Abstract
Purpose
Estrogen receptor α (ERα) is an essential element regulating mammary gland development and it contributes to breast cancer development and progression. Most of the ER negative breast cancers display more aggressive clinical behaviors and are resistant to anti-estrogen therapies. In addition, many ER negative tumors show insensitivity to many chemotherapeutic drugs and radiation therapy, although mechanisms underlying this phenotype are less clear.
Experimental Design
We conducted immunohistochemistry on 296 cases of breast cancer tissues using a variety of antibodies. Based on the clinical data, we performed siRNA knock-down to study the role of ERα on ATM expression in breast cancer cell lines. Further we used antisense oligonucleotides against miRNAs or miRNA overexpression plasmids to study the role of miRNA 18a and 106a on ATM expression. Finally we employed in situ hybridization to assess miRNA 18a and 106a expression in breast cancer tissues.
Results
We found that in ER negative breast cancer tissues, expression of the ATM kinase, a critical DNA damage response protein, is aberrantly up-regulated. We also found that the locoregional recurrence rate after radiotherapy positively correlates with ATM expression. On the cellular level, we demonstrated that ERα, but not ERβ, negatively regulates ATM expression. Furthermore, we identified that ERα activates miRNA- 18a and 106a to down-regulate ATM expression. We also demonstrated that miRNA-18a and 106a were significantly under-expressed in ER negative breast cancer tissues.
Conclusions
We reveal a novel mechanism involving ERα and miRNA 18a and 106a regulation of ATM in breast cancer.
Keywords: ATM, ERα, miRNA
Introduction
About 30% of breast cancer patients do not express estrogen receptor α (ERα) (1). ERα negative tumors are a part of the triple negative breast cancer (TNBC), a subset of the breast cancer population which typically is associated with aggressive clinical phenotypes and poor prognosis (2). For example, ER negative breast cancers are not responsive to anti-estrogen therapies. In addition, many ER negative patients show less sensitivity to many DNA damaging agents (3), except in some cases, such as with a Her2 positive status, patients showed variable sensitivity to chemotherapeutic drugs such as anthracycline (4, 5). At the cellular level, ERα regulates a variety of processes in mammary gland development and it also contributes to the development and progression of breast cancer (6). By binding to estradiol (E2), ERα mediates transcription by interacting directly to specific estrogen response elements located in the promoter or enhancer region of target genes. In addition, ERα may indirectly associate with nuclear proteins such as AP1 and SP1 to stimulate transcription (7). Recent data have also demonstrated that ERα plays a critical role in regulating (either suppress or stimulate) micro RNAs (miRNAs) in order to control protein expression (8). Aberrant miRNA expression has been implicated in estrogen-related breast and endometrial cancers in the clinical setting (9). It has been reported that several phosphorylation events are required for a variety of cell specific functions of ERα (10). Despite the extensive studies on ERα function in regulation of transcription, it is less clear the functional mechanism driving most of ER-negative breast cancer cells resistant to many chemotherapeutic agents as well as radiotherapy.
Cellular sensitivity to DNA damaging agents such as radiotherapy and radiomimetic drugs are regulated by a cascade of DNA damage response proteins. Among them is the ATM kinase (11). ATM is mutated in the autosomal recessive disorder Ataxia-Telangiectasia, manifested by progressive neuronal degeneration, cancer predisposition, immunodeficiency and hypersensitivity to radiotherapy (12). ATM functions in the DNA damage response by phosphorylation and activation of a series of downstream targets to regulate cell cycle checkpoints in response to DNA double strand breaks. ATM hyperactivation has been observed in many stages of tumor tissues (13–16). ATM activation in the early stage of tumorigenesis is oncogene driven and represents the antitumor function of the kinase (14–16). However, hyperactivation of ATM in late stages of breast cancer contributes to breast cancer metastasis (13).
In the current study, we report that ER negative breast cancer tissues show elevated ATM expression. We demonstrate that ERα, but not ERβ, negatively regulates ATM expression in breast cancer cells. Furthermore we have identified two miRNAs (miR-18a and -106a) that are targets of ERα to regulate ATM expression. Finally we show that miRNAs 18a and 106a expressions are aberrantly reduced in ER negative breast cancer tissues.
Materials and Methods
Cell culture and materials
Breast cancer cells lines MCF-7 and MDA-MB-231, obtained from American Type Culture Collection (ATCC, Manassas, VA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS (Invitrogen, Carlsbad, CA) and 50 mg/ml of penicillin/streptomycin (Invitrogen) in humidified 37°C chambers with 5% CO2. β-Estradiol was purchased from Sigma-Aldrich (St. Louis, MO). Cell line authentication was originally done by Short Tandem Repeat profiling at ATCC. After purchase, the cell lines were not authenticated by authors. Ionizing radiation was delivered by X-Rad 320 X-ray irradiator (Precision X-Ray Inc. N. Branford. CT). All the cellular experiments performed in this manuscript include at least three independent experiments.
Antibodies
Rabbit anti-ATM and mouse anti-phospho-ATM (S1981p) antibodies were purchased from Cell Signaling (Beverly, MA). Mouse anti-ATM, mouse anti- ERα) and rabbit anti-ERβ antibodies were purchased from Abcam (Cambridge, MA). All horseradish-peroxidase conjugated secondary antibodies and β-actin antibody were obtained from Santa Cruz biotechnology (Santa Cruz, CA).
