This study investigates the role of the putative splicing regulator and transcriptional cofactor PRMT5 in mammalian development. Using mouse models, Bezzi et al. show that PRMT5 deletion affects splicing through its ability to reduce Sm protein methylation. The authors identify the Mdm4 pre-mRNA as a key PRMT5 target; Mdm4 pre-mRNA senses defects in the spliceosomal machinery and consequently activates the p53 response. This study uncovers a new role for PRMT5 in splicing and identifies Mdm4 pre-mRNA as a potential anti-cancer target.
Keywords: PRMT5, arginine methylation, development, splicing, p53, MDM4
Abstract
The tight control of gene expression at the level of both transcription and post-transcriptional RNA processing is essential for mammalian development. We here investigate the role of protein arginine methyltransferase 5 (PRMT5), a putative splicing regulator and transcriptional cofactor, in mammalian development. We demonstrate that selective deletion of PRMT5 in neural stem/progenitor cells (NPCs) leads to postnatal death in mice. At the molecular level, the absence of PRMT5 results in reduced methylation of Sm proteins, aberrant constitutive splicing, and the alternative splicing of specific mRNAs with weak 5′ donor sites. Intriguingly, the products of these mRNAs are, among others, several proteins regulating cell cycle progression. We identify Mdm4 as one of these key mRNAs that senses the defects in the spliceosomal machinery and transduces the signal to activate the p53 response, providing a mechanistic explanation of the phenotype observed in vivo. Our data demonstrate that PRMT5 is a master regulator of splicing in mammals and uncover a new role for the Mdm4 pre-mRNA, which could be exploited for anti-cancer therapy.
Arginine methylation is a post-translational modification known to play a key role in both transcription and post-transcriptional RNA processing by mediating epigenetic control of chromatin and functionally regulating RNA-binding proteins and components of the splicing machinery (Cheng et al. 2007; Kouzarides 2007; Bedford and Clarke 2009; Migliori et al. 2010).
Initial attempts to identify arginine-methylated proteins have generated lists of putative protein arginine methyltransferase (PRMT) targets (Boisvert et al. 2003; Ong et al. 2004). These studies failed to identify residues methylated by specific PRMT family members and to distinguish between symmetric and asymmetric dimethylation. However, they did shed light on the relevance of arginine methylation in splicing regulation by identifying as targets key components of the constitutive splicing machinery (e.g., Sm proteins) as well as several additional regulators of alternative splicing (e.g., FUS/TLS, SF2, and members of the heterogeneous nuclear ribonucleoprotein [hnRNP] family).
The splicing complexity occurring in the mammalian brain is a remarkable product of evolution and distinctively distinguishes the human species from others (Barbosa-Morais et al. 2012; Dillman et al. 2013). Proper functioning of all splicing-associated proteins allows a significant increase in the complexity of the cell proteome. On the other hand, mutations in proteins involved in RNA processing have been causally linked to neurodegenerative disorders such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS), to mention a few (Zhang et al. 2008; Vance et al. 2009; Da Cruz and Cleveland 2011). These disease-causing mutations emphasize the pivotal role of RNA processing in higher vertebrate brain development, urging researchers to investigate the role of novel splicing regulators in the CNS.
PRMT5 is a type II arginine methyltransferase able to symmetrically dimethylate several nuclear and cytoplasmic proteins (Bedford and Clarke 2009; Karkhanis et al. 2011). In the cytoplasm, PRMT5 acts together with pICln and WDR77 as part of the methylosome, which mainly methylates Sm proteins (SmB/B′, SmD1, and SmD3), increasing their affinity for the Tudor domain of spinal motor neuron 1 (SMN1) (Friesen et al. 2001a,b, 2002; Meister et al. 2001). SMN1 deficiency results in early lethality (embryonic day 3.5 [E3.5]) in the mouse embryo, similarly to Prmt5 constitutive deletion (Hsieh-Li et al. 2000; Tee et al. 2010), while lack of SMN1 in SMA mouse models leads to splicing defects in not only spinal motor neurons, but several organs (Zhang et al. 2008). Given the direct connection between PRMT5 and SMN1 and the relevance of arginine methylation in regulating splicing proteins, it is of extreme relevance to assess the role, if any, of PRMT5 in the CNS.
Here we demonstrate that selective deletion of PRMT5 in the CNS leads to the death of the animal 14 d after birth. We first show that genetic deletion of p53 in a Prmt5-null background partially rescues the developmental defects. Second, we show that the absence of PRMT5 in neural stem/progenitor cells (NPCs) leads to defects in the core splicing machinery. This results in reduced constitutive splicing and the alternative splicing of specific mRNAs, which have weak 5′ donor sites. We identify Mdm4 as one of these mRNAs that acts as a sensor of the splicing defects. Specifically, the Mdm4 alternative splicing event results in the generation of the unstable Mdm4s product, the reduction of the full-length protein, and the transduction of the p53 signaling cascade. We finally expand our findings to other cell types and tissues, demonstrating that Mdm4/Mdm4s alternative splicing senses the absence of PRMT5 also in mouse embryonic fibroblasts (MEFs) in several organs during embryo development and in human cancer cell lines.
