Abstract
The geographic origins of breeds and genetic basis of variation within the widely distributed and phenotypically diverse domestic rock pigeon (Columba livia) remain largely unknown. We generated a rock pigeon reference genome and additional genome sequences representing domestic and feral populations. We find evidence for the origins of major breed groups in the Middle East, and contributions from a racing breed to North American feral populations. We identify EphB2 as a strong candidate for the derived head crest phenotype shared by numerous breeds, an important trait in mate selection in many avian species. We also find evidence that this trait evolved just once and spread throughout the species, and that the crest originates early in development by the localized molecular reversal of feather bud polarity.
Since the initial domestication of the rock pigeon in Neolithic times (1), breeders have selected striking differences in behavior, vocalizations, skeletal morphology, feather ornaments, colors, and color patterns to establish over 350 breeds (2). In many cases, the number and magnitude of differences among breeds are more characteristic of macroevolutionary changes than of changes within a single species (2, 3). Indeed, Charles Darwin was so fascinated by domestic pigeons that he repeatedly called attention to this dramatic example of diversity within a species to communicate his ideas about natural selection (3, 4).
The genetic architecture for many derived traits in pigeons is probably relatively simple (5, 6), likely more so than interspecific trait variation among many wild species, as breeders often focus on qualitative rather than quantitative variation; this increases the chance of identifying genes responsible for differences among breeds. Additionally, several morphological traits show similar patterns of variation in different breeds, making it possible to test whether the same or different genes underlie similar phenotypes. Despite these advantages, the pigeon is underused as a model for the molecular genetic basis of avian variation due to the paucity of genetic and genomic resources for this bird.
We examined genomic diversity, genetic structure, and phylogenetic relationships among domestic breeds and feral populations (free-living birds descended from escaped domestics) of the rock pigeon. The pigeon reference genome was sequenced from a male Danish tumbler with the Illumina HiSeq2000 platform, and we also resequenced 40 additional C. livia genomes to 8-to 26-fold coverage (38 individuals from 36 domestic breeds and two feral pigeons) (7). Genome-wide nucleotide diversity in the rock pigeon (π=3.6×10−3) and the mutation rate estimate in the pigeon lineage (1.42×10−9 substitutions site−1 year−1 ± 2.60×10−12 SE) are comparable to other avian species (8, 9). Observed heterozygosity indicates a large effective population size for the rock pigeon of Ne≈521,000; demographic inferences based on the allele frequency spectrum indicate that, aside from a very recent bottleneck, Ne has been remarkably stable over the past 1.5 million generations (7).
