Abstract
Previous studies have shown conflicting data regarding Cyclin D1/Cdk2 complexes and, considering the widespread overexpression of Cyclin D1 in cancer, it is important to fully understand their relevance. While many have shown Cyclin D1/Cdk2 complexes to form active complexes, others have failed to show activity or association. Here, using a novel p21-PCNA fusion protein as well as p21 mutant proteins, we show that p21 is a required scaffolding protein, with Cyclin D1 and Cdk2 failing to complex in its absence. These p21/Cyclin D1/Cdk2 complexes are active and also bind the trimeric PCNA complex, with each trimer capable of independently binding distinct Cyclin/Cdk complexes. We also show that increased p21 levels due to treatment with chemotherapeutic agents result in increased formation and kinase activity of Cyclin D1/Cdk2 complexes, and that Cyclin D1/Cdk2 complexes are able to phosphorylate a number of substrates in addition to Rb. Nucleophosmin and Cdh1, two proteins important for centrosome replication and implicated in the chromosomal instability of cancer are shown to be phosphorylated by Cyclin D1/Cdk2 complexes. Additionally, PSF is identified as a novel Cdk2 substrate, being phosphorylated by Cdk2 complexed with either Cyclin E or Cyclin D1, and given the many functions of PSF, it could have important implications on cellular activity.
Keywords: Cyclin D1, Cdk2, breast cancer, kinase, p21, PCNA, NPM, Cdh1, PSF, SFPQ
Introduction
The canonical partners for Cyclin D1 are considered to be Cyclin-dependent kinase 4 (Cdk4) and Cyclin-dependent kinase 6 (Cdk6). The resulting complexes are inhibited selectively by INK family proteins that do not act on Cdk1 or Cdk2.1–4 Some studies have suggested that Cyclin D1 does not form complexes with Cdk2 (D1/K2)5 or that these complexes do form, but do not possess kinase activity because D1/K2 complexes are not phosphorylated by the Cdk-Activating Kinase (CAK).6, 7 However, studies indicate that D1/K2 complexes are present in cells in culture and are phosphorylated on the Cdk2 activating site T160,8 and that D1/K2 levels rival D1/K4 abundance in normal tissues and breast cancers in vivo in murine models.9 These latter observations suggest that D1/K2 complexes likely carry out biological functions.
Several observations suggest that Cyclin D1/Cdk2 complexes may carry out important functions during mammary tumorigenesis. First, there is strong evidence from mouse models that Cyclin D1/Cdk2 complexes are of comparable abundance to Cyclin D1/Cdk4 complexes in the normal mouse mammary gland as well as in breast cancers.10 In this publication by Sicinski and colleagues, a Flag-tagged Cyclin D1 construct was knocked into the endogenous Cyclin D1 locus so that it would be expressed at physiologically relevant levels, locations, and developmental intervals. In some tissues examined Cyclin D1/Cdk2 complexes were more abundant than Cyclin D1/Cdk4 complexes. Thus, the idea that Cyclin D1/Cdk4 complexes are typical and that Cyclin D1/Cdk2 complexes are noncanonical needs to be reassessed and reinterpreted. For example, it was shown that knock out of either Cyclin D1 or Cdk4 blocked the formation of breast tumors in MMTV-Her2/neu mice,11, 12 and this was taken as evidence of a critical role for Cyclin D1/Cdk4 complexes in Her2-mediated tumorigenesis. However, a more recent study demonstrates that Cdk2 knockout also blocks the formation of breast tumors in MMTV-Her2/neu mice.13
A second compelling argument for the relevance of Cyclin D1/Cdk2 complexes in breast cancers is the observation made a number of years ago that nine out of thirteen cell lines, including human breast cancer cell lines and a nontransformed human mammary epithelial cell line, contain Cyclin D1/Cdk2 complexes14
Third, recently published data further support the contention that Cyclin D1/Cdk2 complexes may play an important role in oncogenic transformation of mammary epithelial cells. Jackson and colleagues used our Cyclin D1-Cdk2 fusion protein construct (D1K2) to show that D1K2 was sufficient to replace either p53 inactivation or c-Myc overexpression in the anchorage-independent growth of human mammary epithelial cells (HMECs).15 Thus, in addition to the publications from our laboratory,8, 16–18 these other publications from other laboratories support the contention that Cyclin D1/Cdk2 complexes may play an important biological function during mammary tumorigenesis.
We previously constructed a Cyclin D1-Cdk2 fusion protein (D1K2) to model the biological functions of D1/K2 complexes. The D1K2 protein transforms cells and overrides the antiproliferative effects of TGFβ and rapamycin,8, 16 and transgenic expression of D1K2 in the mouse mammary gland induces the formation of breast tumors.17 These cancers exhibit similarities with human basal-like breast tumors,19 and display extensive chromosomal instability.18 Despite the probable importance of D1/K2 complexes in cellular function, little is known about their assembly or substrate specificity. Here we show that the assembly of D1/K2 complexes is highly dependent on p21 levels and that p21 abundance not only controls D1/K2 assembly, but also regulates its kinase activity in a biphasic manner.
Considering the large numbers of cellular proteins phosphorylated by Cdks,20 overactivation of Cdk2 due to high levels of Cyclin D1 likely causes cellular effects through other substrates in addition to Rb. As we have previously observed centrosome amplification and chromosomal instability in tumors initiated by a D1/K2 fusion protein (D1K2),17, 18 we investigated potential phosphorylation targets that regulate centrosome replication.
The process of centrosome duplication is quite complex, involving a large number of proteins throughout the cell cycle.21 Nucleophosmin (NPM) plays a crucial part in this process, binding to unreplicated centrosomes and dissociating upon phosphorylation by Cyclin E/Cdk2 on T199, thus permitting centrosome duplication.22 Interestingly, D1K2 appears capable of phosphorylating this site, comparably to the endogenous Cyclin E/Cdk2 complex, in vitro.
We also show that D1K2 is capable of phosphorylating the activating subunit of the APC/C, Cdh1. Cdh1 is inactivated when phosphorylated and its active form mediates the degradation of many proteins responsible for centrosome duplication23 and cytokinesis.24 Hyperphosphorylation of Cdh1 by D1K2 could lead to centrosome amplification directly through over-amplification, or through failed cytokinesis. Indeed, not only do we find centrosome amplification in cells expressing D1K2, but also instances of failed cytokinesis.18
The D1K2 fusion protein is not only useful for identifying the roles of overactive Cdk2 in tumor formation and progression, but also as a tool to help identify novel Cdk substrates. There are likely numerous Cdk substrates that are not yet known and their identification will greatly improve our knowledge of cell biology. In this study we show that polypyrimidine tract binding protein-associated splicing factor (PSF), also known as splicing factor-proline/glutamine rich (SFPQ), is a novel substrate for Cyclin E/Cdk2 and D1/K2 complexes. PSF is a multifunctional protein that is usually found in complexes with the PSF homolog, p54nrb.25
PSF is a 100 kDa protein that contains an RNA binding RGG domain, a large proline and glutamine rich segment, two RNA recognition motifs, and a DNA binding domain. It binds to polypyrimidine tracts in pre-mRNA and is indispensable for splicing26, 27, serving as a member of the spliceosome28 along with p68, also known as DDX5.29 PSF also functions in regulating transcription, binding to the thyroid hormone and retinoid x receptor control elements to activate transcription,30 as well as termination of transcription and the 3′ processing of subsequent RNAs.31 Additionally, the PSF/p54nrb complex is involved in the DNA double strand break repair mechanism,32 migrating to sites of DNA damage.33
Normally located in sub-nuclear speckles, PSF distribution is controlled by multiple mechanisms. Phosphorylation of c-terminal tyrosine residues by the Breast Tumor Kinase (BRK) induces a cytoplasmic relocalization that is thought to lead to cell cycle arrest.34 Likewise, phosphorylation of serine and threonine residues by an unknown kinase during apoptosis results in cytoplasmic localization and changes in protein interactions.35 Alterations in splicing regulation can also result from phosphorylation, with glycogen synthase kinase 3 phosphorylating threonine residues that cause alternate splicing of CD45 mRNA in T-cells.36
In this study, we show that both Cyclin E/Cdk2 and D1/K2 complexes are capable of phosphorylating PSF. The consequences of this phosphorylation are not yet known, however identifying these effects is of great interest since D1K2 expressing cells show a dramatic increase in PSF phosphorylation.