We believe our data provide an underlying mechanism for many observations on PRMT5 biology (Jansson et al. 2008; Scoumanne et al. 2009) and, more in general, on perturbation of the splicing machinery (Allende-Vega et al. 2013) and their link to the p53 pathway that were previously ignored.
Results
PRMT5 deficiency in the CNS results in early postnatal lethality
To address the effect of PRMT5 depletion in mammals, we made use of a conditional knockout mouse (White et al. 2013) harboring LoxP (F/F) sequences flanking exon 7 in the Prmt5 gene and studied the effect of its conditional deletion in the CNS. We used a Nestin-Cre (Nes) transgenic mouse strain that expresses Cre recombinase under a neural-specific enhancer of the Nestin promoter, leading to an efficient recombination event in precursors of neurons and glia starting at E10.5 (Graus-Porta et al. 2001). All of the Prmt5F/FNes mice were obtained from Prmt5F/F × Prmt5+/FNes crosses, and, as expected, the Prmt5+/FNes mice were viable and fertile, and we could not observe any evident defects. Single-site insertion was verified by Southern blotting, and CNS-specific deletion of PRMT5 was confirmed by genomic PCR and Western blotting (Supplemental Fig. S1).
Prmt5F/FNes transgenic mice were born at the expected Mendelian frequency but displayed balance disorders, tremors, and akinesis and all died within 14 d after birth. CNS development was impaired, as evident from differences in brain size and weight, which was detectable starting at E17.5 (Fig. 1A). At postnatal day 10 (P10), the external granular layer (EGL) of the cerebellum, an actively proliferating area at this age, was missing in mutant mice, as evident from both sagittal and coronal sections. The lateral ventricles were morphologically enlarged and disrupted, and the thickness of the cortex was reduced in size (Fig. 1B). We next focused on two earlier developmental stages: E15.5 and P0. The cortex of P0 Prmt5F/FNes brains had a lower cellularity count in both the cortical plate (CP) and the ventricular zone/subventricular zone (VZ/SVZ) (Fig. 1C) and a lower number of SOX2/Ki67-positive proliferating NPCs (Fig. 1D) as opposed to controls (PRMT5F/F). We could not observe any appreciable differences in E15.5 brains (Supplemental Fig. S1E).
Figure 1.
PRMT5 deficiency in the CNS results in early postnatal lethality. Nestin-Cre-induced deletion of the PRMT5 gene in the CNS. (A) Weight in milligrams of wild-type (Prmt5F/F) and Prmt5-deleted (Prmt5F/FNes) brains at three different time points (E17.5, P0, and P10). Brain sizes of P10 Prmt5F/F and Prmt5F/FNes mice are shown as an example in the right panel. (B) Hematoxylin and eosin (H&E)-stained sagittal and coronal sections of P10 cerebrum (Cr) and the cerebellum (Cb) from Prmt5F/F (right) and Prmt5F/FNes (left). (C) Coronal sections of P0 brains. Cellularity of the cortical plate (CP) and VZ/SVZ are indicated in wild-type (wt; black) and mutant (red) brains. (MZ) Marginal zone; (SP) subplate; (IZ) intermediate zone. (D) SOX2 and Ki67 immunohistochemistry (IHC) staining in P0 brains. (E) Cleaved Caspase 3 (CC3) staining is shown in both the cortex and the gangliomic eminence.
The data suggested that the absence of PRMT5 resulted in either premature differentiation, cell death of NPCs, or a combination of the two. To test the former hypothesis, we extracted proteins from Prmt5F/F and Prmt5F/FNes P0 and P10 brains and tested the expression of NPC markers (SOX2) and intermediate progenitor markers (TBR2) as well as neuronal and glia markers (TBR1/TuJ and GFAP, respectively). We did observe a significant decrease of SOX2 and TBR2 levels upon PRMT5 deletion, while the levels of differentiated neurons and glia markers were similar in both control and mutant brains (Supplemental Fig. S1F). To test the occurrence of cell death, we stained brain sections for cleaved Caspase 3 (CC3) (Kuida et al. 1996). Despite the fact that changes in brain size are not evident at E15.5 (data not shown), we did detect apoptotic death, specifically in the VZ/SVZ and the ganglionic eminence, both areas containing proliferating NPCs, suggesting that this could be the cause for the reduced brain size of Prmt5F/FNes animals (Fig. 1E).