Plasmids, siRNAs and transfection
MicroRNA overexpression plasmids (pCMV-miR18a, 106a, 101 and 421) and ATM-3’UTR plasmid were purchased from Origene (Rockville, MD), ERα plasmid (pEGFP-C1-ERα) was purchased from Addgene (Cambridge, MA). Plasmids were transfected into cells with Fugene HD reagents (Roche, South San Francisco, CA). Control, ERα, ERβ and ATM siRNAs were purchased from Santa Cruz biotechnology (Santa Cruz, CA) and transfected into cells by oligofectamine reagent (Invitrogen) according to the manufacturer’s instruction. Forty-eight hours after transfection, cells were analyzed. MiRNA antisense oligos against has-miR18a, 106a, 101, and 421 were synthesized from Integrated DNA Technologies (Coralville, Iowa). Scrambled sequences were used as negative controls. All the oligos were transfect into cells also using the oligofectamine reagent. The sequences of all the oligos are listed in the following table.
| miRNA | antisense | scrambled |
|---|---|---|
| miR18a | 5' ctgcactagatgcacctta3' | 5' gtaccacgatcatatctgc3' |
| miR 101 | 5' ttcagttatcacagtactgta3' | 5' agaacagacttattctttgtc3' |
| miR 106a | 5' acctgcactgtaagcacttt 3' | 5' ctacttccgatcgaagttac 3' |
| miR 421 | 5' cgcccaattaatgtctgttga3' | 5' ggcgctatttacaactattcg3' |
Luciferase reporter assay
MDA-MB-231 cells were seeded in triplicates in 48-well plates. Luciferase reporter plasmids (ATM 3’UTR or pMirTarget vector) (Origene) were cotransfected with 50 nM control, miR-18a, miR-106a plasmids or both. Cells were harvested 48h after transfection, and lysed for luciferase assays by a Dual-Glo assay kit (Promega) according to the manufacturer’s protocol.
Human breast cancer tissue samples
Paraffin-embedded materials from 296 invasive breast cancer patients with operable breast cancer treated between 01/01/2003–05/31/2003 were provided by the Department of Breast Pathology and Research Laboratory, Cancer Hospital of Tianjin Medical University, Tianjin, China, with the approval of Tianjin Medical University Institutional Review Board (IRB). All cases were female with 29–75 years of age (the mean age is 51.8 years). All patients were treated either with modified radical mastectomy (n=176) or breast-conserving therapy (n=120). Locoregional radiotherapy was administered to the patients underwent with the breast-conserving therapy. The outcome of radiotherapy was evaluated by locoregional recurrence with a follow-up period from 9–96 (mean 74.63) months.
Immunohistochemistry
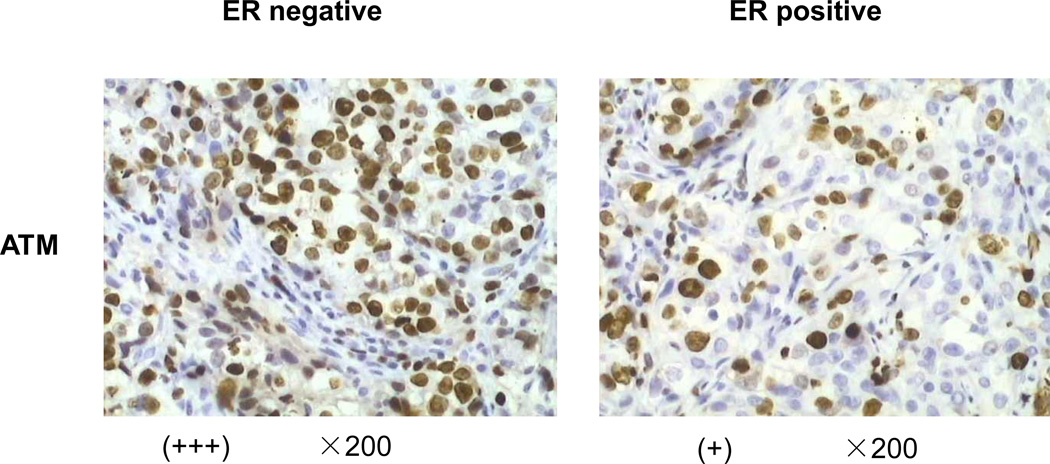
Immunohistochemistry using the avidin-biotin-immunoperoxidase technique was performed for ER and ATM in the 296 cases of clinical samples. Sections of formalin-fixed tissues from all cases were performed using a standard protocol. Briefly, 4 µm tissue sections on coated slides were heated for antigen retrieval, pretreated with a 3% solution of hydrogen peroxide for 5–10 minutes, rinsed and incubated with 10% normal goat serum as a blocking agent. The sections were then incubated sequentially with the primary antibody (ER in a 1:100 dilution and ATM in a 1:200 dilution), a biotinylated secondary antibody and avidin-peroxidase conjugate. All steps were preceded by rinsing of sections with PBS (pH 7.6). The chromogen was 3,3'-diaminobenzidine (DAB). The immunoreaction for ER and ATM in the nucleus of tumor cells was evaluated independently by two experienced pathologists. ER was determined positive if finding of ≥ 1% of tumor cell nuclei were immunoreactive; and ATM was scored as previously described (13). (−) = no positive cells, (+) = 1–10% of the cells stained, (++) = 11–50% of the cells stained, and (+++) = 51–100% of the cells stained.