Materials and Methods
Transient Transfection, Transduction Using Recombinant Adenoviruses, and Stable Cell lines
Transient transfections were performed using 293T cells and Lipofectamine (Invitrogen, Carlsbad, CA) as described previously.37 Recombinant adenoviruses encoding the kinase active and kinase dead Cyclin D1-Cdk2 fusion proteins (D1K2, D1K2(KD), and D1K2(KE)), Flag-Cyclin D1, Flag-Cyclin E, Cdk1-His6, Cdk2-His6, and Cdk4-His6 as well as the various p21 mutants have been described8, 38, 39 and infection of cell cultures was performed as described previously.8 The MCF10A stable cell lines expressing D1K2 and D1K2(KD) were produced and characterized previously.18
Expression Vectors and the Construction of Genes Encoding the p21-PCNA, Cyclin D1-Cdk4 (D1K4), and Cyclin E-Cdk2 (EK2) Fusion Proteins
Expression vectors encoding Myc-tagged Cdh1 (Addgene plasmid 11595) and Green Fluorescent Protein-Flag-tagged NPM (Addgene plasmid 17578) were described previously40, 41 and purchased from Addgene Inc., Cambridge, MA. Construction of vectors encoding the Cyclin D1-Cdk4 and Cyclin E-Cdk2 fusion proteins were based on the Cyclin D1-Cdk2 (D1K2) fusion protein described previously.8 The Cyclin E cDNA described previously38 was used to replace the Cyclin D1 encoding region of D1K2 produce the Cyclin E-Cdk2 (EK2) fusion protein. Likewise, the Cdk4 cDNA described previously38 was used to replace the Cdk2 coding region of D1K2 to produce the Cyclin D1-Cdk4 (D1K4) fusion protein. The cDNA sequences encoding the EK2 and D1K4 proteins were synthesized by Bio Basic Canada Inc. (Markham, Ontario, Canada), subcloned into the EcoRI and XbaI sites of pcDNA3, and sequence verified. The cDNA sequences corresponding to the D1K4 and EK2 fusion proteins are listed in Supplemental Materials.
The p21-PCNA fusion protein was constructed by amplifying wild type human p21 cDNA using primers to attach N-terminal Hemagglutinin (HA) and His6 affinity tags, and to fuse the C-terminus of p21 to the flexible linker used in the construction of the D1K2 fusion protein.8 PCNA was amplified using primers to fuse the same flexible linker to its N-terminus. The central Bam HI site of the p21 and PCNA linkers was cut and used to fuse the corresponding cDNAs in frame. The resulting fusion construct was cloned into the pcDNA3 and pAd-Track-CMV plasmids for transient transfection and the construction of a p21-PCNA adenoviral vector, respectively. The complete p21-PCNA constructs were verified by DNA sequencing and the cDNA sequence is given in Supplemental Materials.
Affinity Purification and Analysis of Cyclin/Cdk-Containing Complexes, and Immunoprecipitation
Purification of D1K2 and D1K2(KD) was performed as described8 by sequential chromatography using Talon resin (BD Biosciences Clontech, Palo Alto, CA) with elution by imidazole followed by anti-FLAG-agarose resin (Sigma-Aldrich, St. Louis, MO) with elution by FLAG peptide (Sigma-Aldrich). Immunoprecipitations were performed using 4 Ig/tube of the Myc-tag antibody 9E10 (Santa Cruz Biotechnology, Inc.) or p-TP (Cell Signaling Technology, Inc., Danvers, MA) and the immunoprecipitates were collected using Protein G-sepharose (Invitrogen). The immunoprecipitates were washed and analyzed by immunoblot.
Immunoblot Analysis
Immunoblotting was performed as described,37 employing antibodies to p-TP (Cell Signaling Technology) or PSF (Santa Cruz Biotechnology). All other antibodies used were purchased from commercial sources and listed previously.8, 16, 17, 20, 37
NPM and Cdh1 In Vitro Kinase Assays
Kinase assays were performed as described.37 GST-Rb was purchased from Santa Cruz Biotechnology, Inc. GST-NPM and GST-Cdh1 were purchased from Abnova (Taipei, Taiwan). Two micrograms each of GST, GST-Rb, GST-NPM, and 1 microgram of GST-Cdh1 were present in each reaction. The amount of each kinase assay subjected to autoradiography represents 38% of the total reaction volume. The amount of each kinase assay analyzed by immunoblot was 10% of each reaction.
PSF Expression Constructs
Plasmids encoding WT-PSF, p54nrb, and DDX5 were obtained from OriGene Technologies, Inc. (Rockville, MD). The His6-PSF construct was a gift from Dr. Stefan Stamm (University of Kentucky, Chandler Medical Center). Nucleotide sequence-optimized constructs encoding FLAG-PSF and FLAG-PSF(12M) were synthesized and subcloned into the pcDNA3 vector by Bio Basic Canada, Inc. Amino acid mutations were made using the following primers for QuickChange PCR:
S33A Forward 5′-GGCCTCCACGACTTCCGTGCTCCGCCGCCCGGCATGGGC-3′
Reverse 5′-GCCCATGCCGGGCGGCGGAGCACGGAAGTCGTGGAGGCC-3′;
S379A Forward 5′-CTTTCTGTTCGTAATCTTGCACCTTATGTTTCCAATGAAC-3′,
Reverse 5′-GTTCATTGGAAACATAAGGTGCAAGATTACGAACAGAAAG-3′;
T687A Forward 5′-GGAATGGGGCCTGGAGCTCCAGCAGGATATGGTAG-3′,
Reverse 5′-CTACCATATCCTGCTGGAGCTCCAGGCCCCATTCC-3′;
A33S Forward 5′-CATCCCGGGTGGAGGAGACCTAAAGTCATGCAG-3′,
Reverse 5′-CTGCATGACTTTAGGTCTCCTCCACCCGGGATG-3′;
A379S Forward 5′-GCGTGAGAAATTTGTCGCCCTATGTCTCCAATG-3′,
Reverse 5′-CATTGGAGACATAGGGCGACAAATTTCTCACGC-3′;
A687T Forward 5′-CCATAACCTGCGGGAGTCCCGGGGCCCATACC-3′,
Reverse 5′-GGTATGGGCCCCGGGACTCCCGCAGGTTATGG-3′.
His6-PSF Bacterial Protein Expression
BL21(DE3) E. coli were transformed with the pET21a His6-PSF vector and cloned on ampicillin-agar plates. The bacteria were grown in LB + ampicillin overnight and then inoculated in LB + ampicillin to an OD600 0.1 – 0.15 and incubated at 37 °C with shaking until the OD600 reached 0.4 – 0.5. Protein expression was induced with 0.4 mM IPTG for 3 hours at 37 °C.
PSF In Vitro Kinase Assays
Bacteria expressing His6-PSF were lysed by sonication in TALON extraction buffer (10 mM MOPS pH 7, 10% glycerin, 100 mM KCl, 10 nM microcystin, 5 mM MgCl2, 1% Triton X-100, 1 mM Na3VO4, 33 mM Na4O7P2), isolated using TALON resin (Clontech Laboratories Inc., Mountain View, CA), and washed with TALON wash buffer (10 mM MOPS pH 7, 10% glycerin, 100 mM KCl, 10 nM microcystin, 5 mM MgCl2, 0.1% CHAPS, 50 mM imidazole). Beads were suspended in kinase assay buffer (50 mM Hepes pH 7.5, 10 mM MgCl2, 2.5 mM EGTA, 1 mM DTT, 0.1 mM NaF, 0.1 mM Na3VO4) and incubated at 37 °C with Cyclin/Cdk complex, 0.08 μM ATP, and 3.875 μCi [γ-32P]-ATP. Cyclin/Cdk complexes contained either 0.157 ng of Cyclin B/Cdk1, Cyclin E/Cdk2, or Cyclin D1/Cdk4 (Cell Signaling Technology, Inc.) or D1K2 purified from the D1K2-T2, CL6 cell line using anti-FLAG agarose. Samples were then boiled in SDS sample buffer and resolved on 12% polyacrylamide gels. Bands were imaged with autoradiography film and excised and quantitated using a Beckman Coulter LS600SC scintillation counter (Beckman Coulter, Inc., Brea, CA).