Patterns of linkage disequilibrium (LD) are indicative of haplotype sizes and genome-wide recombination rates, and inform decisions about genetic mapping strategies. Using genotype data from the 40 resequenced C. livia genomes, we found that mean “useful LD” (10) (r2>0.3) decays in 2.2 kb (Fig. S10J). This suggests that we should expect little LD between typical pairs of genes in an analysis across breeds; thus, the pigeon is well suited for association-mapping strategies.
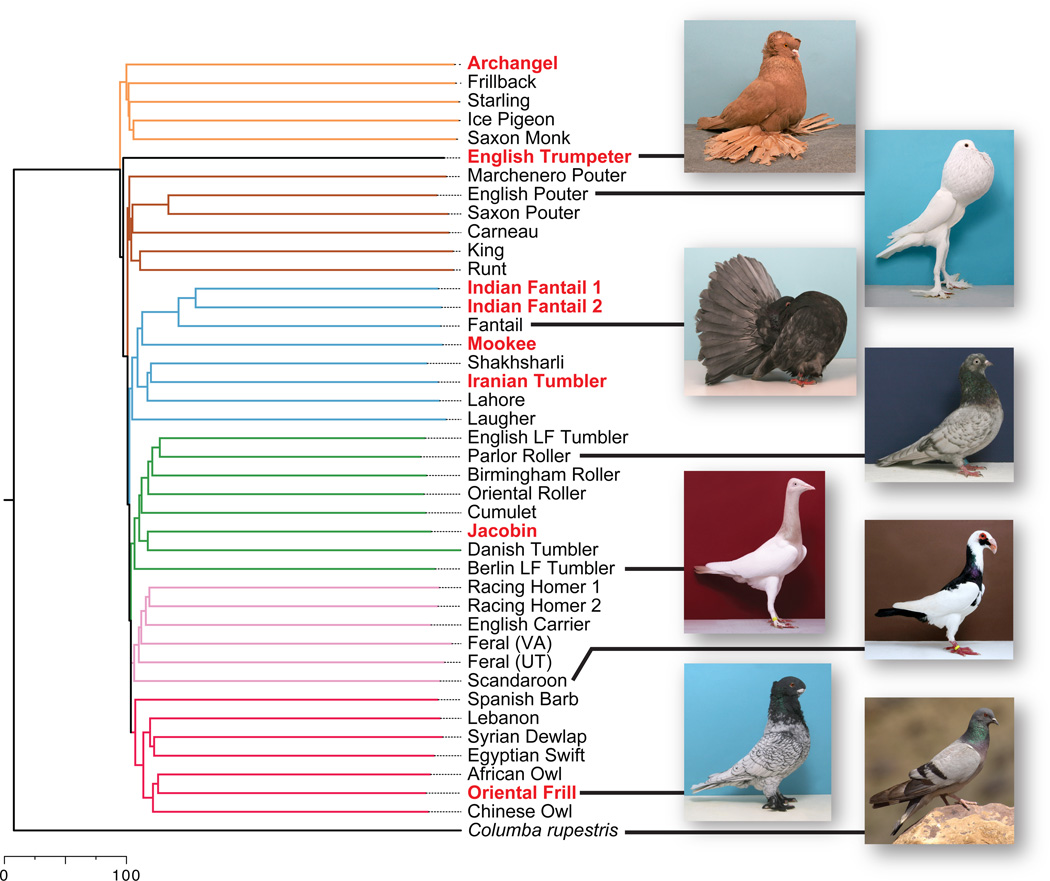
We leveraged our whole-genome data to determine breed relationships using 1.48 million variable loci. A neighbor-joining tree rooted on C. rupestris, the sister species of C. livia (11), yielded several well-supported groups (Figs. 1, S16). Notably, the two feral pigeons grouped with the wattle and homer breeds (Fig. 1, pink branches), supporting the idea that escaped racing homers are probably major contributors to feral populations (12). As with many domesticated species, pigeon evolution is probably not exclusively linear or hierarchical (12). We therefore examined genetic structure among breeds by analyzing 3,950 loci with ADMIXTURE (13), and found a best model fit at K=1 (a single population, where K is the number of assumed ancestral populations). However, higher values of K can also be biologically informative (Figs. S17–S20). Our analysis includes some of the oldest lineages of domestic pigeons and breeds that were not exported from the Middle East until the late nineteenth or early twentieth centuries (14), providing information about likely geographic origins of breeds and their exchange along ancient trade routes (7).
Fig. 1.
Relationships among rock pigeons and the hill pigeon Columba rupestris. Consensus neighbor-joining tree based on 1.48 million genomic SNPs and 1000 bootstrap replicates (see Fig. S16 for bootstrap support). Branches are colored by traditional breed group (12) and/or geographic affinities: orange, toy breeds; brown, pouters and utility breeds; light blue, Indian and Iranian breeds; green, tumblers and highflyers; pink, homers and wattle breeds; red, Mediterranean and owl breeds; black, voice characteristics (14). Bold, red lettering indicates breeds with the head crest phenotype. Scale bar, Euclidean distance. Photo credits: T. Hellmann (domestic breeds) and M.V. Shreeram (C. rupestris).