Western Blot Analysis
Cell lysates were obtained by treatment with the lysis buffer (Thermo Scientific Pierce, Rockford, IL) containing the protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase I and II inhibitors (Sigma), and the protein concentration was determined using the Brad-ford method (Bio-Rad, Hercules, CA). Equal volumes of cell lysates were loaded into 4–15% Mini-protean TGX precast gels (Bio-Rad, Hercules, CA) for electrophoresis. Proteins were then transferred from gels to the nitrocellulose membrane (Bio-Rad). Following incubation with 5% of non-fat milk (LabScientific, Livingston, NJ) for 30 min, the membrane was subsequently incubated with primary and horseradish-peroxidase conjugated secondary antibodies. Signals were detected by adding the Pierce chemiluminescent reagents (Thermo Scientific Pierce).
RNA Extraction, Reverse Transcription (RT) and real time PCR
Total RNA was extracted from cultured cells using TRizol (Life Technologies). cDNA was obtained from 5 ng of total RNA using the M-MLV Reverse transcription kit (Promega, Madison, WI), and the expression levels of miR-18a, miR-106a, miR-101, and miR-421 were quantified using PCR with specific primers listed in the following table.
| Primer’s name | Sequences |
|---|---|
| MiR 18a-F | 5’- TGTTCTAAGGTGCATCTAGTGC -3’ |
| MiR-18a-R | 5’- TGCCAGAAGGAGCACTTAGGG -3’ |
| MiR-101-F | 5’- TGCCCTGGCTCAGTTATCAC -3’ |
| MiR-101-R | 5’- TGC CAT CCT TCA GTT ATC ACA GTA-3’ |
| MiR-106a-F | 5’- CTTGGCCATGTAAAAGTGCTTACA-3’ |
| MiR-106a-R | 5’- CCATGGTAATGTAAGAAGTGCTTCA -3’ |
| MiR-421-F | 5’- GCACATTGTAGGCCTCATTAAATG -3’ |
| MiR-421-R | 5’- GAGATCACAGAGCAGGCGCCCAA -3’ |
Real-time PCR with the SYBR green detection was performed using an ABI Prism 7700 thermocycler with fluorescence detection (Applied Biosystems). The thermal cycling conditions were composed of 50°C for 2 min followed by an initial denaturation step at 95°C for 10 min, 45 cycles at 95°C for 30s, 60°C for 30s and 72°C for 30s. The experiments were carried out in duplicate for each data point. The relative quantification in gene expression was determined using the 2-ΔΔCt method (17). Using this method, we obtained the fold changes in gene expression normalized to an internal control gene GAPDH.
Detection of miRNA expression by in situ hybridization in human breast cancer tissues
The miRCURY LNA™ miRNA ISH kit and miRNA detection has-miR18a, 106a and control probes were purchased from Exiqon (Woburn, MA). The miRNAs in situ hybridization was carried out in the 296 cases of human breast cancer samples. Briefly, breast cancer tissue slides were deparaffinized and incubated with proteinase-K for 10min at 37 °C. After dehydrated, slides were incubated with the hybridization mix and hybridized for 1 hour at 50–60 °C. The slides were then subsequently incubated with the blocking solution (15 min at room temperature), the anti-DIG reagent (60 min at room temperature), and with AP substrate (2 hours at 30 °C). The sections were counter stained with nuclear fast red (Vector laboratories, Burlingame, CA). The stained sections were then scored for expression of has-miR18a and 106a miRNAs under the microscopy (Olympus). The sections were evaluated independently by two experienced pathologists according to the percentage of stained cells (18), with less than 20% of the cells stained was designated low expression of miRNA (+), and more than 20% of the cells stained was designated as high expression of miRNA (++).
Statistics
Data were analyzed by Chi-Square test, Student t-test and Pearson Correlation test and p values ≤ 0.05 were considered significant.
Results
ATM expression is elevated in ER negative breast cancer tissues
We recently reported ATM hyperactivation in breast cancer primary tissues with lymph node metastasis (13). When we further analyzed this set of the clinical data, we surprisingly found that ER negative breast cancer tissues have a much higher expression level of ATM. In the total of 296 samples, we found that the ATM expression level was negatively correlated with ER status (Figure 1A, Table 1) (P<0.001, Chi-square test). However, the ER status showed less correlation with expression of the active form of ATM—pS1981-ATM (P=0.155, chi-square test, Table 1). Since these observations were contradictory to the data reported in a previous publication by Tommiska et al (19), we repeated the immunohistochemistry (IHC) experiment and the analysis done by two independent pathologists confirmed our original observation.
Figure 1. ATM expression is upregulated in ER negative breast cancer tissues.
Immunohistochemistry was performed using the anti- ATM antibody in 296 human breast invasive ductal carcinoma (IDC) tissues. Positive was defined for ER if finding of ≥ 1% of tumor cell nuclei are immunoreactive. The number of positively or negatively stained cases of pan-ATM or pS1981-ATM was listed in Table 1.
Table 1.