Results
p21 serves as an assembly factor for Cyclin D1/Cdk2 complexes
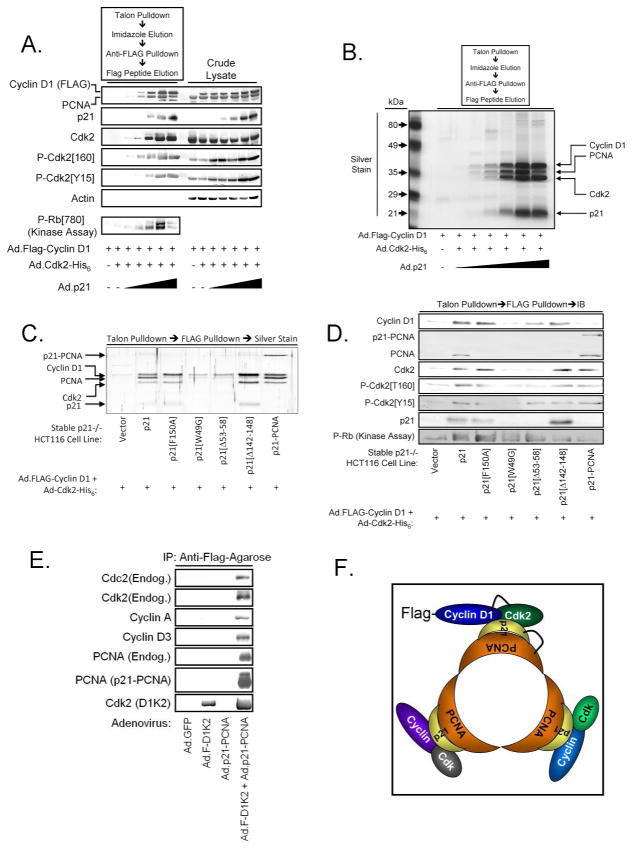
HCT116 colon cancer cells lacking the gene encoding p2142 were employed as a model system to examine the role of p21 in the formation of D1/K2 complexes. Infection of these cells with recombinant adenoviruses encoding epitope-tagged Cyclin D1 and Cdk2 did not result in the formation of D1/K2 complexes (Figure 1A). However, steadily elevating p21 expression using increasing multiplicities of infection of a recombinant adenovirus encoding p21 facilitated the formation D1/K2 complexes. In contrast, p27 was a very poor assembly factor for D1/K2 complexes (Supplemental Figure S1). The D1/K2 complexes stabilized by p21 expression catalyzed Rb phosphorylation in a biphasic manner that was observed in multiple experiments. Interestingly, D1/K2 assembly correlated directly with Cdk2 phosphorylation on T160 suggesting that complex formation is required for recognition by CAK or a CAK-like enzyme. Cdk2 phosphorylation on its inhibitory site, Y15, lagged behind T160 phosphorylation and could explain the biphasic kinase activity observed.
Figure 1.
p21 promotes D1/K2 complex assembly. (A) Immunoblot analysis of crude lysates and affinity purified D1/K2 complexes from p21−/− HCT116 cells infected with adenoviruses encoding Cyclin D1, Cdk2, and increasing amounts of p21. (B) Silver stained SDS-PAGE gel of affinity purified D1/K2 complexes from p21−/− HCT116 cells infected with adenoviruses encoding Cyclin D1, Cdk2, and increasing amounts of p21. (C) Silver stained SDS-PAGE gel of affinity purified D1/K2 complexes from p21−/− HCT116 cells infected with adenoviruses encoding Cyclin D1, Cdk2, and the indicated p21 constructs. (D) Immunoblot analysis of affinity purified D1/K2 complexes from p21−/− HCT116 cells infected with adenoviruses encoding Cyclin D1, Cdk2, and the indicated p21 constructs. (E) Immunoblot analysis of affinity purified Cyclin D1 complexes from p21−/− HCT116 cells infected with adenoviruses encoding the indicated proteins. (F) Model of trimeric p21/PCNA complexes containing multiple Cyclin/Cdk complexes.
Silver stain analysis of the resulting complexes showed a constant ratio between Cyclin D1, PCNA, Cdk2, and p21 at low p21 levels suggesting a 1:1:1:1 stoichiometry within the complex (Figure 1B). This is expected because p21 is able to simultaneously bind PCNA and Cyclin/Cdk complexes via distinct domains.39, 43, 44 At the highest p21 levels, the presence of PCNA in the complexes decreased.
These observations suggested the possibility that the presence of PCNA in the complex may be required for Cdk2 kinase activity. This issue was addressed by employing cell lines stably expressing previously described p21 point or deletion mutants39 that are defective in either binding to PCNA (F150A, Δ142-148) or to Cyclin/Cdk complexes (W49G, Δ53-58). Wild type p21 and the mutants unable to bind PCNA effectively assembled D1/K2 complexes (Figure 1C). Kinase complexes stabilized by wild type p21 and the F150A mutant exhibited similar kinase activities, indicating that PCNA is not required for D1/K2 kinase activity (Figure 1D). The Δ142-148 p21 mutant was equally effective as the wild type and F150A mutant forms of p21 in facilitating complex formation, but exhibited lower kinase activity. This could be due to larger-scale structural perturbations of p21 because of the more dramatic nature of the mutation, or could result from an altered stoichiometry of binding of the p21[Δ142-148] mutant. As expected, the W49G and Δ53-58 p21 mutants defective in Cyclin/Cdk binding stabilized the formation of D1/K2 complexes only slightly over that observed in the absence of p21.
The crystal structure of PCNA bound to the C-terminus of p21 suggests that p21 may be able to associate with trimeric PCNA and Cyclin/Cdk complexes simultaneously. This could facilitate the access of Cyclin/Cdk complexes with PCNA-associated substrates as shown previously.44 We constructed a novel p21-PCNA fusion protein based on the p21/PCNA complex crystal structure45 in which the C-terminus of p21 is fused to the N-terminus of PCNA in order to examine whether PCNA can simultaneously trimerize and associate with Cyclin/Cdk complexes. This was confirmed by showing that the p21-PCNA fusion protein mediated the formation of complexes with Cyclin D1, Cdk2, and endogenous PCNA (Figure 1C and 1D, last lane). We then reasoned that the Cyclin D1-Cdk2 fusion protein and the p21-PCNA fusion protein could be used as tools to determine whether a trimeric PCNA complex is capable of simultaneously associating with more than one Cyclin/Cdk complex.
Affinity purification of Flag-tagged D1K2 did not reveal D1K2 association with other endogenous Cyclins or Cdks (Figure 1E). However, if the p21-PCNA fusion protein was coexpressed with D1K2, multiple endogenous Cyclins and Cdks were detected. The results obtained using the p21-PCNA fusion protein suggest a model in which trimeric p21/PCNA complexes are capable of simultaneously binding more than one Cyclin/Cdk complex (Figure 1F). p21 and PCNA are known to translocate to sites of DNA repair in response to DNA damage.46 Further, published reports indicate that PCNA recruits proteins such as Fen1, DNA ligase, and DNA polymerases that are involved in DNA repair in a rotary manner, suggesting that recruitment of Cyclin/Cdk2 complexes to PCNA may similarly play a role in regulating DNA repair.47, 48 Consistent with this notion, PCNA facilitates Cdk2-dependent phosphorylation of PCNA associated proteins.49
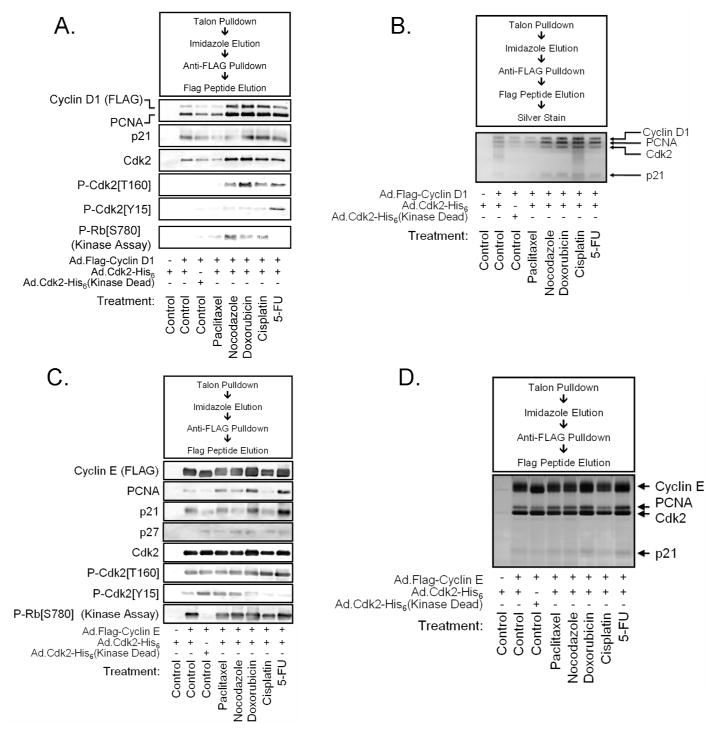
D1/K2 assembly and kinase activity are altered by chemotherapeutic agents
Cellular p21 levels are regulated by multiple signaling pathways, including DNA damage-mediated upregulation of p21 by p53 activation. Therefore, we examined whether different chemotherapy agents increase the formation and kinase activity of D1/K2 complexes in parental HCT116 cells with wild type p21 and p53. Nocodazole, Doxorubicin, Cisplatin, and 5-Fluorouracil (5FU) increased the levels of D1/K2 complexes, while Paclitaxel did not (Figure 2A). The drugs that facilitated the formation of D1/K2 complexes also increased D1/K2 kinase activity and Cdk2 phosphorylation on T160, with the exception of 5-FU. This inhibition of kinase activity is likely a result of 5-FU induced Cdk2 tyrosine phosphorylation on Y15. Silver stain analysis also demonstrated stabilization of D1/K2 complexes resulting from treatment with chemotherapy drugs (Figure 2B). In contrast, the abundance of Cyclin E/Cdk2 were essentially unchanged upon treatment with chemotherapeutic drugs and kinase activity of the complexes was not altered dramatically (Figure 2C, D). Treatment of p21−/− HCT116 cells with chemotherapy agents failed to induce formation of D1/K2 complexes (data not shown).