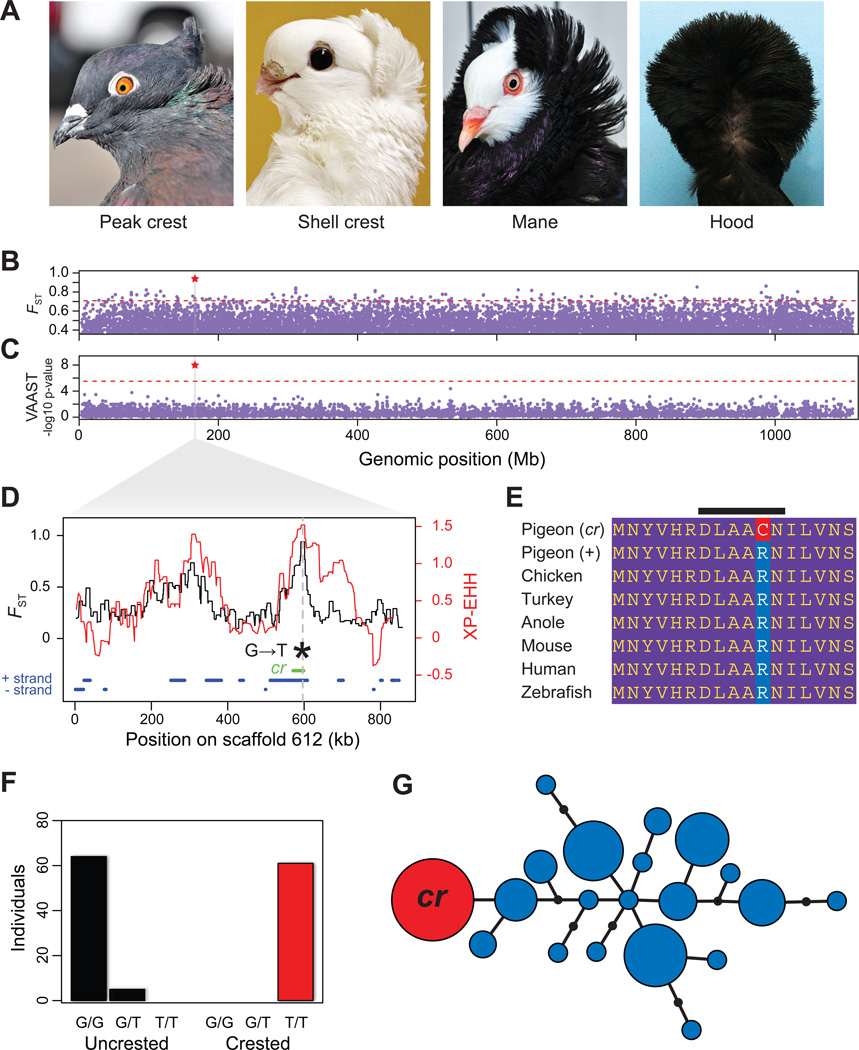
Derived traits in domesticated birds tend to evolve along a predictable temporal trajectory, with color variation appearing in the earliest stages of domestication, followed by plumage and structural (skeletal and soft tissue) variation, and finally behavioral differences (2). One of the genetically simplest derived traits of pigeons is the head crest. Head crests are common ornaments in many bird species (2) and are important display structures in mate selection (15). In pigeons, head crests consist of neck and occipital feathers with reversed growth polarity, such that the feathers grow toward the top of the head instead of down the neck. Crests can be as small and simple as a peak of feathers, or as elaborate as the hood of the Jacobin that envelops the head (Fig. 2A). Remarkably, classical genetics experiments suggest that the head crest segregates as a simple Mendelian recessive trait (6, 14). Moreover, previous studies suggest that the same locus controls the presence of a crest in numerous breeds, either with alternative alleles at this locus or additional modifier loci controlling the extent of crest development (6, 14).
Fig. 2.
EphB2 is associated with the derived head crest phenotype. (A) Head crests are variable among breeds (left to right: Indian fantail, Old German owl, Old Dutch capuchin, Jacobin). (B) FST between crested and uncrested pigeons, maximum value for individual SNPs plotted for non-overlapping 100-kb windows across the genome. Red star, window with the highest score. Dashed red line, top 1% of scores. (C) Genome-wide VAAST scan. Each dot represents a single gene. Red star, gene with the highest score. Dashed red line, genome-wide significance cutoff. (D) Magnification of scaffold 612 in shaded region of (B–C). Black trace, maximum FST between crested and uncrested birds over a 300-SNP window. Red trace, unstandardized cross-population extended haplotype homozygosity (XP-EHH); higher values are evidence of selection (see Fig. S21, genome-wide plot). Dashed vertical line, position of the lone genome-wide significant VAAST hit. Green bar, 27.4-kb haplotype shared by all crested birds, includes only the EphB2 gene. Blue bars, gene predictions on + and − DNA strands. (E) The cr mutation induces a charge-changing amino acid substitution; black bar, highly conserved DLAARN motif of catalytic loop. (F) Genotypes of 159 birds from 79 breeds at the cr locus are perfectly associated with the crest phenotype under a recessive model. (G) Network diagram of the minimal 11-kb haplotype shared by all resequenced rock pigeons with the cr mutation (also see Fig. S23). Many haplotypes contain the + allele (blue), but only one contains the cr SNP (red). Sizes of circles are proportional to the number of chromosomes containing a haplotype. Line segments represent single nucleotide differences. Jacobin photo credit: T. Hellmann.
