Abstract
Purpose
The success of immunotherapy for the treatment of metastatic cancer is contingent on the identification of appropriate target antigens. Potential targets must be expressed on tumors but demonstrate restricted expression on normal tissues. To maximize patient eligibility, ideal target antigens should be expressed on a high percentage of tumors within a histology and potentially in multiple different malignancies.
Design
A Nanostring probe set was designed containing 97 genes, 72 of which are considered potential candidate genes for immunotherapy. Five established melanoma cell lines, 59 resected metastatic melanoma tumors and 31 normal tissue samples were profiled and analyzed using Nanostring technology.
Results
Of the 72 potential target genes, 33 were overexpressed in over 20% of studied melanoma tumor samples. Twenty of those genes were identified as differentially expressed between normal tissues and tumor samples by ANOVA analysis. Analysis of normal tissue gene expression identified 7 genes with limited normal tissue expression that warrant further consideration as potential immunotherapy target antigens: CSAG2, MAGEA3, MAGEC2, IL13RA2, PRAME, CSPG4 and SOX10. These genes were highly overexpressed on a large percentage of the studied tumor samples with expression in a limited number of normal tissue samples at much lower levels.
Conclusion
The application of Nanostring RNA counting technology was used to directly quantitate the gene expression levels of multiple potential tumor antigens. Analysis of cell lines, 59 tumors, and normal tissues identified 7 potential immunotherapy targets for the treatment of melanoma that could increase the number of patients potentially eligible for adoptive immunotherapy.
Keywords: Nanostring, immunotherapy, cancer testis antigens, CSPG4, SOX10
Introduction
The development of successful immunotherapy for metastatic cancer requires the identification of appropriate target antigens. As immunotherapy strategies become increasingly sophisticated and powerful, finding antigens that are overexpressed in malignancies but have restricted expression in normal tissue becomes challenging. To date, the most successful immunotherapy approach is the adoptive cell transfer (ACT) of tumor-infiltrating lymphocytes (TIL), with objective response rates of >70% and complete response rates of approximately 40% reported in trials treating patients with metastatic melanoma. (1) This strategy necessitates the acquisition of tumor specimens for generation of TIL and has primarily shown success in treating melanoma. An alternative approach is the infusion of lymphocytes that have been harvested from the patient and genetically engineered to recognize tumor-associated antigens. (1, 2)
Tumor antigen-reactive T cell receptor (TCR) gene therapies have been employed with success in multiple histologies, however the number of patients that can be treated is somewhat limited as they must express a specific human leukocyte antigen (HLA) (e.g. HLA-A*0201). (2, 3) The restricted expression of certain target antigens in tumor cohorts can also limit a therapy's potential utility. For example, effective TCR therapies have been reported targeting the cancer-testis antigen (CTA) NY-ESO-1, however only around 20-30% of melanomas express this antigen. (4, 5) Clinical trials using non-MHC-restricted chimeric antigen receptor (CAR) therapy can potentially expand the number of patients eligible for ACT if the target antigen is expressed on the cell surface. (2) Recently, CAR therapies have shown success in treating non-melanoma and non-solid organ cancers, specifically the ability of the anti-CD19 CAR to effect regression of advanced B-cell malignancies. (6, 7)
The most effective way to maximize the number of patients potentially eligible for a therapy would be to target an antigen expressed on a high percentage of tumors within a given histology. This study uses Nanostring technology to achieve gene expression profiling of melanoma cell lines, metastatic melanoma tumors, and normal human tissue samples in order to identify potential target antigens for immunotherapy. This robust technology uses unique digital color-coded barcodes that hybridize directly to specific nucleic acid targets and allow for detection and quantitation of hundreds of transcripts in a single reaction. Unlike microarray approaches, it facilitates the direct measurement of mRNA expression levels for subsequent gene expression analysis, and has been shown to be highly reproducible and as sensitive as real-time PCR assays while still allowing for the measurement of multiple genes at one time. (8)
Methods
Sample collection, RNA isolation and cell lines
Patients seen at the Surgery Branch, National Cancer Institute (NCI) (Bethesda, MD) for treatment of metastatic melanoma underwent surgical excision of metastatic lesions for harvest of tumor infiltrating lymphocytes (TIL) in protocols approved by the Institutional Review Board and Food and Drug Administration. Viable-appearing fragments of these tumors were freed from surrounding normal tissue, collected, and either stored in RNAlater (Ambion, Austin, TX) or flash-frozen and stored at −80° Celsius, until RNA isolation was performed using a RNEasy Mini Kit (Qiagen, Valencia, CA). Analyzed tumor samples were collected between September 2007 and December 2012. RNA isolation was performed in the same fashion for established human melanoma cell lines initiated at Memorial Sloan-Kettering (SKmel23) or the NCI Surgery Branch (all others). The lines were grown under standard conditions in Roswell Park Memorial Institute (RPMI) 1640 with 10% fetal bovine serum (FBS) medium at 37°C, 5%CO2. For normal tissues, commercial available RNA samples were used (Agilent, Santa Clara, CA; Ambion, Austin, TX; Biochain, San Leandro, CA; Clontech, Mountain View, CA).
Immunohistochemistry
All tumors samples were confirmed to be metastatic melanoma at the time of harvest by pathological evaluation including immunohistochemistry. Immunohistochemical staining was performed for expression of the antigen NY-ESO-1 (encoded by gene CTAG1B) with the specific anti-NY-ESO-1 monoclonal antibody E978 (Invitrogen, Carslbad, CA). (5) Immunohistochemistry scores were assigned for intensity of staining and percentage of tumors cells that stained positive.