Figure 2.
Treatment with chemotherapy agents alters the assembly and activity of D1/K2 complexes. (A) Immunoblot analysis of affinity purified D1/K2 complexes from parental HCT116 cells infected with adenoviruses encoding the indicated proteins and treated with various chemotherapy agents. (B) Silver stained SDS-PAGE gel of affinity purified D1/K2 complexes from HCT116 cells infected with adenoviruses encoding the indicated proteins and treated with various chemotherapy agents. (C) Immunoblot analysis of affinity purified E/K2 complexes from parental HCT116 cells infected with adenoviruses encoding the indicated proteins and treated with various chemotherapy agents. (D) Silver stained SDS-PAGE gel of affinity purified E/K2 complexes from HCT116 cells infected with adenoviruses encoding the indicated proteins and treated with various chemotherapy agents.
Cellular D1/K2 Complexes can Phosphorylate NPM
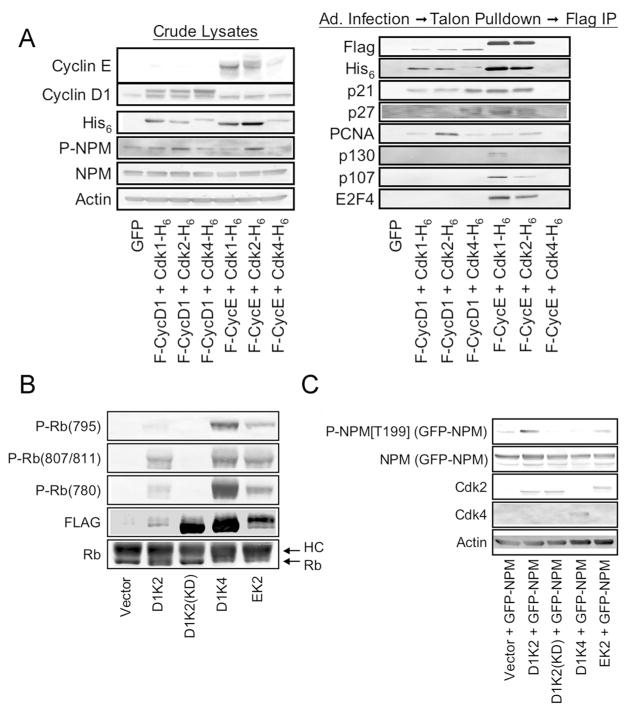
As mentioned above, NPM localizes to centrosomes and plays a critical role in their duplication during the cell cycle. Cdk2 phosphorylates NPM on T199 and phosphorylation of this site initiates centrosome duplication. Cyclin E/Cdk2 complexes are thought to catalyze this phosphorylation event in normal cells.50 Complexes of Cdk6 with the viral Cyclin, K-Cyclin, are also capable of phosphorylating NPM on T199 and inducing centrosome amplification.51 This raises the possibility that other transformation-associated Cyclin/Cdk complexes may be able to perform this function. We executed an experiment to determine which Cyclin D1 or Cyclin E/Cdk complexes are capable of phosphorylating NPM on T199. NMuMG cells were infected with recombinant adenoviruses encoding Flag-tagged Cyclins and His6-tagged Cdks in the indicated combinations (Figure 3A, left panel). Of these combinations, only coexpression of Cyclin D1 and Cdk2 and Cyclin E and Cdk2 significantly increased NPM phosphorylation relative to the control adenovirus treatment. The combinations that did not increase NPM concentration might have been inactive because of the inability of the particular combination to form complexes.
Figure 3.
D1/K2 complexes and the Cyclin D1-Cdk2 fusion protein phosphorylate NPM. (A) Immunoblot analysis of crude lysates (left panel) and affinity purified Cyclin/Cdk complexes (right panel) from NMuMG cells transduced with adenoviruses encoding the indicated proteins. (B) Immunoblot analysis of an in vitro kinase assay using Cyclin/Cdk fusion proteins purified from 293T cells transiently transfected with vectors encoding the indicated proteins. HC represents the heavy chain of the antibody used in the immunoprecipitation. The band labeled Rb below is the GST-Rb substrate present in the kinase assays. (C) Immunoblot analysis of cell lysates obtained from 293T cells transiently transfected with constructs coding for the indicated proteins. “(GFP-NPM)” indicates that the bands shown represent the exogenously expressed GFP-NPM rather than endogenous NPM.
Various Cyclin/Cdk Combinations Form Complexes
Cyclin/Cdk complex formation was assessed by performing sequential purifications using Talon resin to isolate the His6-tagged Cdks followed by purification using anti-FLAG-agarose resin to isolate the FLAG-tagged Cyclins. The results indicate that all possible combinations of Cyclin D1 and Cyclin E with Cdk1, Cdk2, or Cdk4 formed complexes with the exception of Cyclin E and Cdk4 (Figure 3A, right panel). As expected, only the Cyclin E-associated complexes contained p130, p107, and E2F4. D1/K2 complexes bound much more PCNA than the other Cyclin/Cdk complexes. This observation is in line with our previous observations that D1/K2 isolated from NMuMG cells using the same procedure contained stoichiometric amounts of p21 and PCNA.8
Characterization of novel Cyclin/Cdk fusion proteins
Overall, these results underscore the high degree of promiscuity among Cyclins and Cdks and indicate that in breast cancers where individual Cyclins such as Cyclin E, D, or A are overexpressed52–55 it is difficult to determine which of the resulting Cyclin/Cdk complexes mediate the oncogenic effects of the individual Cyclins that are overexpressed. The Cyclin D1-Cdk fusion protein (D1K2) was designed to specifically model the functions of D1/K2 complexes.8 We therefore designed vectors encoding N-terminally FLAG tagged and C-terminally His6-tagged Cyclin D1-Cdk4 (D1K4) and Cyclin E-Cdk2 (EK2) fusion proteins to specifically model the functions of the respective Cyclin/Cdk complexes. These vectors were transfected into 293T cells and the fusion proteins were isolated from the cell lysates using anti-FLAG-agarose and the immunoprecipitates were subjected to Cdk kinase assays using Rb as the substrate. The kinase assays were analyzed by immunoblot with phospho-specific antibodies to Rb (Figure 3B). In these assays the D1K2, D1K4, and EK2 fusion proteins exhibited kinase activity toward Rb and showed preferences in their phosphorylation of different sites. A kinase dead mutant of D1K2 (D1K2(KD)) did not exhibit kinase activity in this assay. These novel Cyclin-Cdk fusion proteins were cotransfected with NPM fused to Green Fluorescent Protein (GFP-NPM) or Myc-tagged Cdh1. In agreement with the results of Figure 3A, D1K2 and EK2 induced NPM phosphorylation on T199 (Figure 3C).
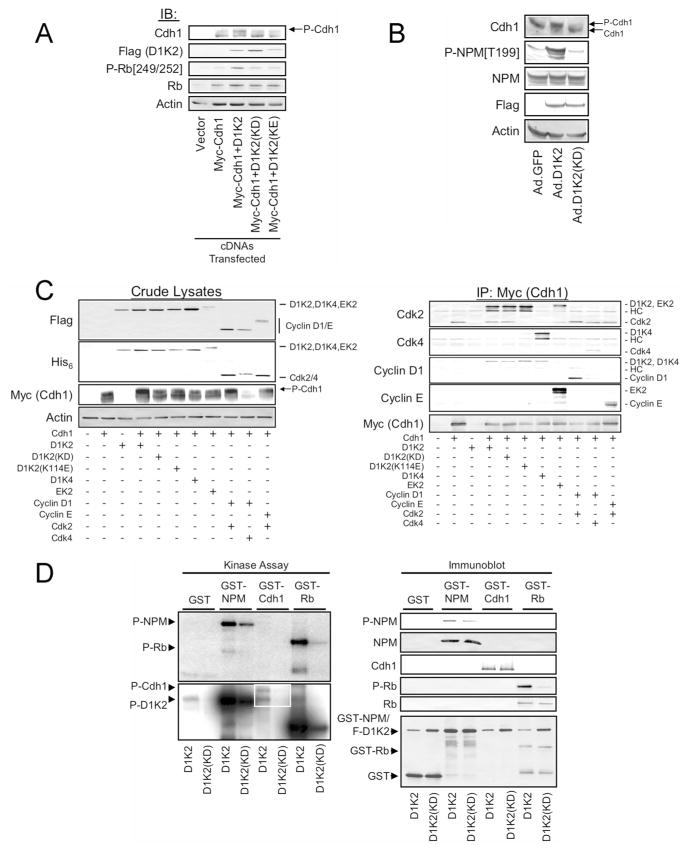
Cellular D1/K2 Complexes can Phosphorylate Cdh1
Like NPM, the Anaphase Promoting Complex/Cyclosome (APC/C) regulatory subunit Cdh1 is a substrate for Cyclin E/Cdk2, and Cyclin E/Cdk2-dependent phosphorylation inhibits its association with the APC/C, thus blocking APC/C activity. Heterozygous deletion of Cdh1 results in a high frequency of tumors, including breast tumors, and chromosomal instability.23 This suggests that Cdh1 phosphorylation by D1K2 could explain its ability to induce breast tumors that exhibit chromosomal instability. Cyclin E/Cdk2-dependent phosphorylation of Cdh1 causes a decrease in its electrophoretic mobility,56 therefore we examined whether D1K2 or kinase dead D1K2 mutants altered the mobility of Cdh1 in coexpression experiments. Transfection experiments in 293T cells were carried out in which Cdh1 was coexpressed with D1K2 or previously described kinase dead mutants of D1K2. D1K2(KD) is catalytically inactive due to a mutation in the ATP binding site of the Cdk2 domain. D1K2(KE) is kinase dead due to a K114E mutation in Cyclin D1. This mutation prevents the intramolecular activation of Cdk2 by the Cyclin D1 domain and prevents regulatory phosphorylation of the Cdk2 domain.8 D1K2 coexpression induced a mobility shift of Cdh1 relative to the control, while both kinase dead D1K2 mutants were without effect (Figure 4A). This shift in Cdh1 mobility correlated with the phosphorylation of endogenous Rb. Cdh1 overexpression increased endogenous Rb expression.