Correlation of ER status with ATM expression in breast cancer tissues
| ER | |||
|---|---|---|---|
| negative | positive | P* | |
| ATM | |||
| − | 16 | 55 | |
| + | 23 | 82 | <0.001 |
| ++ | 26 | 51 | |
| +++ | 30 | 13 | |
| pS1981-ATM | |||
| − | 28 | 66 | |
| + | 24 | 50 | 0.155 |
| ++ | 15 | 47 | |
| +++ | 28 | 38 | |
Chi-Square Test
A retrospective chart review of the 296 cases of breast cancer was also conducted. We focused on 120 cases who received breast-conserving surgery followed by curative radiotherapy in order to assess the clinical relevance of the ATM expression with patient responses to radiotherapy. Our data showed that the local recurrence rate after radiotherapy correlated with ER status (Table 2, P=0.012) and ATM expression (Table 2, P=0.008, Pearson correlation test). Together, these results prompted us to further study a potential connection of ER and ATM expression.
Table 2.
Correlation of ER status and ATM expression with locoregional recurrence after breast-conserving surgery and radiotherapy
| Local recurrence | |||
|---|---|---|---|
| Yes | No | P* | |
| ER | |||
| negative | 11 | 31 | 0.012 |
| positive | 7 | 71 | |
| ATM | |||
| − | 2 | 24 | |
| + | 3 | 35 | 0.008 |
| ++ | 5 | 26 | |
| +++ | 8 | 17 | |
Pearson Correlation Test
ERα, but not ERβ, negatively regulates ATM expression in breast cancer cells
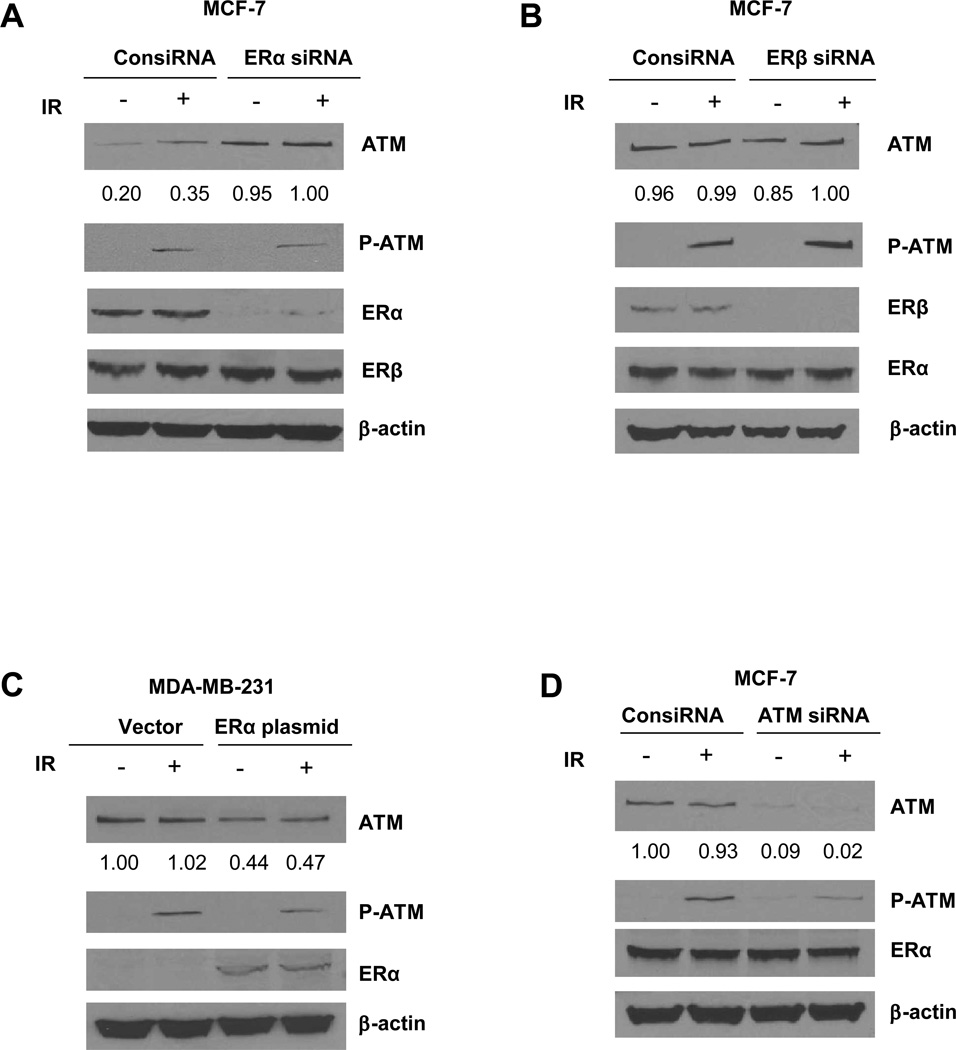
We hypothesized that ER might inhibit ATM expression. To test this notion, we first utilized an ER positive breast cancer cell line MCF-7. We transiently knocked-down ERα (Figure 2A) or ERβ (Figure 2B) in these cells. Interestingly we found that ATM expression level significantly increased in ERα knock-down cells (Figure 2A). On the contrary, ERβ knock-down did not show an effect on ATM expression (Figure 2B). We also assessed ionizing radiation (IR) induced ATM activation using the phospho-ATM Serine 1981 antibody in cells in ER α or β knock-down cells. Unlike the effect of ERα on the basal ATM expression, IR-induced ATM activation was unaffected by either ER α or β knock-down. This observation is consistent with the IHC data shown in Table 1.