Nanostring analysis
Using the Nanostring nCounter Analysis System (Nanostring Technologies, Seattle, WA), gene expression analysis was performed for each sample as previously described using a custom designed codeset containing 97 genes. (8) Each reaction contained 250 ng of total RNA in a 5 μl aliquot, plus reporter and capture probes, and 6 pairs of positive control and 8 pairs of negative control probes. Analysis and normalization of the raw Nanostring data was performed using nSolver Analysis Software v1.1 (Nanostring Technologies). Raw counts were normalized to internal levels of 7 reference genes: CNOT2, GAPDH, HPRT1, PHGDH, SUMO2, SYS1 and WDR45L. A background count level was estimated using the average count of the 8 negative control probes in every reaction plus 2 standard deviations.
Gene expression analysis
Principal component analysis (PCA) and ANOVA analysis were performed using Partek Genomic Suite (Partek Incorporated, St. Louis, MO). PCA was used to characterize samples based on their gene expression profiles. ANOVA analysis was used to identify differentially expressed genes (significant p-value <0.05) and samples were clustered by hierarchical clustering.
Flow cytometry and RT-PCR
Flow cytometry (FACS) was performed using conjugated mAb specific for human chondroitin sulfate proteoglycan 4 (CSPG4) according to manufacturers’ recommendations (R&D Systems, Minneapolis, MD). Reverse transcription (RT) was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY). Quantitative PCR was performed with the TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Grand Island, NY) by the use of a 7500 Fast Real-Time PCR System (Applied Biosystems, Grand Island, NY). Copy numbers were generated using a standard curve from CSPG4 plasmid and results were normalized against β-actin (ACTB).
Results
Samples and Nanostring probeset
Gene expression profiling using Nanostring technology was performed on RNA from 5 melanoma lines (mel1300, mel526, mel624.38, mel888, SKmel23), 59 resected metastatic melanoma tumor deposits and 31 normal tissue samples. A total of 97 genes were included in the probeset (Table 1). Immune-related genes were included to investigate the immune characteristics of melanoma samples, however these would not be considered as potential immunotherapy target antigens. After elimination of the control genes (n=7) and the immune genes (n=18), 72 candidate genes remain. The candidate genes were grouped as follows for organizational purposes: melanoma-related genes, cancer testis antigen (CTA) genes, glioblastoma-related genes and other tumor-related genes. The sites of resection for the tumor deposits were primarily subcutaneous tissue (46%), lymph node (24%), lung (15%) and liver (8%). One tumor was resected from each of the following sites: adrenal, pelvic mass, retroperitoneum and small bowel. Before detailed analysis of the data we used PCA to determine if anatomical location of the tumor influenced gene expression profiling. PCA did not demonstrate differentiation of gene expression profiles based on the site of tumor harvest (Supplemental Figure S1). All 59 patients who underwent tumor resection had Stage III or IV melanoma. They ranged in age from 19-66 years (average 47±13 years) and were 71% male.
Table 1. Genes included in Nanostring probeset.
Gene name and corresponding GenBank Accession Numbers for the 97 genes included in the probeset.
Validation of Nanostring data
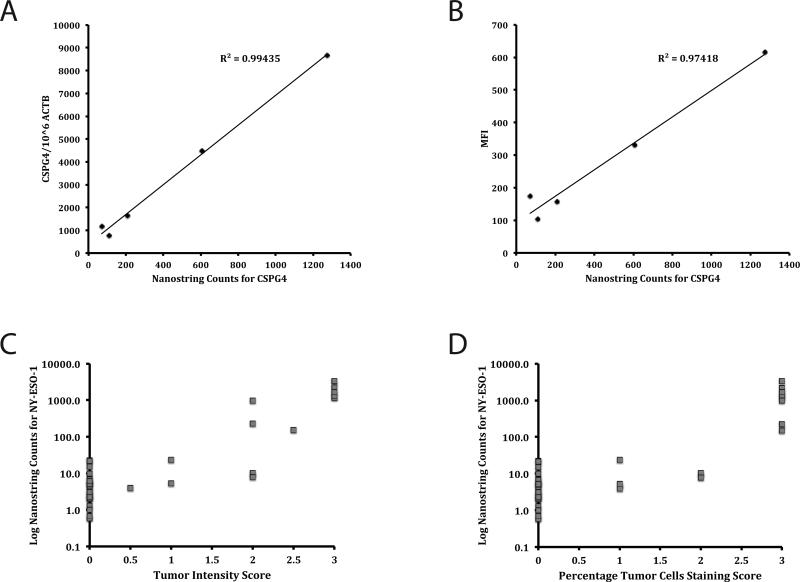
To compare the sensitivity of Nanostring to other methods of gene expression analysis, we used quantitative real time PCR (RT-PCR) and fluorescence antibody staining. Nanostring counts for the high molecular weight melanoma-associated antigen (HMW-MAA) gene CSPG4 for the 5 melanoma lines demonstrated high correlation with CSPG4 copy numbers generated by RT-PCR (R2 = 0.99435) (Figure 1A) and mean fluorescence intensity (MFI) values generated by flow cytometric analysis (R2 = 0.97418) (Figure 1B). Of the 59 tumors samples, 41 had immunohistochemistry staining performed for NY-ESO-1 at the time of resection. Despite the fact that immunohistochemical analysis was performed by multiple individuals over several years, results of both the intensity of staining and the percentage of tumor cells that stained were shown to compare well with Nanostring RNA counts for NY-ESO-1 (Figures 1C and 1D).
Figure 1.