Figure 4.
D1/K2 complexes and the Cyclin D1-Cdk2 fusion protein physically interact with and phosphorylate Cdh1. (A) Immunoblot analysis of cell lysates obtained from 293T cells transiently transfected with vectors encoding the indicated proteins. (B) Immunoblot analysis of cell lysates from NMuMG cells infected with adenoviruses encoding the indicated proteins. Flag staining denotes the presence of the D1K2 and D1K2(KD) proteins. (C) Immunoblot analysis of crude lysates (left panel) and affinity purified Cdh1 complexes (right panel) obtained from 293T cells transiently transfected with constructs encoding the indicated proteins. The identities of the bands are indicated to the right of some panels. (D) Autoradiograms of short (top left panel) and long (lower left panel) exposures and immunoblot analysis (right panel) of kinase assays using D1K2 and D1K2(KD) isolated from NMuMG cells infected with recombinant adenoviruses, and GST-NPM, GST-Cdh1, or GST-Rb as substrates, or GST alone as a control.
D1K2 Phosphorylates NPM and Cdh1 in Nontransformed Cells
We wanted to determine whether similar results would be obtained in nontransformed NMuMG mammary epithelial cells, therefore these cells were infected with control adenovirus (Ad.GFP) or adenoviruses encoding D1K2 or D1K2(KD). D1K2 expression induced a shift in the mobility of endogenous Cdh1 that was not observed with expression of D1K2(KD) (Figure 4B). Similarly, D1K2 but not D1K2(KD) increased the phosphorylation of endogenous NPM on T199 even though both fusion proteins were expressed at equivalent levels based on immunostaining with the FLAG antibody.
Various Cyclin/Cdk Complexes Bind to Cdh1
We next wanted to investigate if different Cyclin/Cdk complexes and the D1K2, D1K4, and EK2 fusion proteins could form stable complexes containing Cdh1. Myc-tagged Cdh1 was coexpressed with various Cyclin/Cdk complexes or the Cyclin-Cdk fusion proteins and the lysates were subjected to immunoblot analysis (Figure 4C, left panel) or immunoprecipitated using an antibody to the Myc epitope tag followed by immunoblot analysis of the immunoprecipitates (Figure 4C, right panel). The ratio of the upper Cdh1 band to the lower Cdh1 band in the crude lysates was highest in the samples co-expressing D1K2, and D1/K2 and Cyclin E/Cdk2 complexes. Cdh1 coimmunoprecipitated with D1K2, D1K2(KD), D1K2(KE), D1K4, EK2, and D1/K2, Cyclin D1/Cdk4, and Cyclin E/Cdk2 complexes (Figure 4C, right panel). Interestingly, endogenous Cdk2 coimmunoprecipitated with Cdh1 when it was expressed alone, and also when Cdh1 was coexpressed with various other Cyclin/Cdk complexes and fusion proteins. Endogenous Cyclin E, Cyclin D1, or Cdk4 were not observed in Cdh1 immunoprecipitates suggesting that Cdh1 has the highest affinity for Cdk2.
D1K2 Directly Phosphorylates NPM and Cdh1 In Vitro
The results in Figures 3 and 4 suggest that the D1K2 fusion protein can phosphorylate NPM and Cdh1, but it is possible that D1K2 induces the phosphorylation of these proteins through indirect mechanisms. We performed in vitro kinase assays using purified GST-NPM and GST-Cdh1 as the substrate for D1K2, or as a control, D1K2(KD) to determine whether D1K2 could directly phosphorylate Cdh1 and NPM. The kinase assays were analyzed by autoradiography (Figure 4D, left panels) and immunoblot (Figure 4D, right panel). D1K2 phosphorylated NPM. Some phosphorylation was observed in the D1K2(KD) control, but this was less than that observed with D1K2, and the immunoblots indicate that three to four fold more D1K2(KD) was present in the kinase assays than D1K2. D1K2 catalyzed robust phosphorylation of Rb and very little phosphorylation was observed with D1K2(KD). This suggests that D1K2 can directly phosphorylate NPM, but that another NPM kinase may also coimmunoprecipitate with D1K2. A phospho-T199 NPM immunoblot showed a pattern similar to the autoradiogram suggesting that the phosphorylation occurs on this site during the kinase reactions, although this does not rule out the phosphorylation of other sites as well.
Cdh1 kinase assays were limited by low concentrations of Cdh1, but a longer exposure of the autoradiogram showed that D1K2, but not D1K2(KD) was able to phosphorylate GST-Cdh1. A band that migrates just beneath GST-Cdh1 was observed in all of the lanes containing D1K2, but not in the lanes containing D1K2(KD). This band migrates at the same molecular size as D1K2 and is likely due to D1K2 autophosphorylation. Immunoblot with a GST antibody shows the relative levels of the GST fusion proteins in the assays. GST and GST-NPM are present in similar abundance (note that GST-NPM comigrates with FLAG-tagged D1K2). GST-Rb is present at lower levels than GST-NPM and GST-Cdh1 is barely detectable by immunoblot with an antibody to GST.
Identification of PSF as a Substrate of Cdk2
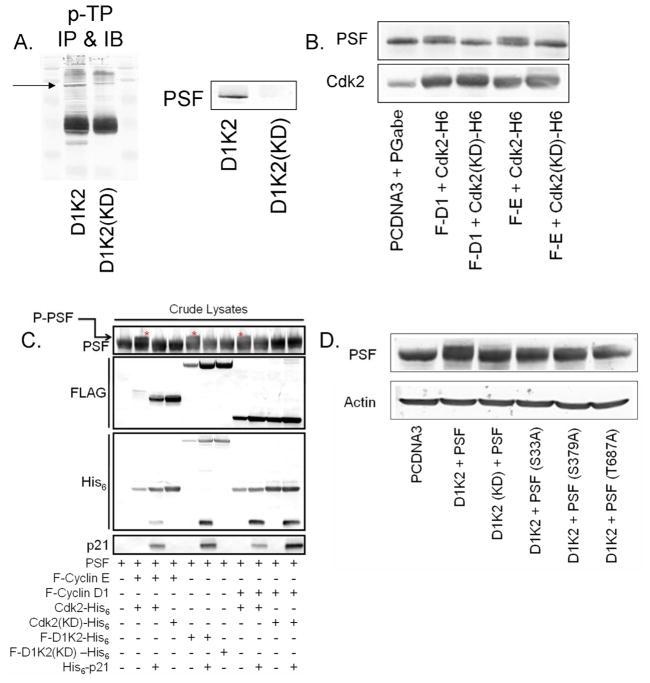
In an effort to identify the cellular substrates of D1K2, we utilized the previously described MCF10A cell lines expressing D1K2 or D1K2(KD).18 Immunoprecipitation followed by immunoblot using an antibody that recognizes phosphorylated threonine residues followed by a proline, potential Cdk2 phosphorylation sites, identified proteins in cell lysates that were preferentially phosphorylated in the cells expressing the kinase active D1K2 (Figure 5A left panel). A protein running slightly below the 120 kDa marker (denoted by an arrow) was identified by mass spectrometry as PSF. Further immunoblot analysis confirmed the differential levels of the protein in the samples (Figure 5A, right panel).