Figure 2. ERα negatively regulates ATM expression.
(A and B). MCF-7 cells were transfected with control, ERα (A) or ERβ -siRNA (B). 48 hours after transfection, total cell lysates were harvested and subjected to immunoblotting using indicated antibodies. (C). MDA-MB-231 cells were transiently transfected with either vector only or ERα before they are assessed for expression of ATM, ATM S1981p, ERα and β-actin using in immunoblotting in the total cell lysates. (D). MCF-7 cells were transfected with control or ATM siRNA. 48 hours after transfection, total cell lysates were harvested and immunoblotting was performed using indicated antibodies. Image J was used for quantification for ATM expression. The numbers shown represent normalized ratios of ATM and β-actin.
We then overexpressed ERα in the ER negative breast cancer cell line MDA-MB-231 and found that ATM expression was reduced with ERα re-introduction (Figure 2C). On the contrary, ERα expression was not affected when ATM was knocked-down by siRNA in MCF-7 cells (Figure 2D). Together, these data indicate that ERα functions as an upstream component of the signaling network that can negatively regulate ATM expression. We also measured the ATM mRNA level in MCF-7 cells when ERα was knocked-down, however, we did not observe a change in ATM mRNA (Supplemental Figure S1), indicating that ERα regulation of ATM is not at the transcriptional level.
ERα regulates ATM expression through miRNA18a and miRNA106a
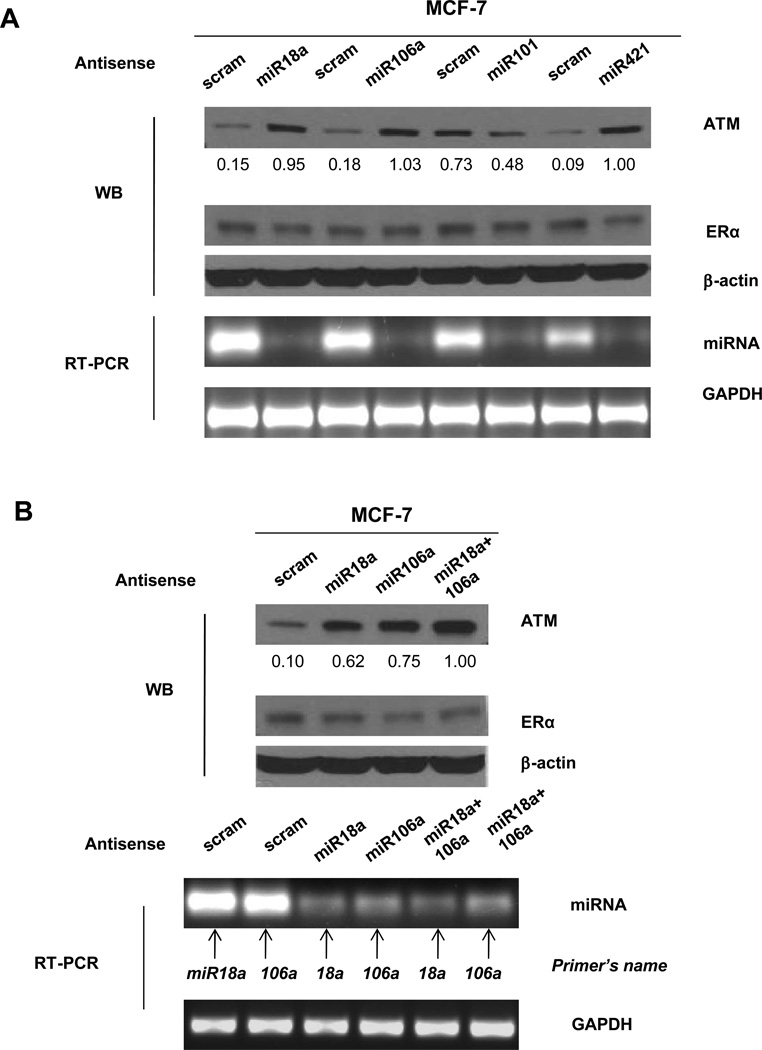
We suspected that ERα regulation of ATM might be through ERα-regulated miRNAs. To test this notion, we searched the available database containing the ERα regulated miRNA signature. There are 19 identified miRNAs that can be transcriptionally activated by ERα (8). We explored the possibility that some of these miRNAs might regulate ATM expression. Using the TargetScan program, we searched the 3’UTR region of the ATM gene for miRNA binding motifs. We found that approximately 7 nucleotides at the 5’ end of three miRNAs (miRNA- 18a, 101 and 106a) showed complementary sequences to the 3’UTR of the ATM gene (Supplemental Figure S2). To test whether these miRNAs indeed regulate ATM expression, we designed antisense oligonucleotides against each of the three miRNAs. Since miRNA421 was previously shown to regulate ATM expression (20), we also included the antisense oligonucleotide against miRNA 421 as the positive control. These oligos were transfected into MCF-7 cells. As shown in Figure 3A, we found that inhibiting miRNA 18a and miRNA 106a expression resulted in a significant increase of ATM expression, similar to the effect of inhibiting miRNA-421. However, knocking down miRNA-101 showed an opposite effect as ATM expression was reduced in antisense miRNA-101 expressing cells. Moreover, simultaneously knocking-down miRNAs-18a and -106a by antisense miRNAs resulted in a more marked increase in ATM expression (Figure 3B).