PRMT5 is required for NPC homeostasis
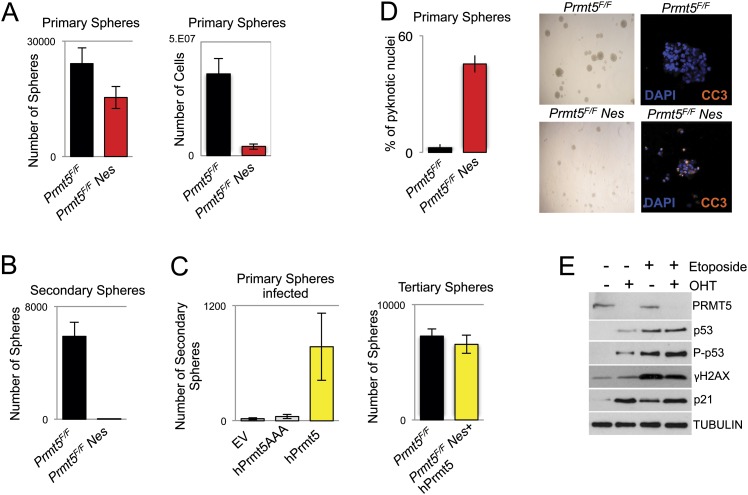
To further define whether PRMT5 is required for normal cell cycle regulation and survival and to protect cells from apoptosis, we derived NPCs from the dorsal telencephalon of E14.5 mice. NPCs can be efficiently grown in vitro as neurospheres. The number of primary neurospheres was significantly reduced in Prmt5F/FNes as opposed to controls. Furthermore, the number of cells in Prmt5F/FNes neurospheres was markedly reduced (Fig. 2A), and, importantly, their self-renewal potential was impaired, as highlighted by the fact that, following replating, virtually no secondary neurospheres could be derived (Fig. 2B). To test whether PRMT5 catalytic activity was necessary for the observed phenotype, we infected primary NPCs derived from Prmt5F/FNes mice with wild-type human PRMT5 (hPRMT5) or a catalytically inactive mutant (hPRMT5AAA). Only cells infected with hPRMT5 were able to grow and could be propagated into secondary neurospheres, and when expanded into tertiary neurospheres, cells expressing hPRMT5 grew as efficiently as NPCs derived from Prmt5F/F control litters (Fig. 2C). To confirm the results obtained in vivo, we first counted the percentage of pyknotic nuclei and then stained Prmt5F/FNes-derived NPCs for CC3 to verify that they were undergoing apoptosis, confirming the requirement for PRMT5 to suppress cell death (Fig. 2D).
Figure 2.
PRMT5 is required for NPC homeostasis. (A) Number of primary neurospheres and total number of cells from cultures of E14.5 dorsal telencephalon NPCs derived from Prmt5F/F and Prmt5F/FNes embryos. Each bar represents an average of at least three experiments. (B) Number of secondary neurospheres, as in A. (C) Primary neurospheres (left panel) from Prmt5F/FNes mice infected with empty vector (EV), wild-type PRMT5 (hPRMT5), or a catalytically inactive PRMT5 mutant (hPRMT5AAA) and passaged to derive secondary and tertiary neurospheres (right panel). (D) Neurospheres derived from Prmt5F/F or Prmt5F/FNes NPCs were stained with DAPI and CC3, and the percentage of pyknotic nuclei was counted. (E) Protein levels upon treatment with OHT and subsequent PRMT5 depletion for 4 d. The antibodies used are indicated on the right of each panel. As a positive control, p53 and the DDR were induced by treating cells with 10 μM etoposide for 2 h.
To understand the molecular mechanism underpinning the observed apoptotic phenotype, we next performed a gene expression analysis of Prmt5F/FNes NPCs. Approximately 2500 genes were differentially expressed when compared with control, showing up-regulation of the p53 pathway and down-regulation of genes involved in cell cycle progression and replication (Supplemental Fig. S2).
We then generated a second conditional knockout strain by crossing the Prmt5F/F mice to the ROSA26:CreERt2 (ER) mice, which allowed the triggering of a recombination event in both live animals and, ex vivo, primary cells by using 4-hydroxytamoxifen (OHT). We switched to the Prmt5F/FER system for three main reasons: First, it allowed us to look at cell-autonomous defects. Second, we could derive a much larger number of cells amenable for further mechanistic studies. Third, it allowed us to focus on early time points after PRMT5 depletion in order to detect causal defects. In all experiments described hereafter in which Prmt5F/FER-derived cells, tissues, or embryos were analyzed, we always used the ER counterparts as negative controls, making sure that the addition of OHT or tamoxifen (TAM) was not toxic (data not shown).
p53 is a transcription factor that drives the expression of several downstream targets in response to a variety of stimuli, including activation of the DNA damage response (DDR) (Lane 1992). Much is known about the regulation of p53 by post-translational modifications, and many of them, including phosphorylation and acetylation, are known to regulate its protein stability, leading to transcriptional activation. We first checked whether, upon PRMT5 deletion, we could detect DDR activation and whether p53 would be stabilized. We did observe a modest p53 protein stabilization and p53 phosphorylation (P-p53) as well as basal levels of H2AX phosphorylation (γH2AX). As a positive control, we used a DNA-damaging agent (etoposide), which, as expected, greatly stabilized p53 and increased the levels of γH2AX. Notably, despite a minor activation of the DDR, the absence of PRMT5 caused an even greater induction of the p53 target gene p21 as compared with etoposide (Fig. 2E, cf. lanes 2 and 3).