Figure 5.
Cdk2 phosphorylates PSF. (A) Immunoprecipitation of lysates from MCF10A cells stably expressing D1K2 or D1K2(KD) using a phospho-TP (p-TP) antibody and immunoblotted for p-TP (left) and PSF (right). (B) Immunoblot analysis showing a PSF mobility shift in 293T cells transiently transfected with PSF and various Cyclins/Cdks. (C) Immunoblot analysis of 293T lysates showing a PSF mobility shift in the presence of different Cyclin/Cdk complexes +/− p21. (D) Immunoblot analysis of 293T lysates expressing D1K2 and wild type PSF or PSF with mutations in potential phosphorylation sites to visualize the PSF mobility shift.
To confirm phosphorylation due to Cdk2, we overexpressed PSF in addition to its binding partners p54nrb and DDX5, along with different combinations of Cyclins and Cdks via transient transfection in the 293T cell line. The coexpression of Cyclin E or Cyclin D1 and Cdk2, induced the appearance of a second, lower mobility band in SDS-PAGE (Figure 5B). This was not seen with a kinase dead mutant of Cdk2. This mobility shift has been used as an indicator of phosphorylation in the past35 and confirms that Cdk2 can induce phosphorylation of PSF. These results were duplicated in Figure 5C, with Cyclin E/Cdk2, D1/K2, and D1K2 all inducing a mobility shift. Additionally, this shift is inhibited by co-expression of p21. In all three cases, p21 expression eliminated this shift, presumably by blocking phosphorylation through inhibition of Cdk2 activity.
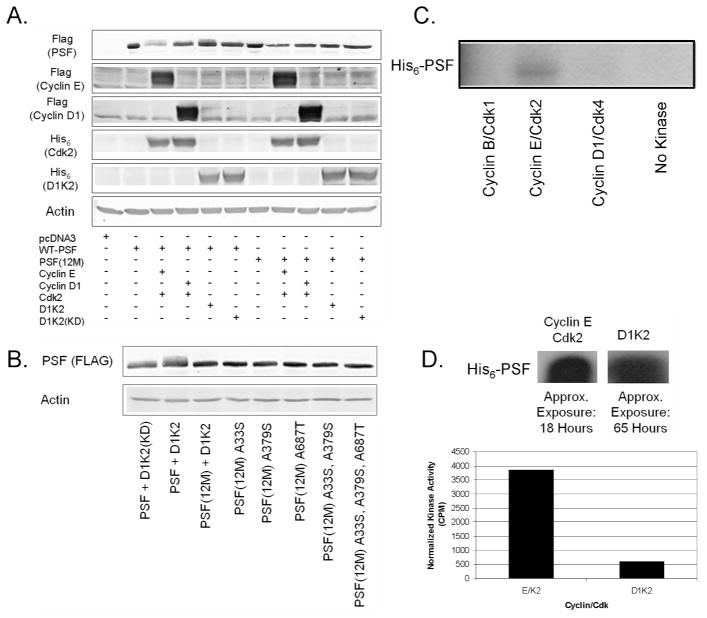
Previous biochemical analyses have identified three serine or threonine residues followed by a proline in PSF that are phosphorylated by unknown kinases: S33, S379, and T687.57 Mutation of any of these potential Cdk2 phosphorylation sites to alanine results in a notable decrease in the upper, phosphorylated band, but not a total elimination of it (Figure 5D). This is complicated by the fact that the endogenous, wild type protein is still present. What is clear is that each site is likely phosphorylated, perhaps leading to other phosphorylation events by Cdks or other kinases.
In order to obviate this problem, we designed a FLAG-tagged PSF construct, allowing the analysis of only the exogenous PSF. Again, Cyclin E/Cdk2 or D1K2 induced a mobility shift and an analogous construct in which all twelve serine and threonine residues followed by a proline residue were mutated to alanine residues failed to show a mobility shift regardless of kinase expression (Figure 6A). The fact that this mutant, lacking any potential Cdk2 phosphorylation sites, is not phosphorylated is further evidence that PSF is directly phosphorylated by Cdk2. Reverse mutations of A33S, A379S, and A687T, restoring potential phosphorylation sites, failed to restore the shift (Figure 6B). These data along with the previous mutation data suggest that these sites are required, but not sufficient, for the observed mobility shift.
Figure 6.
Analysis of PSF phosphorylation. (A) Immunoblot analysis of FLAG-PSF or non-phosphorylatable PSF(12M) expressed with different Cyclin/Cdk complexes. (B) Immunoblot analysis of FLAG-PSF containing various mutations of potential phosphorylation sites expressed with D1K2. (C) Autoradiography of His6-PSF [γ-32P]-phosphorylated with different Cyclin/Cdk complexes. (D) Autoradiography of His6-PSF [γ-32P]-phosphorylated with either Cyclin E/Cdk2 or D1K2 (left panel) and quantitation of the same bands using a scintillation counter (right panel).
Cdk2 Directly Phosphorylates PSF In Vitro
An in vitro kinase assay utilizing bacterially expressed His6-PSF protein isolated on Talon resin showed that commercial purified Cyclin E/Cdk2, but not Cyclin B/Cdk1 or Cyclin D1/Cdk4 is capable of phosphorylating PSF (Figure 6C). D1K2 protein isolated from the previously described D1K2-T2 CL6 cell line17 was also shown to be capable of phosphorylating PSF (Figure 6D, top panel). Cyclin E/Cdk2 showed a much higher level of phosphorylation, even after normalizing based on the amount of kinase present, as measured by immunoblot (Figure 6D, bottom panel).
Discussion
Being overexpressed in approximately 50% of all breast cancers, it is important to thoroughly understand the effects Cyclin D1 has on cells. In addition to forming complexes with its canonical partners Cdk4 and Cdk6, Cyclin D1 also complexes with Cdk2. The relevance of these D1/K2 complexes has previously been debated and the current study provides a possible explanation for previous work showing that D1/K2 complexes are inactive.6 As was previously observed, expression of Cyclin D1 and Cdk2 in p21−/− HCT116 cells failed to produce D1/K2 complexes. However, coexpression of p21 induced formation of these complexes in a concentration-dependent manner, indicating that p21 is required to serve as a scaffolding protein. These complexes showed kinase activity and activating phosphorylation on T160 of Cdk2.
Biochemical analyses indicated formation of a complex of Cyclin D1, Cdk2, p21, and PCNA at a 1:1:1:1 stoichiometric ratio and utilization of a p21-PCNA fusion protein showed that D1K2 was capable of associating with other Cyclin/Cdk complexes. This could be explained by a model in which each monomer in a trimeric p21/PCNA complex is able to independently bind different Cyclin/Cdk complexes.
As our data suggested that slightly higher p21 levels increase D1/K2 kinase activity, and p21 levels rise upon treatment with many chemotherapy agents, we investigated whether chemotherapy agents lead to increased D1/K2 activity. Indeed, the DNA damaging agents Doxorubicin and Cisplatin increased both D1/K2 complex formation and kinase activity while the spindle poison Paclitaxel did not. Given the cellular effects of D1/K2 complexes,17–19 this raises important questions about treating Cyclin D1 overexpressing tumors with DNA damaging agents and puts forward the possibility that co-treatment with a Cdk inhibitor may prove clinically beneficial.
p21 is regulated by phosphorylation on a number of sites including Thr57, Tyr76, Ser78, Ser123, Ser130, Thr145, and Ser146 58–63. These sites control multiple aspects of p21 function including its stability, intracellular localization, inhibition of Cdk activity, and p21 cytoplasmic anti-apoptotic function. Additional studies are required to determine whether p21 phosphorylation on these sites or others controls its ability to assemble Cyclin/Cdk complexes and to regulate their kinase activity. The interplay between the actions of the different phosphorylation sites is likely to be complicated as has been observed with p27 phosphorylation 64–72.
These D1/K2 complexes have been known to phosphorylate Rb for some time,8 but their other targets have not been investigated. Our data indicate that Cdk2 complexed with Cyclin D1 or E phosphorylates other substrates as well. Here we have shown that NPM, which controls the process of centrosome replication, is phosphorylated on T199 by D1/K2 and Cyclin E/Cdk2 complexes, but not by Cyclin D1/Cdk1 or Cyclin D1/Cdk4. Similarly, these results were recapitulated using fusion proteins allowing the Cyclin/Cdk complex composition to be precisely controlled. The D1K2 and EK2 fusion proteins were capable of phosphorylating NPM, but the D1K4 and D1K2(KD) proteins were not.
When Cyclin/Cdk complexes were analyzed for their ability to phosphorylate Cdh1, an activating subunit of the APC/C, comparable results were obtained. It was shown that D1K2 phosphorylates Cdh1 and that all Cyclin/Cdk combinations and fusion proteins tested bind Cdh1 in cells, with the exception of Cyclin D1/Cdk4. The ability of D1K2 to phosphorylate NPM and Cdh1 was also confirmed through an in vitro kinase assay.