We resequenced 8 individuals with head crests to directly test if the same mutation controls crest development in different breeds. We sorted genomic variants from birds with and without head crests into separate bins, and calculated allele frequency differentiation (FST) across the genome (Fig. 2B). We identified a region of high differentiation between crested and uncrested birds in the pigeon ortholog of Ephrin receptor B2 (EphB2; FST=0.94, top hit genome wide; Fig. S22A) (Fig. 2D). The role of EphB2 in feather growth is not known, but it plays important roles in tissue patterning and morphogenesis, and is a member of a receptor tyrosine kinase family that mediates development of the feather cytoskeleton (16, 17). All 8 crested birds were homozygous for a T nucleotide at scaffold 612, position 596613 (hereafter, “cr” allele), while uncrested birds were heterozygous (n=3) or homozygous (n=30, including the uncrested outgroup C. rupestris) for the putatively ancestral C nucleotide (“+” allele). These results were consistent with the known simple recessive architecture of the trait, and implicated a common polymorphism associated with head crest development in multiple breeds with different genetic histories (Fig. 1). This trend extended well beyond our resequencing panel: we genotyped an additional 61 crested birds from 22 breeds, and 69 uncrested birds from 57 breeds, and found a perfect association between cr/cr genotype and the crest phenotype (Fig. 2F). By treating the genomes of crested and uncrested birds as separate populations, we also found suggestive evidence for positive selection around the cr allele using cross-population extended haplotype homozygosity analysis (Figs. 2D, S21,S22B).
We then used the Variant Annotation, Analysis, and Search Tool (VAAST (18)) to interrogate the pigeon genomes for additional coding changes associated with the head crest phenotype. This identified one gene with genome-wide significance: EphB2, and specifically the cr SNP (Pgenome = 2.0×10−8) (Fig. 2C,D). The cr allele has a predicted charge-changing arginine (basic) to cysteine (polar uncharged) transition in the catalytic loop of the intracellular tyrosine kinase domain of EphB2 (Fig. 2E). This amino acid position is invariant among other vertebrates suggesting strong purifying selection for conserved protein function. Notably, the same DLAARN to DLAACN motif change we observe in EphB2 is sufficient to abrogate kinase activity in human and mouse orthologs of the protein tyrosine kinase ZAP-70, and in both mammals and pigeons the mutant phenotypes are inherited recessively (19). Hence, the pigeon cr mutation probably abrogates kinase activity in EphB2 and disrupts downstream signal propagation, consistent with the high VAAST score for this gene. EphB2 is therefore a convincing candidate for the cr locus of classical pigeon genetics (5–7, 14).
In several wild and domesticated species, the repeated evolution of a derived trait has occurred by selection on the same gene, possibly due to the repeated selection on the same allele or haplotype (20–22). Similarly, the cr SNP is part of a 27.4-kb haplotype that is shared by all crested pigeons, suggesting that the mutation occurred just once and spread to multiple breeds by introgression among domestic breeds, or was selected repeatedly from a standing variant in wild rock pigeons (Figs. 2G, S23; the core haplotype containing the cr mutation is reduced to 11 kb with the inclusion of uncrested heterozygotes). The only gene present in the shared cr haplotype is EphB2 (Fig. 2D, green bar), although at this time we cannot rule out the presence of regulatory variants that might alter the expression of another gene. Crested members of the toy, fantail, Iranian, Jacobin, and owl breed groups are not more closely related to each other than to uncrested breeds (Fig. 1). Nevertheless, members of these groups had head crests hundreds of years ago (14), so some of these introgression events must have occurred in the distant past. Breeds with a wide variety of crest phenotypes share the same derived allele; therefore, allelic variation at the cr locus alone does not control all aspects of crest development (14). Other genetic and developmental factors beyond this locus must contribute to variation in crest morphology, akin to the presumed complex genetic architecture of species-level divergence in feather ornaments (2).