p53 deletion partially rescues Prmt5F/FNes developmental defects
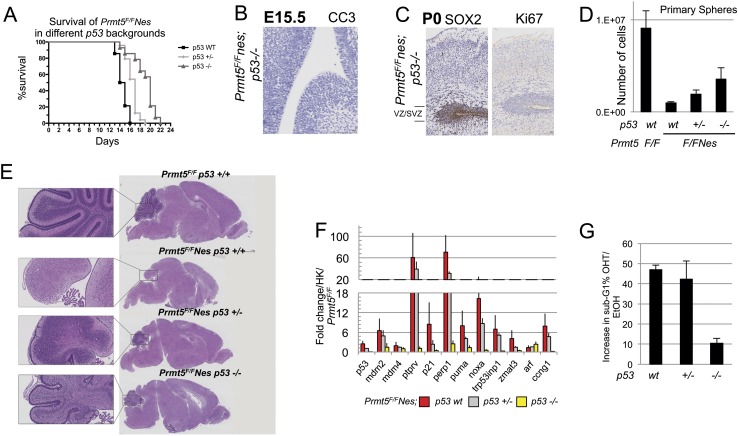
The data indicated that PRMT5 deficiency triggered a p53 response and that the phenotypic outcome in NPCs led to cell death. To formally prove this conclusion, we crossed Prmt5F/FNes mice into a p53-null background. Prmt5F/FNes;p53−/− mice survived, on average, 1 wk longer than Prmt5F/FNes;p53wt, while mice heterozygous for p53 (Prmt5F/FNes;p53+/−) displayed an intermediate phenotype (Fig. 3A). When stained for activated Caspase 3, E15.5 Prmt5F/FNes;p53−/− embryos showed a complete rescue of the apoptotic response, with levels of staining similar to wild type (Fig. 3B). Importantly, the number of SOX2-positive cells in the VZ/SVZ of Prmt5F/FNes;p53−/− embryos was increased when compared with Prmt5F/FNes;p53wt brains (Fig. 3C). However, we did not observe a significant rescue of proliferating KI67-positive cells, suggesting a p53-independent impairment in cell cycle progression, which most likely accounts for the lethality of the animals 20–22 d after birth (Fig. 3C, right panel). Indeed, when we derived NPCs from mice with different p53 backgrounds and cultured them as neurospheres, p53 deficiency led to a significant but not complete rescue in the number of proliferating cells (Fig. 3D). The same was true in P10 brains, where defects in EGL morphogenesis in the cerebellum and, in general, in brain development were only partially rescued in the absence of p53 (Fig. 3E). We can conclude that p53 plays an important role in regulating the apoptotic response in Prmt5F/FNes cells. The fact that we still observed death of the animals, although significantly delayed, however, pointed at additional proliferative defects in targeted cells. To gain further insight, we first checked by RT-qPCR the level of transcriptional up-regulation of p53 targets in both Prmt5F/FNes and Prmt5F/FER in a p53−/− background. Activation of cell cycle inhibitor p21, proapoptotic Noxa, Puma (Akhtar et al. 2006), and all other target genes was completely muted in the absence of p53, excluding compensation by other transcription factors such as p53 family members p63 and p73 (Fig. 3F; Supplemental Fig. S3A). In the absence of p53, PRMT5 depletion led to the reduction of the number of BrdU-positive cells and their exit from the cell cycle, and, consistently, we observed a reduction in the levels of apoptotic cells (Fig. 3G; Supplemental Fig. S3B). These data confirm that, despite inactivation of the p53 response, a second checkpoint mechanism prevents these cells from proliferating.
Figure 3.
p53 deletion partially rescues Prmt5F/FNes developmental defects. (A) Kaplan-Meier survival analysis of Prmt5F/FNes mice in a p53wt (n = 14), p53+/− (n = 24), or p53−/− (n = 14) background. (B) Nestin-Cre-induced deletion of the PRMT5 gene in the CNS of p53−/− embryos. Coronal sections of E15.5 brains stained for CC3 (B) and P0 brains stained for SOX2 and Ki67 (C) to identify stem cells and assess their proliferation status. The antibodies used are indicated for each panel. (D) Total number of NPC cells grown as primary neurospheres derived from Prmt5F/FNes;p53wt, Prmt5F/FNes;p53+/−, and Prmt5F/FNes;p53−/− as indicated. (E) H&E-stained coronal brain sections of PRMT5F/FNes mice with different p53 backgrounds. The cerebellum is shown at a higher magnification in the inset. (F) Expression of p53 up-regulated target genes in NPCs from different genotypes as indicated. The activation of the genes is expressed as the average fold change of three embryos/NPCs, normalized against Prmt5F/F;p53wt and HK. (G) NPCs treated with OHT to delete Prmt5 were stained with propidium iodide and subjected to FACS. Bars indicate the increase in sub-G1/apoptotic cell populations, normalized to EtOH-treated cells. P53 genotypes are indicated.