Validation of Nanostring Data. Nanostring counts for CSPG4 gene expression in five melanoma lines were graphed against results from RT-PCR analysis (A) and MFI from FACS with CSPG4-specific mAb (B). Immunohistochemistry results, including scores for intensity of staining (C) and percentage of tumors cells staining (D) for NY-ESO-1 staining in 41 tumors samples are graphed against the log of the Nanostring counts for NY-ESO-1. Immunohistochemistry scores are as follows: for intensity of staining 0=no reactivity, 1=weak reactivity, 2=moderate reactivity, 3=intense reactivity, and for percentage of tumors cells that stained 0=0%, 1=0-5%, 2=5-50%, 3=>50%.
Selection of potential target genes
Based on the counts obtained with the 8 pairs of negative control probes (average = 9.0, STD = 5.8), 20 was chosen as the background level for gene detection. To set a value that would be likely for potential immune recognition (PIR), we used our previously published data on the ability of the engineered T-cells to recognize three tumor antigens: NY-ESO-1 (encoded by gene CTAG1B), MART-1 (encoded by gene MLANA), and CSPG4 (formerly known as HMW-MAA, encoded by gene CSPG4). (9-11) These reports used the same melanoma lines analyzed in this study as targets and we observed that tumor cell lines with Nanostring counts greater than 100 for a given target gene consistently demonstrated effector cytokine release when they were co-cultured with genetically-modified peripheral blood lymphocytes (PBL), whereas lines with counts lower than 100 did not reproducibly demonstrate such reactivity (Supplemental Table S1 and S2).
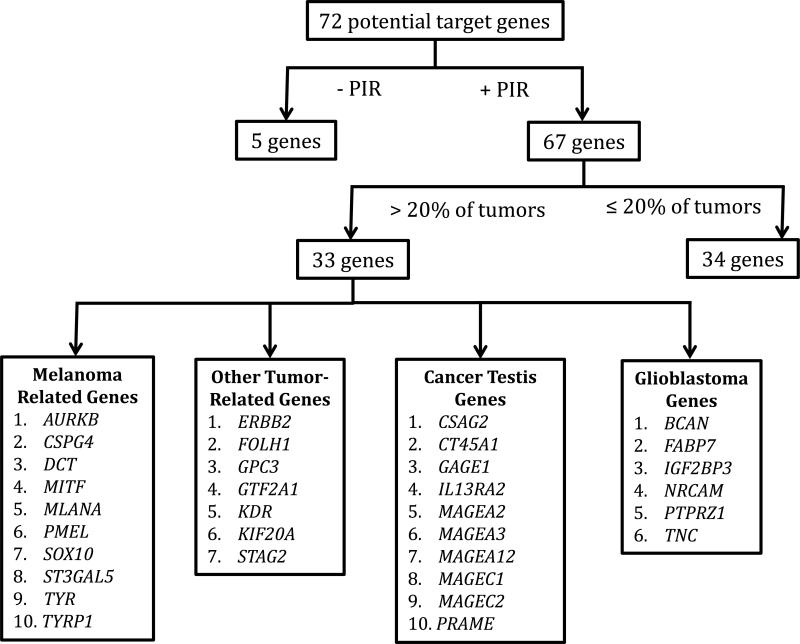
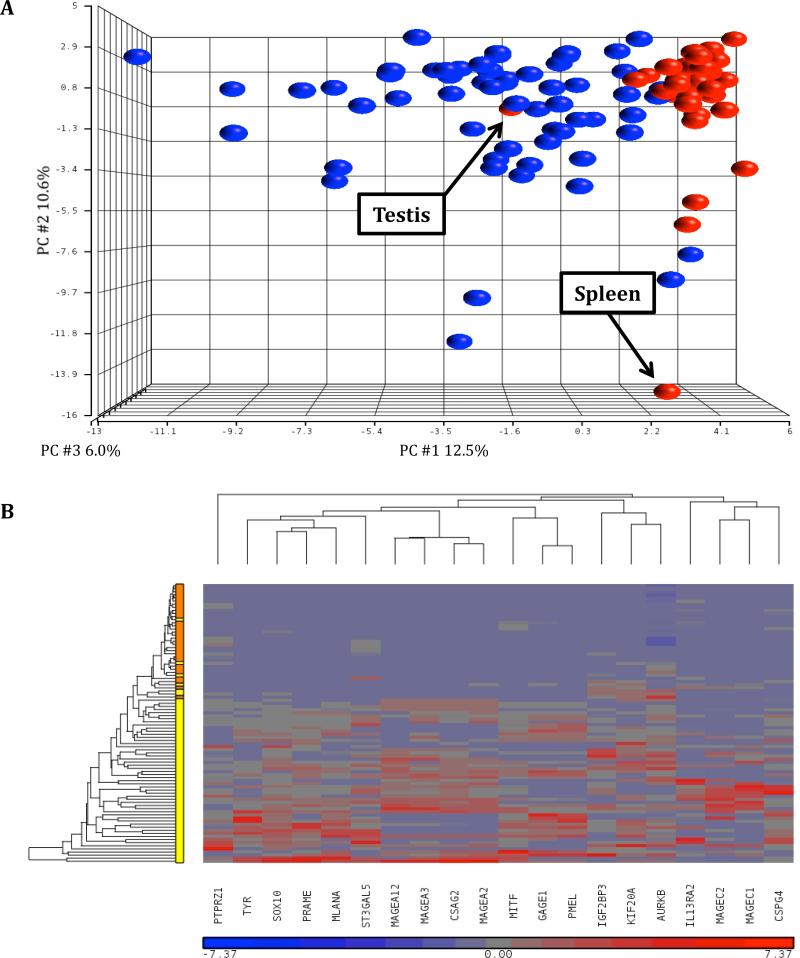
For each gene, the percentage of tumors demonstrating PIR positivity (PIR+, defined as a Nanostring count > 100) was determined as was the average Nanostring count of the PIR+ tumors (Table 2, Supplemental Table S3). Of the 72 potential target genes, 5 were not expressed in any of the tumor samples (Figure 2, Supplemental Table S3). Of the remaining 67 genes, 33 were PIR+ in more than 20% of tumor samples. 20% was chosen as a cutoff value based on our previous experience targeting NY-ESO-1. (5) RNA from 31 normal tissues was then subject to Nanostring Analysis with the same codeset (Supplemental Table S4). Analysis of these data by PCA demonstrated clear differences in gene expression profiles between normal tissues and tumor samples (Figure 3A). The positioning of the normal tissues testis and spleen were likely attributable to the expression of the cancer testis genes and immune genes, respectively. We then performed hierarchical clustering of the combined tumor and normal tissue data sets and observed that 27 of the initial 72 candidate genes were differentially expressed between tumors and normal tissues with at least a two-fold higher expression in