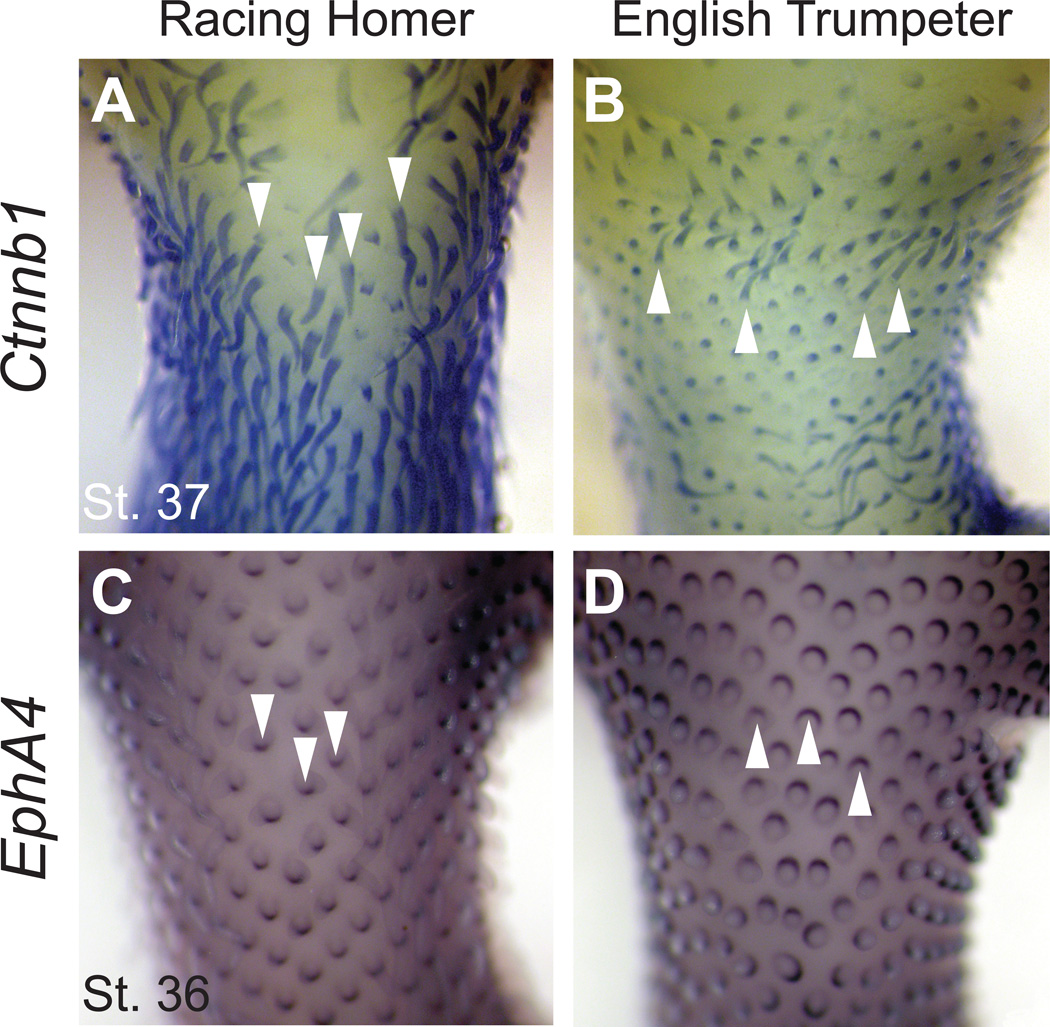
In crested pigeons, feather placode polarity and bud outgrowth are inverted during embryogenesis (Fig. 3). Expression of EphB2 is not polarized in early placodes (Fig. S26), so the effects of the cr mutation on feather polarity are probably exerted earlier in development. Why might the crest phenotype be limited to the head and neck? In Naked neck chicken mutants, regionalized production of retinoic acid allows uniform upregulation of Bmp7 expression to change skin phenotypes in the neck but not the body (23). Similarly, the head crests of several chicken breeds, in which feathers are elongated but do not have a reversed growth trajectory as in pigeons, are localized to the top of the head probably due to ectopic expression of Hox positional cues (24). Together, these examples provide evidence for regionalization of the developing head and neck skin in the chicken. We propose that analogous mechanisms might underlie skin regionalization in the pigeon and allow cr to change feather polarity in the occiput and neck, but not elsewhere.
Fig. 3.