PRMT5 loss leads to malfunction of the constitutive splicing machinery and to alternative splicing events
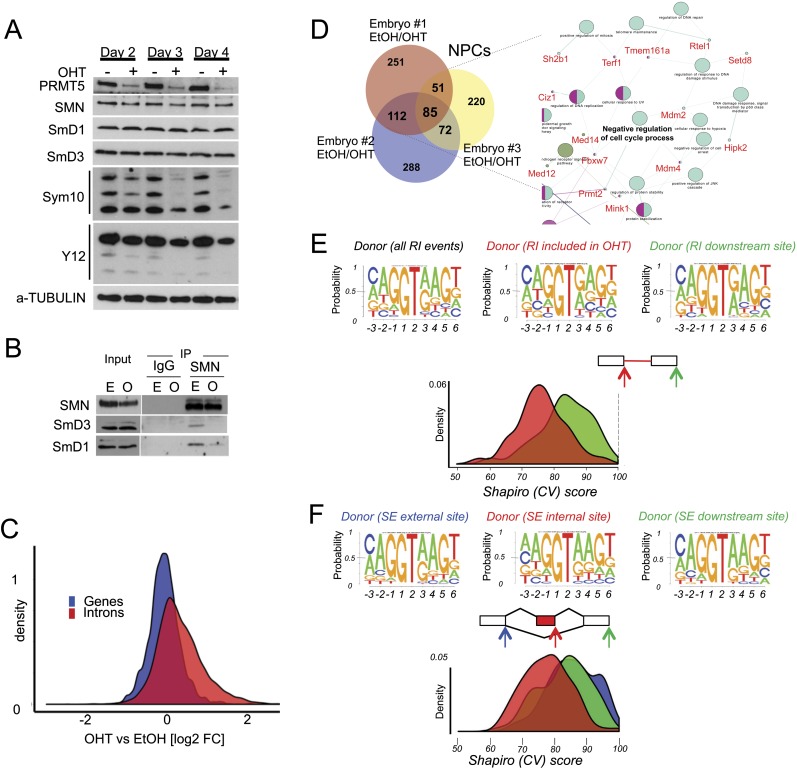
We looked for defects that could mechanistically underpin both the activation of the p53 pathway and the additional proliferation defects. In Drosophila and HeLa cells, PRMT5 is known to symmetrically dimethylate Sm proteins (Gonsalvez et al. 2006, 2007; Deng et al. 2010; Sanchez et al. 2010). We first tested whether this was also relevant during mammalian development. We treated Prmt5F/FER NPCs with either ethanol (EtOH = wild-type PRMT5 levels) or OHT (OHT = PRMT5-depleted) and observed that despite constant levels of SmD1 and SmD3 proteins (Fig. 4A, top panels), there was a reduction in the levels of symmetric arginine dimethylation by day 4, as detectable by two independent antibodies (SYM10 and Y12) (Fig. 4A, bottom panels). We analyzed cells at this early time point for further experiments. Consistent with the fact that the SMN1 Tudor domain binds arginine-methylated SmB/B′, SmD1, and SmD3, we observed a reduced binding of SMN1 to SmD1 and SmD3 (Fig. 4B), suggesting that PRMT5-depleted NPCs would have suboptimal small nuclear ribonucleoprotein (snRNP) maturation. This is indeed the case, since we observed a clear reduction of assembled Sm proteins by 35S pulse-chase assay (Supplemental Fig. S4A). In order to mechanistically understand what could link the splicing defects to apoptosis, we then generated libraries for pair-end RNA sequencing (RNA-seq) from samples treated with EtOH or OHT in order to delete PRMT5. We identified 2416 genes being differentially expressed between the two conditions (Supplemental Table S1). Consistently, the functional annotation of the up-regulated and down-regulated genes looked similar to the one from Prmt5F/FNes cells and showed the activation of the p53 pathway as the top up-regulated category (Supplemental Fig. S2B).
Figure 4.
PRMT5 loss leads to malfunction of the constitutive splicing machinery and to alternative splicing events. (A) PRMT5, SmD1, SmD3, and SMN1 levels were assessed in Prmt5F/FER NPC cells depleted of PRMT5 after 2, 3, and 4 d after OHT treatment. Levels of symmetric arginine dimethylation were assessed by staining SmB/B′, SmD1, and SmD3 with SYM10 and Y12 antibodies. (B) Coimmunoprecipitation between SMN, SmD3, and SmD1, as indicated, in the presence ([E] EtOH) or absence ([O] OHT) of PRMT5. (C) Total number of reads in introns (red) or genes (blue) expressed as fold change of the events in NPCs lacking PRMT5 over control (wild-type PRMT5). A smooth density estimate is drawn as calculated by a Gaussian kernel. (D) Number of genes affected by alternative splicing events in each NPC population (derived from independent embryos). (Right panel) (Snapshot; full figure is in Supplemental Fig. S4B.) Network representation of the differentially spliced genes upon Prmt5 deletion in NPCs. The gene ontology (GO) terms are represented as nodes based on their κ scores. The edges represent the relationships between the GO terms and the shared genes. (E) Shapiro (CV) score of 5′ donor sites of the RI events in NPCs identified by MATS. A smooth density estimate is drawn as calculated by a Gaussian kernel. The top panels depict the sequence logo of the 5′ donor of all RI events (left) and the 5′ donor of the RI events detected upon PRMT5 deletion (right, indicated by the red arrow). The CV score of the downstream donor site is displayed for direct comparison. (F) Same as in E. Shapiro (CV) score of 5′ donor sites of the SE events (in red). The CV scores of the exclusion site (left, indicated by the blue arrow) and the downstream donor site (right, indicated by the green arrow) are displayed for direct comparison.