tumors (Supplemental Figure S2). Of these 27 genes, 20 were PIR+ in more than 20% of tumors samples (Table 2, Figure 3B). Of those 20, MAGEA12 was eliminated from further consideration because of severe reported toxicities in a previous clinical trial. (12) The melanocyte differentiation antigens MLANA, PMEL and TYR were eliminated because they were targeted in previous trials and although clinical responses were achieved, patients suffered associated skin, eye and ear toxicities. (13, 14) 16 genes remained as potential target antigens for consideration (Table 3).
Table 2. Expression of potential target genes (n=72) in tumors.
The percentage of tumors (n=59) that were PIR+ (potential immune recognition+, defined as a Nanostring count >100) for each gene in the codeset is shown as is the average Nanostring count of the tumors which were PIR+ for that gene.
| Gene | Percentage of Tumors PIR+ | Average Count |
|---|---|---|
| Melanoma Related Genes | ||
| AURKB | 98 | 461 |
| B4GALNT1 | 15 | 222 |
| CSPG4 | 92 | 2406 |
| DCT | 76 | 10124 |
| ERBB4 | 7 | 172 |
| GRIN2A | 3 | 120 |
| GRM3 | 0 | n/a |
| MITF | 90 | 3665 |
| MLANA | 83 | 7923 |
| PMEL | 85 | 84888 |
| SOX10 | 90 | 902 |
| ST3GAL5 | 100 | 2077 |
| ST8SIA1 | 20 | 202 |
| TYR | 76 | 9014 |
| TYRP1 | 47 | 23414 |
| Cancer Testis Genes | ||
| CSAG2 | 71 | 1046 |
| CT45A1 | 24 | 2478 |
| CTAG1B | 19 | 1787 |
| CTAG2 | 17 | 585 |
| CTCFL | 10 | 210 |
| CXorf48 | 0 | n/a |
| GAGE1 | 29 | 186 |
| GAGE4 | 17 | 6062 |
| IL13RA2 | 32 | 1468 |
| MAGEA1 | 20 | 251 |
| MAGEA2 | 49 | 237 |
| MAGEA3 | 75 | 2567 |
| MAGEA4 | 15 | 220 |
| MAGEA5 | 0 | n/a |
| MAGEA8 | 7 | 250 |
| MAGEA9 | 19 | 570 |
| MAGEA10 | 20 | 443 |
| MAGEA11 | 15 | 580 |
| MAGEA12 | 51 | 308 |
| MAGEB1 | 14 | 289 |
| MAGEB2 | 14 | 528 |
| MAGEB3 | 2 | 171 |
| MAGEB6 | 2 | 130 |
| MAGEC1 | 27 | 693 |
| MAGEC2 | 39 | 1699 |
| POTEF | 8 | 213 |
| PRAME | 86 | 2899 |
| SAGE1 | 2 | 109 |
| SPANXN3 | 20 | 139 |
| SPANXA1 | 2 | 135 |
| SSX1 | 12 | 629 |
| SSX2 | 5 | 239 |
| SSX3 | 20 | 796 |
| SYCP1 | 0 | n/a |
| TSPY1 | 7 | 4996 |
| Glioblastoma Related Genes | ||
| BCAN | 56 | 2143 |
| CHI3L2 | 19 | 211 |
| EGFRvIII | 0 | n/a |
| FABP7 | 47 | 1106 |
| IGF2BP3 | 61 | 437 |
| NRCAM | 37 | 317 |
| PTPRZ1 | 78 | 2437 |
| TNC | 92 | 2249 |
| Other Tumor-Related Genes | ||
| CEACAM5 | 2 | 618 |
| ERBB2 | 75 | 255 |
| FOLH1 | 41 | 539 |
| GPC3 | 54 | 1308 |
| GTF2A1 | 100 | 1026 |
| KDR | 92 | 310 |
| KIF20A | 69 | 255 |
| MSLN | 7 | 124 |
| MUC1 | 20 | 297 |
| PSCA | 12 | 328 |
| STAG2 | 100 | 1004 |
| TG | 3 | 128 |
| TKTL1 | 10 | 1443 |
| WT1 | 3 | 140 |
Figure 2.
Selection of potential target genes based on PIR positivity in tumor samples. 67 of the 72 potential candidate genes were PIR+ (defined as Nanostring count >100) in some the tumor samples (n=59). 33 genes were PIR+ in over 20% of the tumor samples.
Figure 3.