Figure 3. Knocking down miRNAs 18a and 106a increases ATM expression in breast cancer cells.
(A). MCF-7 cells were transfected with antisense oligos against miRNAs 18a, 106a, 101 or 421. A scrambled oligo was designed as a negative control. Immunobloting was conducted using antibodies against ATM, ERα or β-actin. RT-PCR was conducted to measure the expression of miRNAs, and GAPDH was used as a loading control. (B). MCF-7 cells transfected with scrambled, antisense miRNA 18a, 106a or both (miR18a+106a) were subjected to immunoblotting using the indicated antibodies or to RT-PCR using indicated primers. Image J was used for quantification for ATM expression. The numbers shown represent normalized ratios of ATM and β-actin.
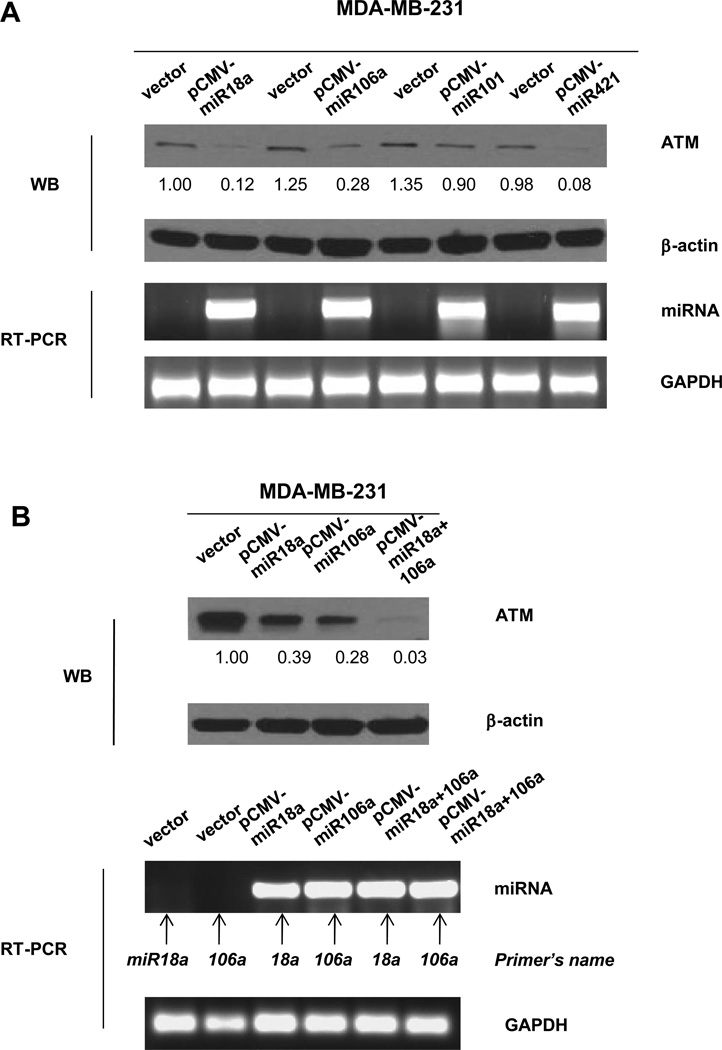
To further test ATM expression regulated by miRNA-18a and -106a, we utilized pCMV retroviral based expression constructs containing vector or miRNA-18a, -106a, -101 or -421. We infected MDA-MB-231 cells with these constructs and measured the change of ATM expression. We found that over-expression of miRNA -18a or -106a reduced ATM expression (Figure 4A). However, expressing miRNA 101 did not show any effects on ATM expression. Furthermore, we found that co-expression of miRNAs 18a and 106a resulted in a more significant reduction of ATM expression (Figure 4B). We also utilized a luciferase reporter assay following co-transfection of the luciferase reporter plasmid with miRNA-18a or miRNA-106a into MDA-MB-231 cells. Significant reductions in the luciferase activity from the reporter construct containing ATM 3’UTR were observed in the presence of miR-18a or miR-106a (Supplemental Figure S3). Taken together, we conclude that ERα mediates ATM expression through miRNAs 18a and 106a in breast cancer cells.
Figure 4. Overexpression of MiRNAs 18a and 106a suppresses ATM expression.
(A). MDA-MB-231 cells were infected with vector-only, pCMV-miRNA 18a, pCMV-miRNA106a or pCMV-miRNA421 followed by immunoblotting and PT-PCR using indicated antibodies or primers. (D). MDA-MB-231 cells were infected with vector, pCMV-18a, pCMV-106 or co-infected with pCMV18a and 106a. 48 hours after infection, immunoblotting and RT-PCR were conducted. Image J was used for quantification for ATM expression. The numbers shown represent normalized ratios of ATM and β-actin.
miRNA-18a and 106a are under-expressed in ER negative breast cancer tissues
To test the clinical connection of ER, miRNAs -18a and -106a, and ATM expression, we conducted in situ hybridization in the 296 cases of breast cancer tissues. We found that both miRNAs showed significant reduced expression levels in ER negative tissues (P=0.008 and 0.033 for miRNA-18a and miRNA-106a, respectively) (Supplemental Figure S4, Table 3). Furthermore, expression of miRNAs 18a and 106a negatively correlated with ATM expression (P<0.001 and P=0.001 for miRNA-18a and miRNA-106a, respectively) (Table 3). However, we did not observe any correlation of expression miRNAs 18a and 106a with Serine 1981 phosphorylated form of ATM (Table 3). Combining with the data presented in Figure 1 and Table 1 on the negative correlation of ER status and ATM expression, these observations further demonstrate the involvement of miRNAs -18a and -106a in ERα dependent ATM regulation.
