenous IFN. This notion is consistent with the concept that a higher baseline ISG expression is associated with IFN non-responsiveness and the relative change in ISG expression is more important than the absolute level in determining downstream effects. As shown in other signaling cascades involving STAT[21], a tight regulation of IFN signaling may be necessary to prevent IFN-refractoriness[22]. One possibility is that the enhanced sensitivity to IFN may be driven by a RBV-induced change in the inflammatory milieu, as reflected by the association of biochemical response to RBV with improved response to subsequent PEG-IFN treatment. As our gene expression studies focused on IFN signaling, we cannot rule out an effect of RBV on other cytokines or signaling cascades.
Our current study can be contrasted with the previous one by Feld et al[9], which showed a significant effect of RBV on hepatic gene expression when added to PEG-IFN. Part of the discrepancy can be explained by differences in study design and analysis. First, in our study we used 4 weeks of RBV monotherapy to ensure achievement of near steady-state RBV blood levels[16, 23], as opposed to only 3 days by Feld et al. Since ALT and viral levels declined during the 4 week period, hepatic gene expression including ISGs could have changed as well, a possibility that is supported by decreasing serum IP10 levels with RBV monotherapy (Figure 6a, Supplementary Figure 7a). This opposing effect could offset a direct positive effect of RBV on ISG expression, particularly when our biopsy was obtained at week 4. Second, Feld et al examined liver biopsies taken 24 hours after the initial PEG-IFN injection, while in this study, a 6-hour time point was used, to coincide with peak induction of ISGs[24] (as can also be seen in PBMC samples in Supplementary Figure 6). Theoretically, if RBV augmented hepatic induction of ISGs by affecting the duration of induction and not the peak level, our biopsies would have failed to detect such an effect.
In lieu of performing serial liver biopsies, we studied the effect of RBV over time on the mRNA expression of 5 selected ISGs in PBMCs[25, 26] and on the serum protein levels of IP-10, a known ISG. We did not find any effect of RBV on the magnitude and temporal pattern of IFN-induced ISG expression in the PBMCs. IP-10 levels declined during treatment, likely driven by the decline in viral levels, but here as well, the IFN-induced changes were not affected by RBV pre-treatment. Finally, the biochemical but not virological response to RBV was associated with host IL28B polymorphism, suggesting a possible link between RBV effect and IFN response in treatment of hepatitis C.
When given in combination, it is very difficult to separate the effects of RBV from those of IFN. We therefore used RBV monotherapy as a tool to understand the mechanism of action of RBV and dissect its specific contribution to combined treatment. We have not observed a clinical benefit with RBV pre-treatment but our study was not powered, nor was it designed to detect a small clinical effect. However, there was also no effect of pre-treatment on early viral kinetics and serum IP10 once PEG-IFN was started. Taken together, these suggest that the biochemical response to RBV did not prime patients to respond to subsequent combination but instead, served as a marker to identify a subset of “RBV responders” that could not be identified directly without the use of monotherapy. Whether the weak effect of RBV on hepatic ISG expression is the mechanism by which RBV exerts its biochemical effect remains unclear.
A small uncontrolled pilot study, published in abstract form only[27], examined 4 weeks of RBV pre-treatment in 10 genotype 1 patients who previously failed PEG-IFN and RBV treatment. They observed a similar decline in HCV-RNA levels after 4 weeks of treatment and demonstrated a clinical benefit, with 50% of patients achieving SVR, a rate that is higher than what we achieved in our treatment-naïve genotype 1 patients, irrespective of RBV pre-treatment. The discrepant clinical results could be due to different demographics.
In conclusion, our findings support the existence of more than one mechanism for RBV in treatment of chronic hepatitis C, including a direct antiviral effect and an indirect biochemical effect, potentially through reduction of liver injury through an unknown mechanism. We show that the contribution of RBV to combination treatment seems to be related more to its indirect effect than to its antiviral activity, possibly by resetting the liver’s inflammatory state and making it more responsive to exogenous PEG-IFN. Further studies are necessary to elucidate this indirect effect, which may provide crucial insight into the mechanism of action of RBV in treatment of hepatitis C.
Supplementary Material
SUMMARY BOX.
What is known about this subject?
Addition of ribavirin improves rates of sustained virological response in patients with chronic hepatitis C treated with interferon or pegylated interferon
Ribavirin alone exerts a weak, and not always sustained, antiviral effect on hepatitis C virus
In vitro data suggests ribavirin may augment stimulation of gene expression by interferon
What are the new findings?
Virological response to ribavirin monotherapy does not predict response to peginterferon/ribavirin combination
Biochemical response to ribavirin is not correlated with the virological response and is associated with clinical response to peginterferon/ribavirin combination, suggesting two separate mechanisms
Hepatic expression of interferon-stimulated genes is weakly enhanced with peginterferon/ribavirin combination compared to peginterferon alone
How might it impact clinical practice in the foreseeable future?
Better understanding of the mechanism of action may allow selective use of ribavirin in the direct-acting antivirals era
Acknowledgments
Support: The study was supported by the intramural research program of NIDDK; NIH grants R01-OD011095, R37-AI028433, R01-AI078881, and P20-RR018754; and the University of Illinois Walter Payton Liver Center GUILD.