Differentiation of gene expression between normal tissues and tumors. (A) PCA for normal tissues (n=59) and tumor samples (n=31). The color coding is as follows: red = normal tissue, blue = tumor. (B) Hierarchical clustering of 20 genes that differentiate tumors from normal tissues with at least a two-fold higher expression in tumors, and are overexpressed in more than 20% of tumors. Vertical axis shows normal tissues (orange) and tumors (yellow).
Table 3.
Nanostring counts for potential candidate genes (n=16).
| Gene | Average Count Normal Tissues | Average Count Tumors | Ratio (Tumors: Normal Tissues) |
|---|---|---|---|
| Melanoma Related Genes | |||
| AURKB | 397 | 461 | 1.2 |
| CSPG4 | 367 | 2406 | 6.6 |
| MITF | 433 | 3665 | 8.5 |
| SOX10 | 195 | 902 | 4.6 |
| ST3GAL5 | 672 | 2077 | 3.1 |
| Cancer Testis Genes | |||
| CSAG2 | – | 1046 | – |
| GAGE1 | – | 186 | – |
| IL13RA2 | 332 | 1468 | 4.4 |
| MAGEA2 | – | 237 | – |
| MAGEA3 | 453* | 2567 | 5.7 |
| MAGEC1 | 165* | 693 | 4.2 |
| MAGEC2 | 650* | 1699 | 2.6 |
| PRAME | 600* | 2899 | 4.8 |
| Glioblastoma Related Genes | |||
| IGF2BP3 | 267 | 437 | 1.6 |
| PTPRZ1 | 573 | 2437 | 4.3 |
| Other Genes | |||
| KIF20A | 192 | 255 | 1.3 |
denotes an average normal tissue count that is reflective solely of expression on testis.
(–) denotes no overexpression on any normal tissues. The average Nanostring counts for all normal tissues that were PIR+ (potential immune recognition+, defined as a Nanostring count >100) for each gene are provided as are the average Nanostring counts for tumors that were PIR+. The ratio of average expression in tumors to average expression in normal tissues is also shown.
MITF was expressed in tumors at an average level that is 8.5 times higher than the average expression in normal tissues (Table 3), however high levels of expression (counts > 1000) in diaphragm, muscle and uterus (Supplemental Table S4) eliminated it from consideration. ST3GAL5 was eliminated secondary to expression at very high levels in multiple tissues including brain, adrenal, thyroid, spleen and artery (Supplemental Table S4). Likewise, PTPRZ1 was eliminated because of high levels of expression in brain, brainstem and artery samples (Supplemental Table S4). AURKB, IGF2BP3 and KIF20A all demonstrated low levels of expression on limited normal tissues, however the average level of expression detected on tumors was sufficiently low (ratio < 2.0) to render them non-ideal targets (Table 3). Of the remaining 10 genes, 8 encode cancer testis antigens and exhibit little-to-no normal tissue expression outside of testis. Of these 8 genes, the highest levels of tumor RNA expression (all >1000) were seen for CSAG2, IL13RA2, MAGEA3, MAGEC2 and PRAME, all of which warrant further consideration as possible targets (Table 3). The remaining non-CTA candidate genes were CSPG4 and SOX10. Both exhibited low levels of expression in a number of normal tissues (skin, trachea, vein, heart, lung, diaphragm, muscle, adipose, uterus, prostate, thymus, spleen, bone marrow and gastrointestinal organs for CSPG4 and brain, brainstem, trachea, spleen, artery and breast for SOX10), however the average levels of expression in tumors for both were substantially higher than in normal tissues (6.6 times higher for CSPG4 and 4.6 times higher for SOX10). Therefore, 7 genes are identified as potential immunotherapy targets: CSAG2, MAGEA3, MAGEC2, IL13RA2, PRAME, CSPG4 and SOX10.
Discussion
The identification of target antigens for immunotherapy is a complex process that involves assessing the expression of antigens on tumors and normal tissues. Cancer testis antigens are of particular interest as immunotherapy targets because they are expressed in multiple cancers of diverse histological origin, including breast cancer, prostate cancer, non-small cell lung cancer, gastrointestinal cancers such as colon and esophageal, bladder cancer, and melanoma. Aside from expression in male germ cells, CTA expression in normal human tissues is relatively restricted. (15) Previous studies have examined gene expression profiles of this antigen group in melanoma and other cancers using microarray technology and RT-PCR. (16, 17) Herein we identified five genes encoding cancer-testis antigens which are expressed at high levels on a large percentage of melanoma tumor samples studied: CSAG2, MAGEA3, MAGEC2, IL13RA2 and PRAME.