Table 3.
Correlation of miRNA -18a and -106a with ER status and ATM expression in breast cancer tissues
| miR18a | P* | miR106a | P* | |||
|---|---|---|---|---|---|---|
| + | ++ | + | ++ | |||
| ER | ||||||
| negative | 29 | 66 | 0.008 | 34 | 61 | 0.033 |
| positive | 34 | 167 | 48 | 153 | ||
| ATM | ||||||
| − | 13 | 58 | <0.001 | 18 | 53 | 0.001 |
| + | 13 | 92 | 24 | 81 | ||
| ++ | 16 | 61 | 17 | 60 | ||
| +++ | 21 | 22 | 23 | 20 | ||
| pS1981-ATM | ||||||
| − | 23 | 71 | 0.142 | 21 | 73 | 0.376 |
| + | 11 | 63 | 20 | 54 | ||
| ++ | 18 | 44 | 18 | 44 | ||
| +++ | 11 | 55 | 23 | 43 | ||
Chi-Square Test
Discussion
In this report, we demonstrate in both cellular models and clinical samples that ERα negatively regulates expression of the ATM kinase. ATM is critical to determine the cellular survival to DNA damaging agents. In ER negative breast cancer tissues, we observed aberrant upregulation of ATM expression. We elucidated that ERα suppressed ATM expression through regulating miRNAs 18a and 106a. These observations highlight a novel mechanism linking ERα and ATM which might explain the resistant phenotype of many ER negative cancers to DNA damaging agents.
It is noted that there is discrepancy of our observations with a previous report that ATM expression is reduced in Brca1-/2 deficient and TNBC cells (19). Although it is not clear why the results differ, one possible explanation is that the clinical samples we used were all from sporadic breast cancer patients, and in the previous paper, most of the non-BRCA tumors samples were from familial breast cancer patients.
It is interesting to observe that despite its regulation on ATM expression, ERα does not affect ATM activation in response to IR. Although it is widely accepted that ATM Serine 1981 phosphorylation is a marker of ATM activation in response to IR, it is much less clear to what extent ATM activation might affect the radiation response in human tissues. Furthermore, ATM Serine 1981 phosphorylation induced by 6Gy of IR is not directly relevant to the clinical responsiveness to radiotherapy. Our data shown in Figure 1E has suggested that ATM expression appears to be more relevant to the therapeutic outcome of breast cancer radiotherapy. It is possible that other mechanisms of ATM regulation (such as the alterations of ATM expression due to ERα and its regulated miRNAs) contribute to the outcome of locoregional recurrence. However, we do not rule out that other factors regulated directly by ER alpha or regulated through ER alpha induced miRNAs are important for locoregional recurrence.
Unlike on ATM expression, ERα can directly regulate the DNA damage responsive proteins. For example, a recent report showed that ERα activated DNA-PKcs, a family member of the PI-3 like kinases (21). In addition, p53 can be a direct target of ERα (22). However, how these regulation is associated with the clinical responsiveness of ER negative tumors to DNA damaging agents are not clear. These regulatory cascades might be related to antiestrogen resistance as hyperactivation of the PI3K pathway, the most frequently mutated pathway in breast cancer, promotes antiestrogen resistance (23).
During our study, the link of miRNA18a-ATM has been reported (24). Our data provide a more comprehensive picture of miRNA regulation of ATM expression in breast cancer tissues. In addition to miRNA 18a, we found miRNA 106a also regulates the expression of ATM. More interestingly, deletion of both miRNAs results in a more significant effect on ATM expression, highlighting that multiple miRNAs regulates ATM in breast cancer tissues. Since miRNAs serve as fine-tuning regulators of protein expression, we do not rule out that other mechanisms might regulate ATM expression in breast cancer tissues. For example, ATM promoter methylation shown in locally advanced breast cancer (25) might contribute low ATM expression in ER positive tissues.
Since most of the ER-negative breast cancers are more aggressive and unresponsive to anti-estrogen therapies, other targeted therapies are urgently needed. Because ATM expression is up-regulated in ER negative cancers, ATM might represent a more plausible drug target in these tumors. Using specific ATM inhibitors might achieve more clinical benefits as this might specifically increase tumor sensitivities to many of the chemotherapeutic drugs as well as radiotherapy.