In contrast to what was reported in plants and Drosophila, where PRMT5 regulates splice site selection without greatly affecting constitutive pre-mRNA splicing (Sanchez et al. 2010), we observed that the compiled number of reads in introns was elevated in the absence of PRMT5, with 1682 introns being significantly affected (Fig. 4C). We then proceeded to characterize the splicing defects in more detail using Multivariate Analysis of Transcript Splicing (MATS) (Fig. 4D–F; Supplemental Fig. S4B; Shen et al. 2012). Prmt5F/FER mice were not on a pure C57BL/6 genetic background; hence, we sequenced three independent NPC populations and first checked the variability in splicing among embryos. In the absence of PRMT5, we observed an overlap of 320 genes affected by alternative splicing in two out of three embryos. These alternatively spliced genes are not random but belonged to specific biological pathways. Importantly, network analysis revealed that these genes are involved in post-transcriptional RNA processing, membrane organization, and negative regulation of cell cycle processes (Fig. 4D; Supplemental Fig. S4C). The latter included transduction of the p53 signaling pathway, suggesting that early problems with the core splicing machinery can be sensed by key alternatively spliced mRNAs to instruct cell cycle arrest or apoptosis (Fig. 4D).
In the absence of PRMT5, we observed a majority of retained intron (RI) and skipped exon (SE) events in all three embryos (Supplemental Fig. S4B), and we could validate 18 of 20 SE events (Supplemental Fig. S4D) and 21 of 21 RI events (Supplemental Fig. S4E), confirming that despite the observed embryo-to-embryo variability, we identified a robust set of conserved alternatively spliced events. Both the RI (Fig. 4E) and SE (Fig. 4F) events detected in the absence of PRMT5 are characterized by a weak 5′ donor site, as quantified by their low CV score (Shapiro score) (Shapiro and Senapathy 1987), their low MaxEntScan (Yeo and Burge 2004) and H-Bond (Freund et al. 2003) scores (Supplemental Fig. S4F), and an overall randomization of the key bases at positions −1, −2, +4, and +5. What distinguishes SE from RI events is the length of the affected intron, which is significantly shorter in the case of RI events (Supplemental Fig. S4G). Hence, absence of PRMT5 leads to selective retention of introns and skipping of exons with weak donor sites.
The Mdm4 alternative splicing event is a sensor of PRMT5 depletion and defects in the constitutive splicing machinery
MDM4 (also known as MDMX) has been reported to be down-regulated upon direct depletion of spliceosome components (Allende-Vega et al. 2013), and perturbation of its levels stood out as potentially recapitulating the activation of the p53 response that we observed in vivo. Importantly, the phenotype of the Mdm4−/− conditional CNS deletion is remarkably similar to what was observed for Prmt5F/FNes, and the most up-regulated gene in the absence of PRMT5 is Ptprv (Supplemental Fig. S2), which was originally identified as deregulated in Mdm4−/− embryos (Doumont et al. 2005).
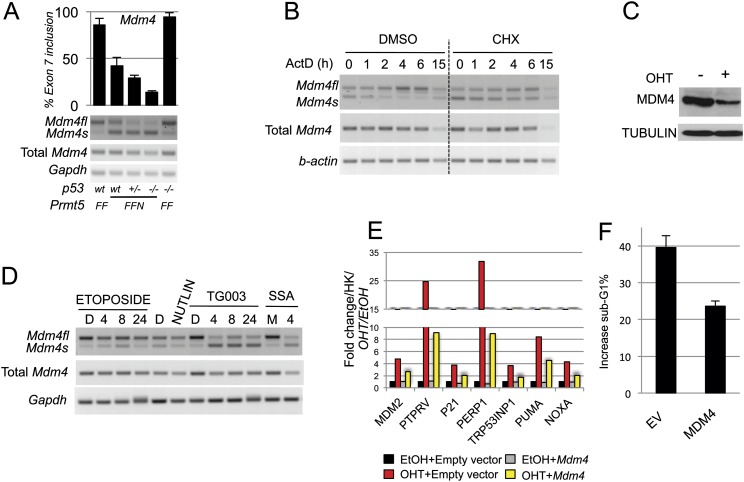
We thus decided to focus our attention on the alternative splicing of Mdm4 (Supplemental Fig. S5) for the rest of the study. MDM4 is a direct regulator of p53 activity; it binds to p53 and inhibits its function by blocking its transactivation capabilities (Francoz et al. 2006; Xiong et al. 2006). Mdm4 undergoes alternative splicing at exon 7 in Prmt5F/FER OHT-treated cells, resulting in the production of a shorter MDM4 isoform that has been previously described as MDM4S (Supplemental Fig. S5A; Rallapalli et al. 2003; Lenos and Jochemsen 2011). Mdm4 exon 7 is located within a 1-kb genomic region that is highly conserved in vertebrates (as assessed by PhyloP) (Supplemental Fig. S5B), suggesting a common mechanism to regulate the abundance of the differentially spliced isoform. To verify that the alternative splicing event of Mdm4/Mdm4s was not a consequence, rather than a cause, of p53 activation and apoptosis, we derived Prmt5F/FNes NPCs with different p53 backgrounds. Reassuringly, the degree of alternative splicing was even greater in p53−/− cells (Fig. 5A). This suggests that the cells in which Mdm4 alternative splicing takes place are rapidly eliminated due to p53 activation. Importantly, all alternative splicing events that we validated were not a consequence of p53 activation or apoptosis (Supplemental Fig. S4D,E).