CSAG2, also known as taxol-resistance-associated gene-3 (TRAG3), was overexpressed on 71% of studied melanoma tumors with no significant expression on any normal tissues. It is overexpressed in multiple other histologies including carcinoma of the bladder, cervix, breast, esophagus, bile duct, stomach, colon and lung, and its expression has been correlated with poor prognosis in both ovarian cancer and osteosarcoma. (18-21) Preclinical studies have identified a potential target CSAG2-directed T-cell epitope capable of inducing cytotoxic lymphocytes (CTL). (22) MAGEA3 is a very appealing target given its overexpression on a high percentage of metastatic melanoma tumors (75% in this study) and its potential as a target for ACT in other histologies such as colorectal cancer, lung cancer, breast cancer, esophageal cancer and glioblastoma. (23, 24) A particular epitope, MAGEA3 112-120, was targeted in TCR gene therapy trials in MAGEA3+ patients with observed cancer regression but severe associated neurologic toxicity and mortality, likely secondary to TCR cross-recognition of this epitope in MAGEA12, which was found to be expressed in brain. (12) The generation of a completely MAGEA3-specific TCR, however, could allow for its targeting in future studies. MAGEC2, formerly known as hepatocellular carcinoma-associated antigen 587 (HCA587), is also overexpressed in a variety of cancers aside from melanoma, including hepatocellular carcinoma, gallbladder carcinoma, medulloblastoma, multiple myeloma and squamous cell cancer of the head and neck. (25-28) It was overexpressed on 39% of tumors in this study with no expression on any normal tissues aside from testis. Several preclinical studies have identified potential target epitopes and have generated CTL that demonstrate functionality against target cells. (29, 30)
IL13RA2 was overexpressed on 32% of studied tumors and demonstrated low levels of expression on the liver and adrenal samples. It has been associated with adrenocortical carcinoma, glioblastoma multiforme and systemic sclerosis, and IL13RA2-specific CARs have demonstrated efficacy against glioma cell targets in preclinical studies. (31-33) PRAME was overexpressed in 86% of melanoma samples and its average count on overexpressing tumors (2899) was the highest among any of the potential CTA targets. It was absent on any normal tissues in this cohort though other studies have reported some expression on normal tissues such as endometrium, ovaries and adrenals. (34, 35) Its expression has been widely reported in other cancers including carcinoma of the lung and kidney, squamous cell carcinoma of the head and neck, sarcomas, mammary cancer, multiple myeloma and acute leukemias. (34) It is one of the few CTA that is commonly expressed in leukemic malignancies. Preclinical studies have generated PRAME-specific T-cells that demonstrated activity across multiple histologies encompassing both solid organ and hematologic malignancies. (36, 37)
CSPG4 and SOX10 are both melanoma-associated genes that were expressed in 92% and 90% of tumors in this study, respectively. Unlike CTA, they do exhibit expression on multiple normal tissues, though at much lower levels than on tumors. This does raise concerns regarding application in the clinical setting and argues for safety measures such as dose escalation trials or the implementation of a suicide gene. (38) CSPG4 is a highly immunogenic cell surface proteoglycan which was identified on melanoma cells in the 1970's, and it has been found on glioblastoma, triple-negative breast cancer, head and neck squamous cell cancer, mesothelioma, renal cell cancer and sarcoma as well. (39-43) It has been targeted with vaccines in clinical trials with no reported toxicity and anti-tumor effects have been reported in preclinical immunotherapy models with melanoma and head and neck squamous cell cancers targets. (9, 40, 44) SOX10 is a transcription factor that is expressed on neural crest cells and melanocytes, and has been shown to be crucial for the maintenance of neoplastic cells. (45, 46) In addition to being widely expressed on melanomas and other lesions such as giant congenital nevi, it has also been identified on cancers of the breast and prostate. (45, 47-49) Naturally occurring anti-SOX10 CTLs were identified in a patient with a dramatic response to immunotherapy. (50)
This study identified genes that have potential as immunotherapy targets based on their expression in a high percentage of studied melanoma tumors with limited expression in normal tissues. One possible study limitation is the correlation between gene expression and antigen expression. We have demonstrated that for CSPG4 and NY-ESO-1 there is a strong association between the level of gene expression as assessed by Nanostring and the degree of antigen expression, as determined by both FACS and immunohistochemistry, however this may not be the case for every gene. Importantly, these data do not directly address issues involving the potential safety of a given target gene.
To have the potential for clinical application, each new target gene would require the generation of reagents (TCR or CAR) that can mediate specific antigen recognition and, the target antigen must not be expressed on a vital tissue. In the case of cancer testis antigens, while it is widely reported that these genes are cancer-specific, this is not universally true. As we recently reported, the MAGEA12 gene is strongly expressed in rare neurons in the human brain and expression in these isolated cells was likely sufficient to initiate a destructive immune response leading to death in some patients. (12) On the other hand, we have used the identical strategy of TCR gene therapy to target NY-ESO-1, and in over 40 patients treated, have not observed any target-related toxicities. Clearly more detailed studies (e.g., multiple tissue immunohistochemistry) would be needed prior to any of these new antigens being targeted in clinical trials, however Nanostring provides a reliable way to test multiple candidate genes at once and select attractive potential targets for further investigation.
Supplementary Material
Statement of Translational Relevance.
Accurate quantitation of RNA levels is an essential step in the identification of potential tumor antigens. Nanostring is solution-based gene expression profiling technology that accurately counts individual RNA molecules in the small amounts of total RNA (250ng or less) that are often obtained from biopsy samples. We used this technology to study potential tumor antigen gene expression in 59 metastatic melanoma samples and identified 7 genes as potential targets for adoptive immunotherapy.
Acknowledgements
The authors would like to thank Arnold Mixon and Shawn Farid for technical support with FACS analysis and the Laboratory of Pathology Department at the National Cancer Institute for their role in the immunohistochemistry staining.