Feather bud polarity is reversed in the cr mutant. (A,B) Expression of the feather structural gene Ctnnb1 reveals the direction of outgrowth of early feather buds. (A) Neck and occipital head expression of Ctnnb1 in an embryo of the uncrested racing homer. Feather buds point downward along the contour of the head and neck (arrowheads). (B) Occipital feathers buds point upward in the equivalent region of the crested English trumpeter, indicating morphological reversal of feather orientation. (C,D) Expression of the polarity marker EphA4 was assayed at an earlier developmental stage to test if feather placodes, the ectodermal thickenings that give rise to feather buds, are also reversed. (C) Polarity marker EphA4 is expressed posteriorly (arrowheads) in feather placodes of the racing homer. (D) Polarity of placodes is reversed in the English trumpeter. Expression of EphB2 in the skin is weak and unpolarized at this stage in both morphs (Fig. S26).
Our study of domestic rock pigeons illustrates how combining comparative genomics and population-based analyses forwards our understanding of genetic relationships and the genomic basis of traits. Many of the traits that vary among pigeon breeds also vary among wild species of birds and other animals (2, 25); thus, pigeons represent a model for identifying the genetic basis of variation in traits of general interest. Moreover, variation in many traits in domestic pigeons, including the head crest phenotype described here, is constructive rather than regressive: breeds derived from the ancestral rock pigeon possess traits that the ancestor does not have. While adaptive regressive traits are important, the genetic basis of constructive traits in vertebrates remains comparatively poorly understood. The domestic pigeon is thus a promising model to explore the genetic architecture of derived, constructive phenotypes in a bird that is amenable to genetic, genomic, and developmental investigation.
Supplementary Material
Acknowledgements
We thank the University of Washington Burke Museum for the C. rupestris tissue sample (UWBM 59803); J. Oldham, K. Wright, Utah Pigeon Club, National Pigeon Association, and A. and H.O. Christiansen for domestic pigeon samples; D. Clayton for feral samples; and D.C., M. Horvath, D. Kingsley, R. Nielsen, and W. Warren for discussion and comments. Supported by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences, NSF CAREER DEB-1149160, University of Utah Research Foundation (M.D.S.); NIH training grant T32GM007464 (S.A.S.); NIH training grant T32HD07491 (E.T.D.); NSF EDEN internship (A.I.V.); NIH/NHGRI R01HG004694, NIH ARRA GO RC2HG005619 (M.Y.); Danish National Research Foundation (M.T.P.G., E.W.). We acknowledge a computer time allocation from the Center for High Performance Computing at the University of Utah.
Footnotes
This whole-genome shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession AKCR00000000 (first version described here is AKCR01000000; raw reads, SRA052637); RNA-seq data for annotation, GSE39333; raw reads for resequenced genomes, SRA054391.
References and Notes
- 1.Driscoll CA, Macdonald DW, O'Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci U S A. 2009 Jun 16;106(Suppl 1):9971. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price TD. Domesticated birds as a model for the genetics of speciation by sexual selection. Genetica. 2002;116:311. [PubMed] [Google Scholar]
- 3.Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 4.Darwin CR. The Variation of Animals and Plants Under Domestication. vol. 1. London: John Murray; 1868. p. 412. [Google Scholar]
- 5.Morgan TH. Notes on two crosses between different races of pigeons. Biological Bulletin. 1911;21:215. [Google Scholar]
- 6.Sell A. Breeding and Inheritance in Pigeons. Germany: Schober Verlags-GmbH, Hengersberg; 1994. p. 202. [Google Scholar]
- 7.Supplementary Online Materials
- 8.Balakrishnan CN, Edwards SV. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata) Genetics. 2009 Feb;181:645. doi: 10.1534/genetics.108.094250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012 Nov 29;491:756. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 10.Aerts J, et al. Extent of linkage disequilibrium in chicken. Cytogenet Genome Res. 2007;117:338. doi: 10.1159/000103196. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KP, et al. A molecular phylogeny of the dove genera Streptopelia and Columba. The Auk. 2001;118:874. [Google Scholar]
- 12.Stringham SA, et al. Divergence, convergence, and the ancestry of feral populations in the domestic rock pigeon. Curr Biol. 2012 doi: 10.1016/j.cub.2011.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009 Sep;19:1655. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levi WM. In: The Pigeon. Second Revised. Sumpter SC, editor. Levi Publishing Co., Inc.; 1986. p. 667. [Google Scholar]
- 15.Amundsen T. Why are female birds ornamented? Trends Ecol Evol. 2000 Apr;15:149. doi: 10.1016/s0169-5347(99)01800-5. [DOI] [PubMed] [Google Scholar]
- 16.McKinnell IW, Makarenkova H, de Curtis I, Turmaine M, Patel K. EphA4, RhoB and the molecular development of feather buds are maintained by the integrity of the actin cytoskeleton. Dev Biol. 2004 Jun 1;270:94. doi: 10.1016/j.ydbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots - a review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009 Feb;20:90. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yandell M, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011 Jun 23; doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elder ME, et al. Distinct T cell developmental consequences in humans and mice expressing identical mutations in the DLAARN motif of ZAP-70. J Immunol. 2001 Jan 1;166:656. doi: 10.4049/jimmunol.166.1.656. [DOI] [PubMed] [Google Scholar]