Figure 5.
Mdm4 alternative splicing event is a sensor of PRMT5 depletion and defects in the constitutive splicing machinery. (A) PCR validation and relative quantification of the alternative splicing event taking place on the Mdm4 mRNA upon PRMT5 deletion in different p53 genetic backgrounds. (B) Semiquantitative PCR of the indicated transcripts upon CHX (100 μg/mL) treatment to block NMD. Cells were pretreated for 3 h and then for the indicated time with 5 μg/mL Actinomycin D to block transcription. (C) MDM4 full-length protein levels are reduced upon PRMT5 deletion. (O) OHT. Tubulin was used as a loading control. (D) PCR detecting both Mdm4 and Mdm4s in wild-type (wt) and mutant NPCs upon inhibition of the core splicing machinery, 100 μM TG003, and 30 ng/mL Spliceostatin A (SSA), or p53 stabilization (Nutlin and 5 μM etoposide). (D) DMSO; (M) MetOH. (E) Full-length Mdm4, re-expressed in PRMT5-depleted NPCs, is able to partially rescue the activation of the p53 response. PCR quantification of p53 target genes upon PRMT5 deletion in cells re-expressing full-length Mdm4 (gray and blue bars) or negative control, empty vector plasmid (black and red bars). A representative experiment of three is shown as an example. (F) NPCs infected with a retroviral vector stably expressing MDM4 or empty vector (EV) control. Prmt5 was deleted (OHT), and cells were stained with propidium iodide and subjected to FACS. Bars indicate the increase in sub-G1/apoptotic cell populations, normalized to EtOH-treated cells.
The literature on the MDM4S protein isoform is quite controversial (Rallapalli et al. 2003; Lenos and Jochemsen 2011), with some reports suggesting its possible role as a potent p53 inhibitor, and others stating that the MDM4S product is unstable. Notably, all of the data are based on forced overexpression experiments and negative results (failure to detect the endogenous protein product). We thus decided to address this issue by looking at the Mdm4s mRNA stability. We performed polysome profiling of cells upon PRMT5 deletion and noted that the full-length Mdm4 product was present in the polysome fractions (F4–F5), while the Mdm4s mRNA was associated with fractions containing significantly fewer polysomes (F3–F4) (Supplemental Fig. S5C). This result suggested two possibilities: either a low level of translation of the Mdm4s RNA or the fact that this RNA would be targeted for nonsense-mediated decay (NMD) (Chiu et al. 2004). To test the latter possibility, we treated cells with cyclohexamide (CHX), an inhibitor of protein synthesis known to block NMD-mediated mRNA degradation, and later with Actinomycin D, which blocks RNA polymerase II (Pol II) transcription. The data in Figure 5B demonstrate that the Mdm4s isoform is less stable than the full-length product and is targeted for NMD.
Our data suggest that upon PRMT5 depletion, the Mdm4 mRNA undergoes alternative splicing, giving rise to the unstable Mdm4s product. Indeed, this leads to the reduction of the full-length MDM4 protein (Fig. 5C). To extend our findings beyond perturbation of the splicing machinery through PRMT5, we treated NPCs with well-characterized splicing inhibitors (TG003 and Spliceostatin A) (Muraki et al. 2004; Kaida et al. 2007) and consistently observed Mdm4/Mdm4s alternatively splicing. As controls, neither p53 activation by Nutlin nor the induction of DNA damage by etoposide generated similar results (Fig. 5D). These results are in contrast with previous reports (Allende-Vega et al. 2013) and provide a direct mechanistic link between perturbation of the splicing machinery and downstream activation of p53.
To confirm our hypothesis, we demonstrated that the p53 transcriptional response (Fig. 5E) and the induction of apoptosis (Fig. 5F) caused by PRMT5 deletion could be rescued by reintroducing full-length MDM4 into NPCs. Not surprisingly, the rescue was only partial due to other alternative splicing events induced by the absence of PRMT5 (Supplemental Fig. S4).
To further prove that loss of PRMT5 leads to reduced levels of spliceosomal snRNPs and that this leads to changes in splicing patterns, we decided to deplete snRNPs alternatively by directly knocking down SmB/B′. We obtained efficient knockdown in NPCs with four independent shRNA-expressing vectors (Supplemental Fig. S5D). Importantly, these phenocopy the loss of PRMT5 and induce reduction of full-length Mdm4 (Supplemental Fig. S5E), activation of p53 target genes (Supplemental Fig. S5F), and increased apoptosis (Supplemental Fig. S5G).