Grant Support
This work is supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Footnotes
Conflicts of Interest:
None declared
References
- 1.Turcotte S, Rosenberg SA. Immunotherapy for metastatic solid cancers. Advances in surgery. 2011;45:341–60. doi: 10.1016/j.yasu.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park TS, Rosenberg SA, Morgan RA. Treating cancer with genetically engineered T cells. Trends Biotechnol. 2011;29:550–7. doi: 10.1016/j.tibtech.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Science translational medicine. 2012;4:127ps8. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai JP, Robbins PF, Raffeld M, Aung PP, Tsokos M, Rosenberg SA, et al. NY-ESO-1 expression in synovial sarcoma and other mesenchymal tumors: significance for NY-ESO-1-based targeted therapy and differential diagnosis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:854–8. doi: 10.1038/modpathol.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:917–24. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kochenderfer JN, Dudley ME, Feldman SA, Wilson WH, Spaner DE, Maric I, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119:2709–20. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England journal of medicine. 2011;365:725–33. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 9.Burns WR, Zhao Y, Frankel TL, Hinrichs CS, Zheng Z, Xu H, et al. A high molecular weight melanoma-associated antigen-specific chimeric antigen receptor redirects lymphocytes to target human melanomas. Cancer Res. 2010;70:3027–33. doi: 10.1158/0008-5472.CAN-09-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wargo JA, Robbins PF, Li Y, Zhao Y, El-Gamil M, Caragacianu D, et al. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol Immunother. 2009;58:383–94. doi: 10.1007/s00262-008-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff SL, Johnson LA, Black MA, Xu H, Zheng Z, Cohen CJ, et al. Enhanced receptor expression and in vitro effector function of a murine-human hybrid MART-1-reactive T cell receptor following a rapid expansion. Cancer Immunol Immunother. 2010;59:1551–60. doi: 10.1007/s00262-010-0882-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer Regression and Neurological Toxicity Following Anti-MAGE-A3 TCR Gene Therapy. J Immunother. 2013;36:133–51. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–46. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seaman BJ, Guardiani EA, Brewer CC, Zalewski CK, King KA, Rudy S, et al. Audiovestibular dysfunction associated with adoptive cell immunotherapy for melanoma. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2012;147:744–9. doi: 10.1177/0194599812448356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer immunity. 2004;4:1. [PubMed] [Google Scholar]
- 16.Segal NH, Blachere NE, Guevara-Patino JA, Gallardo HF, Shiu HY, Viale A, et al. Identification of cancer-testis genes expressed by melanoma and soft tissue sarcoma using bioinformatics. Cancer immunity. 2005;5:2. [PubMed] [Google Scholar]
- 17.Bredenbeck A, Hollstein VM, Trefzer U, Sterry W, Walden P, Losch FO. Coordinated expression of clustered cancer/testis genes encoded in a large inverted repeat DNA structure. Gene. 2008;415:68–73. doi: 10.1016/j.gene.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Zhu B, Wu Y. Expression of TRAG-3 antigen in non-small-cell lung carcinomas. Lung Cancer. 2002;38:101–2. doi: 10.1016/s0169-5002(02)00223-4. [DOI] [PubMed] [Google Scholar]
- 19.Ohta M, Tanaka F, Sadanaga N, Yamaguchi H, Inoue H, Mori M. Expression of the TRAG-3 gene in human esophageal cancer: the frequent synchronous expression of MAGE-3 gene. Oncology reports. 2006;15:1529–32. [PubMed] [Google Scholar]
- 20.Wu YZ, Zhao TT, Ni B, Zou LY, Liu HL, Zhu B. Expression of TRAG-3 in breast cancer. International journal of cancer Journal international du cancer. 2003;107:167–8. doi: 10.1002/ijc.11349. [DOI] [PubMed] [Google Scholar]
- 21.Zou C, Shen J, Tang Q, Yang Z, Yin J, Li Z, et al. Cancer-testis antigens expressed in osteosarcoma identified by gene microarray correlate with a poor patient prognosis. Cancer. 2012;118:1845–55. doi: 10.1002/cncr.26486. [DOI] [PubMed] [Google Scholar]
- 22.Zhu B, Chen Z, Cheng X, Lin Z, Guo J, Jia Z, et al. Identification of HLA-A*0201-restricted cytotoxic T lymphocyte epitope from TRAG-3 antigen. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:1850–7. [PubMed] [Google Scholar]
- 23.Shantha Kumara HM, Grieco MJ, Caballero OL, Su T, Ahmed A, Ritter E, et al. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer immunity. 2012;12:16. [PMC free article] [PubMed] [Google Scholar]
- 24.Chinnasamy N, Wargo JA, Yu Z, Rao M, Frankel TL, Riley JP, et al. A TCR targeting the HLA-A*0201-restricted epitope of MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen superfamily in several types of cancer. J Immunol. 2011;186:685–96. doi: 10.4049/jimmunol.1001775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oba-Shinjo SM, Caballero OL, Jungbluth AA, Rosemberg S, Old LJ, Simpson AJ, et al. Cancer-testis (CT) antigen expression in medulloblastoma. Cancer immunity. 2008;8:7. [PMC free article] [PubMed] [Google Scholar]
- 26.Pabst C, Zustin J, Jacobsen F, Luetkens T, Kroger N, Schilling G, et al. Expression and prognostic relevance of MAGE-C1/CT7 and MAGE-C2/CT10 in osteolytic lesions of patients with multiple myeloma. Experimental and molecular pathology. 2010;89:175–81. doi: 10.1016/j.yexmp.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Riener MO, Wild PJ, Soll C, Knuth A, Jin B, Jungbluth A, et al. Frequent expression of the novel cancer testis antigen MAGE-C2/CT-10 in hepatocellular carcinoma. International journal of cancer Journal international du cancer. 2009;124:352–7. doi: 10.1002/ijc.23966. [DOI] [PubMed] [Google Scholar]