The D1K2 fusion protein is not only useful for identifying known Cyclin E/Cdk2 substrates as D1/K2 substrates as well, but is also a tool capable of helping to identify novel Cdk substrates. While PSF is phosphorylated by BRK, GSK3, and unknown serine/threonine kinases34–36 resulting in changes in intracellular localization and splicing activity, it was not known to be a substrate for Cdk2. We have identified PSF as being highly phosphorylated in MCF10A cells expressing the D1K2 fusion protein and have confirmed the protein as a Cdk2 substrate through coexpression experiments utilizing constructs coding for various Cyclins, Cdks, and Cyclin/Cdk fusion proteins. This phosphorylation was also inhibited by overexpression of p21.
Three potential Cdk2 phosphorylation sites, a serine or threonine followed by a proline, have previously been shown to be phosphorylated by an unknown kinase.57 Phosphorylation was blocked by the individual mutation of these sites: S33A, S379A, or T687A. In all, PSF contains twelve potential phosphorylation sites, and a construct in which all twelve sites have been mutated to alanine does not exhibit any phosphorylation-dependent shift in electrophoretic mobility. Reverse mutation to restore the phosphorylation sites at residues 33, 379, and 687 failed to restore the mobility shift. Using in vitro kinase assays, both Cyclin E/Cdk2 complexes and the D1K2 fusion protein were shown to directly phosphorylate PSF.
It is not currently known what effect phosphorylation of PSF by Cdk2 has on its function as our experiments have not shown a change in intracellular localization or complex formation. It is possible that phosphorylation may modulate the RNA splicing or DNA repair functions of PSF and this will be important to explore in future studies.
Supplementary Material
Acknowledgments
We thank Dr. Stefan Stamm at the University of Kentucky for the gift of the His6-PSF construct and Dr. Bert Vogelstein for the gift of the p21−/− HCT116 cells.
Funding Statement: This work was supported in part by National Institutes of Health Grant R01-CA93651, Florida Department of Health grants 07BB-8 and 09BB-10, and the Susan G. Komen for the Cure grant KG080510 to B.K.L. S.C.J. was supported by the University of Florida College of Medicine Alumni Fellowship. P.E.C. was supported by NIH/NCI T32 training grant in Cancer Biology, CA09126.
Abbreviations
- 5FU
5-Fluorouracil
- APC/C
Anaphase Promoting Complex/Cyclosome
- BRK
Breast tumor Kinase
- CAK
Cdk Activating Kinase
- Cdh1
Cdc20 homology 1
- Cdk
Cyclin dependent kinase
- D1/K2
Cyclin D1/Cdk2 complex
- D1K2
Cyclin D1-Cdk2 fusion protein
- D1K4
Cyclin D1-Cdk4 fusion protein
- EK2
Cyclin E-Cdk2 fusion protein
- HA
Hemagglutinin
- NPM
Nucleophosmin
- PCNA
Proliferating Cell Nuclear Antigen
- PSF
Polypyrimidine tract binding protein-associated Splicing Factor
- SFPQ
Splicing Factor proline/glutamine rich
Footnotes
Supporting Information Available
Supplemental Figure S1 examines the limited ability of p27 to substitute for p21 in D1/K2 complex formation. Supplement S2 contains the DNA sequence information for the novel fusion proteins utilized in this study. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 2.Guan KL, Jenkins CW, Li Y, Nichols MA, Wu X, O’Keefe CL, Matera AG, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 3.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 4.Okuda T, Hirai H, Valentine VA, Shurtleff SA, Kidd VJ, Lahti JM, Sherr CJ, Downing JR. Molecular cloning, expression pattern, and chromosomal localization of human CDKN2D/INK4d, an inhibitor of cyclin D-dependent kinases. Genomics. 1995;29:623–630. doi: 10.1006/geno.1995.9957. [DOI] [PubMed] [Google Scholar]
- 5.Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 6.Higashi H, Suzuki-Takahashi I, Saitoh S, Segawa K, Taya Y, Okuyama A, Nishimura S, Kitagawa M. Cyclin-dependent kinase-2 (Cdk2) forms an inactive complex with cyclin D1 since Cdk2 associated with cyclin D1 is not phosphorylated by Cdk7-cyclin-H. Eur J Biochem. 1996;237:460–467. doi: 10.1111/j.1432-1033.1996.0460k.x. [DOI] [PubMed] [Google Scholar]
- 7.Morisaki H, Ando A, Nagata Y, Pereira-Smith O, Smith JR, Ikeda K, Nakanishi M. Complex mechanisms underlying impaired activation of Cdk4 and Cdk2 in replicative senescence: roles of p16, p21, and cyclin D1. Exp Cell Res. 1999;253:503–510. doi: 10.1006/excr.1999.4698. [DOI] [PubMed] [Google Scholar]
- 8.Chytil A, Waltner-Law M, West R, Friedman D, Aakre M, Barker D, Law B. Construction of a cyclin D1-Cdk2 fusion protein to model the biological functions of cyclin D1-Cdk2 complexes. J Biol Chem. 2004;279:47688–47698. doi: 10.1074/jbc.M405938200. [DOI] [PubMed] [Google Scholar]
- 9.Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, Frampton GM, Cole MF, Odom DT, Odajima J, Geng Y, Zagozdzon A, Jecrois M, Young RA, Liu XS, Cepko CL, Gygi SP, Sicinski P. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463:374–378. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, Kunkel TA, van Harn T, Xia B, Correll M, Quackenbush J, Livingston DM, Gygi SP, Sicinski P. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474:230–234. doi: 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowe DB, Kenney NJ, Adereth Y, Maroulakou IG. Suppression of Neu-induced mammary tumor growth in cyclin D1 deficient mice is compensated for by cyclin E. Oncogene. 2002;21:291–298. doi: 10.1038/sj.onc.1205025. [DOI] [PubMed] [Google Scholar]
- 12.Reddy HK, Mettus RV, Rane SG, Grana X, Litvin J, Reddy EP. Cyclin-dependent kinase 4 expression is essential for neu-induced breast tumorigenesis. Cancer Res. 2005;65:10174–10178. doi: 10.1158/0008-5472.CAN-05-2639. [DOI] [PubMed] [Google Scholar]
- 13.Ray D, Terao Y, Christov K, Kaldis P, Kiyokawa H. Cdk2-null mice are resistant to ErbB-2-induced mammary tumorigenesis. Neoplasia. 2011;13:439–444. doi: 10.1593/neo.101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney KJ, Swarbrick A, Sutherland RL, Musgrove EA. Lack of relationship between CDK activity and G1 cyclin expression in breast cancer cells. Oncogene. 1998;16:2865–2878. doi: 10.1038/sj.onc.1201814. [DOI] [PubMed] [Google Scholar]
- 15.Junk DJ, Cipriano R, Stampfer M, Jackson MW. Constitutive CCND1/CDK2 Activity Substitutes for p53 Loss, or MYC or Oncogenic RAS Expression in the Transformation of Human Mammary Epithelial Cells. PLoS One. 2013;8:e53776. doi: 10.1371/journal.pone.0053776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law M, Forrester E, Chytil A, Corsino P, Green G, Davis B, Rowe T, Law B. Rapamycin disrupts cyclin/cyclin-dependent kinase/p21/proliferating cell nuclear antigen complexes and cyclin D1 reverses rapamycin action by stabilizing these complexes. Cancer Res. 2006;66:1070–1080. doi: 10.1158/0008-5472.CAN-05-1672. [DOI] [PubMed] [Google Scholar]
- 17.Corsino P, Davis B, Law M, Chytil A, Forrester E, Norgaard P, Teoh N, Law B. Tumors initiated by constitutive Cdk2 activation exhibit transforming growth factor beta resistance and acquire paracrine mitogenic stimulation during progression. Cancer Res. 2007;67:3135–3144. doi: 10.1158/0008-5472.CAN-06-3815. [DOI] [PubMed] [Google Scholar]
- 18.Jahn SC, Corsino PE, Davis BJ, Law ME, Norgaard P, Law BK. Constitutive Cdk2 activity promotes aneuploidy while altering the spindle assembly and tetraploidy checkpoints. J Cell Sci. 2013 doi: 10.1242/jcs.117382. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corsino PE, Davis BJ, Norgaard PH, Parker NN, Law M, Dunn W, Law BK. Mammary tumors initiated by constitutive Cdk2 activation contain an invasive basal-like component. Neoplasia. 2008;10:1240–1252. doi: 10.1593/neo.08710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canavese M, Santo L, Raje N. Cyclin dependent kinases in cancer: Potential for therapeutic intervention. Cancer Biol Ther. 2012;13:451–457. doi: 10.4161/cbt.19589. [DOI] [PubMed] [Google Scholar]
- 21.Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20:R816–825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 24.Lindon C. Control of mitotic exit and cytokinesis by the APC/C. Biochem Soc Trans. 2008;36:405–410. doi: 10.1042/BST0360405. [DOI] [PubMed] [Google Scholar]