In summary, we identified the signaling transduction pathway involving ERα-miRNA18a and 106a - ATM that provides an explanation of chemo and radio-resistance in many ER negative breast cancers.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Most of the ERα negative breast tumors are resistant to many DNA damaging agents, although mechanisms underlying this phenotype are less clear. In this report, we demonstrate in cellular models and clinical samples that ERα negatively regulates expression of the ATM kinase through miRNAs 18a and 106a. These observations highlight a novel mechanism linking ERα and ATM which explains the resistant phenotype of ER negative cancers to chemotherapy and radiotherapy. Since ER-negative breast cancers are more aggressive and unresponsive to anti-estrogen therapies, other targeted therapies are urgently needed. Because ATM expression is up-regulated in ER negative cancers, ATM might represent an interesting drug target in these tumors. Using specific ATM inhibitors might achieve more clinical benefits as this might specifically increase tumor sensitivities to many of the chemotherapeutic drugs as well as radiotherapy.
Acknowledgements
We thank all members of the Xu laboratory for help. We thank Dr. Jaideep Thottassery (Southern Research Institute) and Dr. Xinmin Zhang (Temple University Hospital) for helpful comments on the manuscript. This work was supported by NIH grants R01CA133093 and R01ES016354 to Bo Xu, National Natural Science Foundation of China (Grant No. 30930038), Program for Changjiang Scholars and Innovative Research Team (Grant No. IRT0743) and the National 973 Program of China (Grant No. 2009CB521700) to Li Fu, and National Natural Science Foundation of China (Grant No. 81172531) to Xiaojing Guo.
Financial support: NIH grants R01CA133093 and R01ES016354 to Bo Xu, National Natural Science Foundation of China (Grant No. 30930038), Program for Changjiang Scholars and Innovative Research Team (Grant No. IRT0743) and the National 973 Program of China (Grant No. 2009CB521700) to Li Fu, and National Natural Science Foundation of China (Grant No. 81172531) to Xiaojing Guo.
Footnotes
Conflict of Interest: None
References
- 1.Stanford JL, Szklo M, Brinton LA. Estrogen receptors and breast cancer. Epidemiologic reviews. 1986;8:42–59. doi: 10.1093/oxfordjournals.epirev.a036295. [DOI] [PubMed] [Google Scholar]
- 2.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 3.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 4.Fountzilas G, Dafni U, Bobos M, Batistatou A, Kotoula V, Trihia H, et al. Differential response of immunohistochemically defined breast cancer subtypes to anthracycline-based adjuvant chemotherapy with or without paclitaxel. PLoS One. 2012;7:e37946. doi: 10.1371/journal.pone.0037946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheang MC, Voduc KD, Tu D, Jiang S, Leung S, Chia SK, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC.CTG MA.5 randomized trial. Clin Cancer Res. 2012;18:2402–2412. doi: 10.1158/1078-0432.CCR-11-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler AM, Alarid ET. Amping up estrogen receptors in breast cancer. Breast Cancer Res. 2007;9:305. doi: 10.1186/bcr1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marino M, Galluzzo P, Ascenzi P. Estrogen signaling multiple pathways to impact gene transcription. Curr Genomics. 2006;7:497–508. doi: 10.2174/138920206779315737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klinge CM. miRNAs and estrogen action. Trends Endocrinol Metab. 2012;23:223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maggi A. Liganded and unliganded activation of estrogen receptor and hormone replacement therapies. Biochim Biophys Acta. 2011;1812:1054–1060. doi: 10.1016/j.bbadis.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL, et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol. 2012;4(5):304–315. doi: 10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of 'conditional haploinsufficiency'. Cell Cycle. 2007;6:2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- 15.Bartkova J, Bakkenist CJ, Rajpert-De Meyts E, Skakkebaek NE, Sehested M, Lukas J, et al. ATM activation in normal human tissues and testicular cancer. Cell Cycle. 2005;4:838–845. doi: 10.4161/cc.4.6.1742. [DOI] [PubMed] [Google Scholar]
- 16.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 17.Frontini M, Kukalev A, Leo E, Ng YM, Cervantes M, Cheng CW, et al. The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2 activity in maintaining replicative fidelity and neurodevelopment. Dev Cell. 2012;23:356–370. doi: 10.1016/j.devcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian P, Zuo Z, Wu Z, Meng X, Li G, Wu Z, et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71:6463–6474. doi: 10.1158/0008-5472.CAN-11-1322. [DOI] [PubMed] [Google Scholar]
- 19.Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–2506. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- 20.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medunjanin S, Weinert S, Poitz D, Schmeisser A, Strasser RH, Braun-Dullaeus RC. Transcriptional activation of DNA-dependent protein kinase catalytic subunit gene expression by oestrogen receptor-alpha. EMBO Rep. 2010;11:208–213. doi: 10.1038/embor.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger CE, Qian Y, Liu G, Chen H, Chen X. p53, a target of estrogen receptor (ER) alpha, modulates DNA damage-induced growth suppression in ER-positive breast cancer cells. J Biol Chem. 2012;287:30117–30127. doi: 10.1074/jbc.M112.367326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morelli C, Garofalo C, Bartucci M, Surmacz E. Estrogen receptor-alpha regulates the degradation of insulin receptor substrates 1 and 2 in breast cancer cells. Oncogene. 2003;22:4007–4016. doi: 10.1038/sj.onc.1206436. [DOI] [PubMed] [Google Scholar]
- 24.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, et al. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One. 2011;6:e25454. doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vo QN, Kim WJ, Cvitanovic L, Boudreau DA, Ginzinger DG, Brown KD. The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene. 2004;23:9432–9437. doi: 10.1038/sj.onc.1208092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.