- 28.Figueiredo DL, Mamede RC, Spagnoli GC, Silva WA, Jr., Zago M, Neder L, et al. High expression of cancer testis antigens MAGE-A, MAGE-C1/CT7, MAGE-C2/CT10, NY-ESO-1, and gage in advanced squamous cell carcinoma of the larynx. Head & neck. 2011;33:702–7. doi: 10.1002/hed.21522. [DOI] [PubMed] [Google Scholar]
- 29.Wen W, Zhang L, Peng J, Chen J, Hao J, Li X, et al. Identification of promiscuous HLA-DR-restricted CD4(+) T-cell epitopes on the cancer-testis antigen HCA587. Cancer science. 2011;102:1455–61. doi: 10.1111/j.1349-7006.2011.01986.x. [DOI] [PubMed] [Google Scholar]
- 30.Xing Q, Pang XW, Peng JR, Yin YH, Li Y, Yu X, et al. Identification of new cytotoxic T-lymphocyte epitopes from cancer testis antigen HCA587. Biochemical and biophysical research communications. 2008;372:331–5. doi: 10.1016/j.bbrc.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Ranvier GG, Weng J, Yeh RF, Khanafshar E, Suh I, Barker C, et al. Identification of biomarkers of adrenocortical carcinoma using genomewide gene expression profiling. Arch Surg. 2008;143:841–6. doi: 10.1001/archsurg.143.9.841. discussion 6. [DOI] [PubMed] [Google Scholar]
- 32.Granel B, Allanore Y, Chevillard C, Arnaud V, Marquet S, Weiller PJ, et al. IL13RA2 gene polymorphisms are associated with systemic sclerosis. The Journal of rheumatology. 2006;33:2015–9. [PubMed] [Google Scholar]
- 33.Kong S, Sengupta S, Tyler B, Bais AJ, Ma Q, Doucette S, et al. Suppression of human glioma xenografts with second-generation IL13R-specific chimeric antigen receptor-modified T cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5949–60. doi: 10.1158/1078-0432.CCR-12-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 35.van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, et al. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. British journal of haematology. 1998;102:1376–9. doi: 10.1046/j.1365-2141.1998.00982.x. [DOI] [PubMed] [Google Scholar]
- 36.Amir AL, van der Steen DM, van Loenen MM, Hagedoorn RS, de Boer R, Kester MD, et al. PRAME-specific Allo-HLA-restricted T cells with potent antitumor reactivity useful for therapeutic T-cell receptor gene transfer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:5615–25. doi: 10.1158/1078-0432.CCR-11-1066. [DOI] [PubMed] [Google Scholar]
- 37.Quintarelli C, Dotti G, Hasan ST, De Angelis B, Hoyos V, Errichiello S, et al. High-avidity cytotoxic T lymphocytes specific for a new PRAME-derived peptide can target leukemic and leukemic-precursor cells. Blood. 2011;117:3353–62. doi: 10.1182/blood-2010-08-300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Stasi A, Tey SK, Dotti G, Fujita Y, Kennedy-Nasser A, Martinez C, et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N Engl J Med. 2011;365:1673–83. doi: 10.1056/NEJMoa1106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, et al. CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:5352–63. doi: 10.1158/1078-0432.CCR-12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Wang Y, Yu L, Sakakura K, Visus C, Schwab JH, et al. CSPG4 in cancer: multiple roles. Current molecular medicine. 2010;10:419–29. doi: 10.2174/156652410791316977. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. Journal of the National Cancer Institute. 2010;102:1496–512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benassi MS, Pazzaglia L, Chiechi A, Alberghini M, Conti A, Cattaruzza S, et al. NG2 expression predicts the metastasis formation in soft-tissue sarcoma patients. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2009;27:135–40. doi: 10.1002/jor.20694. [DOI] [PubMed] [Google Scholar]
- 43.Svendsen A, Verhoeff JJ, Immervoll H, Brogger JC, Kmiecik J, Poli A, et al. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta neuropathologica. 2011;122:495–510. doi: 10.1007/s00401-011-0867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Katayama A, Wang Y, Yu L, Favoino E, Sakakura K, et al. Functional characterization of an scFv-Fc antibody that immunotherapeutically targets the common cancer cell surface proteoglycan CSPG4. Cancer research. 2011;71:7410–22. doi: 10.1158/0008-5472.CAN-10-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agnarsdottir M, Sooman L, Bolander A, Stromberg S, Rexhepaj E, Bergqvist M, et al. SOX10 expression in superficial spreading and nodular malignant melanomas. Melanoma research. 2010;20:468–78. doi: 10.1097/CMR.0b013e3283403ccd. [DOI] [PubMed] [Google Scholar]
- 46.Shin J, Vincent JG, Cuda JD, Xu H, Kang S, Kim J, et al. Sox10 is expressed in primary melanocytic neoplasms of various histologies but not in fibrohistiocytic proliferations and histiocytoses. Journal of the American Academy of Dermatology. 2012;67:717–26. doi: 10.1016/j.jaad.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 47.Zhong WD, Qin GQ, Dai QS, Han ZD, Chen SM, Ling XH, et al. SOXs in human prostate cancer: implication as progression and prognosis factors. BMC cancer. 2012;12:248. doi: 10.1186/1471-2407-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cimino-Mathews A, Subhawong AP, Elwood H, Warzecha HN, Sharma R, Park BH, et al. Neural crest transcription factor Sox10 is preferentially expressed in triple-negative and metaplastic breast carcinomas. Human pathology. 2012 doi: 10.1016/j.humpath.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nature cell biology. 2012;14:882–90. doi: 10.1038/ncb2535. [DOI] [PubMed] [Google Scholar]
- 50.Khong HT, Rosenberg SA. The Waardenburg syndrome type 4 gene, SOX10, is a novel tumor-associated antigen identified in a patient with a dramatic response to immunotherapy. Cancer research. 2002;62:3020–3. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.