- 20.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005 Mar 25;307:1928. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 21.Sutter NB, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007 Apr 6;316:112. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Laere AS, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003 Oct 23;425:832. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- 23.Mou C, et al. Cryptic patterning of avian skin confers a developmental facility for loss of neck feathering. PLoS Biol. 2011 Mar;9:e1001028. doi: 10.1371/journal.pbio.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, et al. The crest phenotype in chicken is associated with ectopic expression of HOXC8 in cranial skin. PLoS One. 2012;7:e34012. doi: 10.1371/journal.pone.0034012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baptista LF, Gomez Martinez JE, Horblit HM. Darwin's pigeons and the evolution of the columbiforms: recapitulation of ancient genes. Acta Zoológica Mexicana. 2009 Oct 29;25:719. [Google Scholar]
- 26.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010 Jan 21;463:311. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 28.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999 Jan 15;27:573. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalloul RA, et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillier L, Miller W, Birney E, Warren W, Hardison R. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004 Dec 9;432:695. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 31.Warren WC, et al. The genome of a songbird. Nature. 2010 Apr 1;464:757. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004 May;14:988. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25:1105. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010 May;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003 Oct;19(Suppl 2):ii215. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 36.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. Journal of molecular biology. 1997 Apr 25;268:78. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 37.Elsik CG, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apweiler R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004 Jan 1;32:D115. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apweiler R, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001 Jan 1;29:37. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000 Jan 1;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997 Mar 1;25:955. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009 May 15;25:1335. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003 Jan 1;31:439. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006 Jan 1;34:D572. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009 Jan;37:1. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006 May 15;22:1269. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods in molecular biology. 2009;537:113. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 48.Morgulis A, et al. Database indexing for production MegaBLAST searches. Bioinformatics. 2008 Aug 15;24:1757. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2008. [Google Scholar]
- 50.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004 Jan 22;20:289. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81:559. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137. [Google Scholar]
- 53.Rogers AR, Huff C. Linkage disequilibrium between loci with unknown phase. Genetics. 2009 Jul;182:839. doi: 10.1534/genetics.108.093153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990 Oct 5;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 56.Pacheco MA, et al. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 2011 Jun;28:1927. doi: 10.1093/molbev/msr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007 Aug;24:1586. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 58.Nam K, et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010;11:R68. doi: 10.1186/gb-2010-11-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009 Oct;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002 Feb;18:337. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 61.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 62.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007 Nov;81:1084. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000 Oct;9:1657. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 64.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abler LL, et al. A high throughput in situ hybridization method to characterize mRNA expression patterns in the fetal mouse lower urogenital tract. JoVE. 2011 doi: 10.3791/2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fazl A. The art of training pigeons in the East [annotated translation from Ain-i-Akbari, 1590] The Zoologist (London) 1888;12:167. [Google Scholar]
- 67.Tegetmeier WB. Pigeons: Their Structure, Varieties, Habits, and Management. London: George Routledge and Sons; 1868. p. 178. [Google Scholar]
- 68.National Pigeon Association. 2010 National Pigeon Association Book of Standards. Goodlettsville, TN: Purebred Pigeon Publishing; 2010. [Google Scholar]
- 69.Pelak K, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010 Sep;6 doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kent W. BLAT - the BLAST-like alignment tool. Genome Res. 2002;12:656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992 Jun;8:275. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 72.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 Oct;28:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.