Abbreviations
- BMI
body mass index
- EVR
early virological response
- FDR
false detection rate
- HCV
hepatitis C virus
- ISG
interferon-stimulated gene
- PBMC
peripheral blood mononuclear cell
- PEG-IFN
peginterferon
- RBV
ribavirin
- RVR
rapid virological response
- SNP
single nucleotide polymorphism
- SVR
sustained virological response
Footnotes
Author Contributions
Study Concept & Design: YR, JJF, TH, MGG, ED, ET, BR, JHH, TJL
Acquisition of Data: YR, MN, JJF, MW, HH, YJP, SYP, CK, AA, NG, SS, ET, GA, BE, RT, LH, OE
Analysis and Interpretation of Data: YR, MN, JG, HD, ASP, JHH, TJL
Statistical Analysis: YR, JG, HD
Drafting of the Manuscript YR, TJL
Critical Review of the Manuscript: All authors
Guarantor:YR
REFERENCES
- 1.Ghany MG, Nelson DR, Strader DB, et al. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433–1444. doi: 10.1002/hep.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 4.Di Bisceglie AM, Conjeevaram HS, Fried MW, et al. Ribavirin as therapy for chronic hepatitis C. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 1995;123:897–903. doi: 10.7326/0003-4819-123-12-199512150-00001. [DOI] [PubMed] [Google Scholar]
- 5.Brillanti S, Mazzella G, Roda E. Ribavirin for chronic hepatitis C: and the mystery goes on. Dig Liver Dis. 2011;43:425–430. doi: 10.1016/j.dld.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 7.Thomas E, Ghany MG, Liang TJ. The Application and Mechanism of Action of Ribavirin in Therapy of Hepatitis C. Antivir Chem Chemother. 2012 doi: 10.3851/IMP2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas E, Feld JJ, Li Q, et al. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology. 2011;53:32–41. doi: 10.1002/hep.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feld JJ, Nanda S, Huang Y, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 12.Chen L, Borozan I, Feld J, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 13.Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 14.Zeuzem S, Buggisch P, Agarwal K, et al. The protease inhibitor GS-9256 and non-nucleoside polymerase inhibitor tegobuvir alone, with RBV or peginterferon plus RBV in hepatitis C. Hepatology. 2011 doi: 10.1002/hep.24744. [DOI] [PubMed] [Google Scholar]
- 15.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. The New England journal of medicine. 2010;362:1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky JM, Dahari H, Neumann AU, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126:703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Bodenheimer HC, Jr, Lindsay KL, Davis GL, et al. Tolerance and efficacy of oral ribavirin treatment of chronic hepatitis C: a multicenter trial. Hepatology. 1997;26:473–477. doi: 10.1002/hep.510260231. [DOI] [PubMed] [Google Scholar]
- 18.Hoofnagle JH, Ghany MG, Kleiner DE, et al. Maintenance therapy with ribavirin in patients with chronic hepatitis C who fail to respond to combination therapy with interferon alfa and ribavirin. Hepatology. 2003;38:66–74. doi: 10.1053/jhep.2003.50258. [DOI] [PubMed] [Google Scholar]
- 19.Dahari H, Markatou M, Zeremski M, et al. Early ribavirin pharmacokinetics, HCV RNA and alanine aminotransferase kinetics in HIV/HCV co-infected patients during treatment with pegylated interferon and ribavirin. J Hepatol. 2007;47:23–30. doi: 10.1016/j.jhep.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulkowski MS, Shiffman ML, Afdhal NH, et al. Hepatitis C virus treatmentrelated anemia is associated with higher sustained virologic response rate. Gastroenterology. 2010;139:1602–1611. doi: 10.1053/j.gastro.2010.07.059. 11 e1. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Huang J, Dasgupta M, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarasin-Filipowicz M, Wang X, Yan M, et al. Alpha interferon induces longlasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–4851. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slavenburg S, Huntjens-Fleuren HW, Dofferhoff TS, et al. Ribavirin plasma concentration measurements in patients with hepatitis C: early ribavirin concentrations predict steady-state concentrations. Ther Drug Monit. 2011;33:40–44. doi: 10.1097/FTD.0b013e318205f892. [DOI] [PubMed] [Google Scholar]
- 24.Lanford RE, Guerra B, Lee H, et al. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 25.Ji X, Cheung R, Cooper S, et al. Interferon alfa regulated gene expression in patients initiating interferon treatment for chronic hepatitis C. Hepatology. 2003;37:610–621. doi: 10.1053/jhep.2003.50105. [DOI] [PubMed] [Google Scholar]
- 26.Taylor MW, Tsukahara T, Brodsky L, et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from nonresponders to antiviral therapy. J Virol. 2007;81:3391–3401. doi: 10.1128/JVI.02640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brillanti S, Buonfiglioli F, Feletti V, et al. Ribavirin Priming Enhances Efficacy of Chronic Hepatitis C Re-Treatment in Patients Who Had Not Responded to Previous Combination Therapy. Hepatology. 2009;50:317A-A. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.