- 25.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 26.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 27.Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol. 2005;25:6734–6746. doi: 10.1128/MCB.25.15.6734-6746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang S, Lutz CS. p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA. 2006;12:111–121. doi: 10.1261/rna.2213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 33.Ha K, Takeda Y, Dynan WS. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst) 2011;10:252–259. doi: 10.1016/j.dnarep.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukong KE, Huot ME, Richard S. BRK phosphorylates PSF promoting its cytoplasmic localization and cell cycle arrest. Cell Signal. 2009;21:1415–1422. doi: 10.1016/j.cellsig.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Shav-Tal Y, Cohen M, Lapter S, Dye B, Patton JG, Vandekerckhove J, Zipori D. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol Biol Cell. 2001;12:2328–2340. doi: 10.1091/mbc.12.8.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40:126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law BK, Chytil A, Dumont N, Hamilton EG, Waltner-Law ME, Aakre ME, Covington C, Moses HL. Rapamycin potentiates transforming growth factor beta-induced growth arrest in nontransformed, oncogene-transformed, and human cancer cells. Mol Cell Biol. 2002;22:8184–8198. doi: 10.1128/MCB.22.23.8184-8198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cozar-Castellano I, Harb G, Selk K, Takane K, Vasavada R, Sicari B, Law B, Zhang P, Scott DK, Fiaschi-Taesch N, Stewart AF. Lessons from the first comprehensive molecular characterization of cell cycle control in rodent insulinoma cell lines. Diabetes. 2008;57:3056–3068. doi: 10.2337/db08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi M, Robetorye RS, Pereira-Smith OM, Smith JR. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem. 1995;270:17060–17063. doi: 10.1074/jbc.270.29.17060. [DOI] [PubMed] [Google Scholar]
- 40.Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 42.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 43.Zhang H, Hannon GJ, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 44.Koundrioukoff S, Jonsson ZO, Hasan S, de Jong RN, van der Vliet PC, Hottiger MO, Hubscher U. A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol Chem. 2000;275:22882–22887. doi: 10.1074/jbc.M001850200. [DOI] [PubMed] [Google Scholar]
- 45.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 46.Li R, Hannon GJ, Beach D, Stillman B. Subcellular distribution of p21 and PCNA in normal and repair-deficient cells following DNA damage. Curr Biol. 1996;6:189–199. doi: 10.1016/s0960-9822(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 47.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 48.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 49.Koundrioukoff S, Jonsson ZO, Hasan S, de Jong RN, van der Vliet PC, Hottiger MO, Hubscher U. A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol Chem. 2000;275:22882–22887. doi: 10.1074/jbc.M001850200. [DOI] [PubMed] [Google Scholar]
- 50.Tarapore P, Shinmura K, Suzuki H, Tokuyama Y, Kim SH, Mayeda A, Fukasawa K. Thr199 phosphorylation targets nucleophosmin to nuclear speckles and represses pre-mRNA processing. FEBS Lett. 2006;580:399–409. doi: 10.1016/j.febslet.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 51.Cuomo ME, Knebel A, Morrice N, Paterson H, Cohen P, Mittnacht S. p53-Driven apoptosis limits centrosome amplification and genomic instability downstream of NPM1 phosphorylation. Nat Cell Biol. 2008;10:723–730. doi: 10.1038/ncb1735. [DOI] [PubMed] [Google Scholar]
- 52.Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA, Sutherland RL. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- 53.Loden M, Stighall M, Nielsen NH, Roos G, Emdin SO, Ostlund H, Landberg G. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002;21:4680–4690. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- 54.Lindahl T, Landberg G, Ahlgren J, Nordgren H, Norberg T, Klaar S, Holmberg L, Bergh J. Overexpression of cyclin E protein is associated with specific mutation types in the p53 gene and poor survival in human breast cancer. Carcinogenesis. 2004;25:375–380. doi: 10.1093/carcin/bgh019. [DOI] [PubMed] [Google Scholar]
- 55.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Muller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keck JM, Summers MK, Tedesco D, Ekholm-Reed S, Chuang LC, Jackson PK, Reed SI. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1) J Cell Biol. 2007;178:371–385. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2:ra46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 58.Wang Z, Zhang Y, Gu JJ, Davitt C, Reeves R, Magnuson NS. Pim-2 phosphorylation of p21(Cip1/WAF1) enhances its stability and inhibits cell proliferation in HCT116 cells. Int J Biochem Cell Biol. 2010;42:1030–1038. doi: 10.1016/j.biocel.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hodeify R, Tarcsafalvi A, Megyesi J, Safirstein RL, Price PM. Cdk2-dependent phosphorylation of p21 regulates the role of Cdk2 in cisplatin cytotoxicity. Am J Physiol Renal Physiol. 2011;300:F1171–1179. doi: 10.1152/ajprenal.00507.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harms C, Albrecht K, Harms U, Seidel K, Hauck L, Baldinger T, Hubner D, Kronenberg G, An J, Ruscher K, Meisel A, Dirnagl U, von Harsdorf R, Endres M, Hortnagl H. Phosphatidylinositol 3-Akt-kinase-dependent phosphorylation of p21(Waf1/Cip1) as a novel mechanism of neuroprotection by glucocorticoids. J Neurosci. 2007;27:4562–4571. doi: 10.1523/JNEUROSCI.5110-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hukkelhoven E, Liu Y, Yeh N, Ciznadija D, Blain SW, Koff A. Tyrosine phosphorylation of the p21 cyclin-dependent kinase inhibitor facilitates the development of proneural glioma. J Biol Chem. 2012;287:38523–38530. doi: 10.1074/jbc.M112.366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277:29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- 63.Dash BC, El-Deiry WS. Phosphorylation of p21 in G2/M promotes cyclin B-Cdc2 kinase activity. Mol Cell Biol. 2005;25:3364–3387. doi: 10.1128/MCB.25.8.3364-3387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohanty AR, Kan Q, Srivastava S, Uranbileg B, Arakawa-Takeuchi S, Fujita N, Okayama H. Successive phosphorylation of p27(KIP1) protein at serine-10 and C terminus crucially controls its potency to inactivate Cdk2. J Biol Chem. 2012;287:21757–21764. doi: 10.1074/jbc.M112.346254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ou L, Ferreira AM, Otieno S, Xiao L, Bashford D, Kriwacki RW. Incomplete folding upon binding mediates Cdk4/cyclin D complex activation by tyrosine phosphorylation of inhibitor p27 protein. J Biol Chem. 2011;286:30142–30151. doi: 10.1074/jbc.M111.244095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D, He F, Zhang L, Zhang F, Wang Q, Qian X, Pan X, Meng J, Peng C, Shen A, Chen J. The role of p27(Kip1) phosphorylation at serine 10 in the migration of malignant glioma cells in vitro. Neoplasma. 2011;58:65–73. [PubMed] [Google Scholar]
- 67.Short JD, Dere R, Houston KD, Cai SL, Kim J, Bergeron JM, Shen J, Liang J, Bedford MT, Mills GB, Walker CL. AMPK-mediated phosphorylation of murine p27 at T197 promotes binding of 14-3-3 proteins and increases p27 stability. Mol Carcinog. 2010;49:429–439. doi: 10.1002/mc.20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong F, Larrea MD, Doughty C, Kwiatkowski DJ, Squillace R, Slingerland JM. mTOR-raptor binds and activates SGK1 to regulate p27 phosphorylation. Mol Cell. 2008;30:701–711. doi: 10.1016/j.molcel.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 69.Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Viglietto G, Motti ML, Bruni P, Melillo RM, D’Alessio A, Califano D, Vinci F, Chiappetta G, Tsichlis P, Bellacosa A, Fusco A, Santoro M. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med. 2002;8:1136–1144. doi: 10.1038/nm762. [DOI] [PubMed] [Google Scholar]
- 71.Shin I, Yakes FM, Rojo F, Shin NY, Bakin AV, Baselga J, Arteaga CL. PKB/Akt mediates cell-cycle progression by phosphorylation of p27(Kip1) at threonine 157 and modulation of its cellular localization. Nat Med. 2002;8:1145–1152. doi: 10.1038/nm759. [DOI] [PubMed] [Google Scholar]
- 72.Ciarallo S, Subramaniam V, Hung W, Lee JH, Kotchetkov R, Sandhu C, Milic A, Slingerland JM. Altered p27(Kip1) phosphorylation, localization, and function in human epithelial cells resistant to transforming growth factor beta-mediated G(1) arrest. Mol Cell Biol. 2002;22:2993–3002. doi: 10.1128/MCB.22.9.2993-3002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.