PRMT5 depletion triggers Mdm4 alternative splicing and p53 activation in multiple tissues
So far, we dissected the role of PRMT5 in the developing CNS and showed that it plays a key role in ensuring the proper splicing of Mdm4 in proliferating NPCs. We next asked what the effect would be of deleting PRMT5 in other cell types or in different organs in the mouse embryo. First, we could confirm in MEFs most of what was observed in NPCs (Supplemental Fig. S6). The most notable differences were that MEFs displayed less splicing defects when compared with NPCs and phenotypically did not undergo cell death but rather exited the cell cycle. Despite these differences, the overlap of genes with increased intronic reads was remarkable (57%), and the p53-independent exon skipping event on Mdm4 was fully conserved between the two cell types (Supplemental Fig. S6; Supplemental Table S1).
To assess the effect of PRMT5 depletion during organogenesis, Prmt5 was selectively deleted in Prmt5F/FER embryos from pregnant Prmt5F/F females crossed to Prmt5+/FER males following TAM injection. CRE-ER was activated efficiently in different organs (Fig. 6A), and mutant embryos were readily recognizable by their smaller size, pale color, and growth retardation (Fig. 6B). Upon Prmt5 deletion, we did observe a switch in the ratio of the full-length over the Mdm4s isoform in most samples (Fig. 6C, bottom panel). Importantly, the degree of Mdm4 alternative splicing upon PRMT5 deletion correlates with the up-regulation of p53 targets (Fig. 6C, top panel). The effect was more pronounced in actively proliferative organs such as the lung and liver. The latter, at this stage of development, is populated by hematopoietic progenitor cells, recognizable by their dark-purple color in the hematoxylin and eosin (H&E) staining (Fig. 6D), and the impairment of their homeostasis is consistent with the pale color of the embryos (Fig. 6B). Phenotypically, we observed both activation of the apoptotic response (CC3 staining) and exit from the cell cycle (reduced Ki67 staining) (Fig. 6D).
Figure 6.
PRMT5 depletion triggers Mdm4 alternative splicing and p53 activation in multiple organs. (A) Experimental strategy used to delete PRMT5 at mid-gestation (E10.5). Embryos were analyzed at E15.5 and E17.5. Upon TAM injection, no pups were born alive. (Bottom panel) Efficiency of CRE recombination taking place in different organs. (B) Weight of PRMT5 wild-type (EtOH) or PRMT5-deleted (TAM) whole embryos at E15.5 and E17.5. (Right panel) Representative example of E15.5 embryos with wild-type (left) or deleted PRMT5 (right). (C) Quantitative PCR (qPCR) quantification of p53 targets in the indicated organs upon PRMT5 deletion. (Bottom panel) PCR validation and relative quantification of the alternative splicing event taking place on the Mdm4 mRNA upon PRMT5 deletion in the same organs. (D) H&E staining of wild-type and knockout E15.5 lung and liver sections. In the liver, light-purple-stained hepatocytes and dark-purple-stained hematopoietic precursor cells are easily detectable. Note the dramatic loss of the latter and the corresponding loss of ki67 staining. Below each H&E staining are the IHC stainings of lung and liver sections from a representative embryo. CC3 was used to detect apoptotic cells, and Ki67 was used to detect proliferating cells. Mdm4 pre-mRNA senses defects in the spliceosomal machinery in cancer lines. (E) PCR quantification of the alternative splicing event taking place on the Mdm4 mRNA upon PRMT5 knockdown (KD). Scramble shRNA was used as a control (Scr). Treatment with the splicing inhibitor TG003 (or with DMSO vehicle control) was used as an alternative way of perturbing the splicing machinery. The experiments were performed in the indicated human cancer cells (shown at the top). GAPDH was used as a loading control. (F) Quantification of Mdm4fl/Mdm4s splicing levels 4 d after infection and 2-d selection in 1 mg/mL puromycin upon knockdown with three different shRNA lentiviral constructs (Sh1–Sh3) in U2OS cells. (Scr) Scrambled control shRNA.
Finally, we cloned the region surrounding mouse exon 7 into a minigene construct (Supplemental Fig. S7). Exon 7 is skipped upon PRMT5 depletion (OHT) in MEFs, recapitulating what was observed at the endogenous level.
Mdm4 pre-mRNA senses defects in the spliceosomal machinery in cancer lines
Activation of the p53 pathway is important in cell homeostasis as well as in development but is certainly best known for its aberrant deregulation in human cancer. PRMT5 has been described as a potential oncogene in human malignancies (Karkhanis et al. 2011). Given the high degree of conservation of the region around the alternative splicing exon 7 on mouse Mdm4 (Supplemental Fig. S5B), we tested whether the orthologous human exon 6 conserved a similar sensing mechanism. Upon Prmt5 knockdown and treatment with the splicing inhibitor TG003, we observed a similar alternative splicing event occurring on the human Mdm4 transcript in cancer cell lines derived from different tissues (osteosarcoma, gastric, breast, and glioma) (Fig. 6E). Notably, in accordance with what was observed in NPCs (Supplemental Fig. S5D–G), knockdown of SmB/B′ in U2OS cells leads to Mdm4 alternative splicing (Fig. 6F).
Overall, our data uncover a key mechanism of PRMT5 function through the regulation of Mdm4 splicing that is conserved in mammalian cells during development and relevant to human cancer (Fig. 7).
Figure 7.
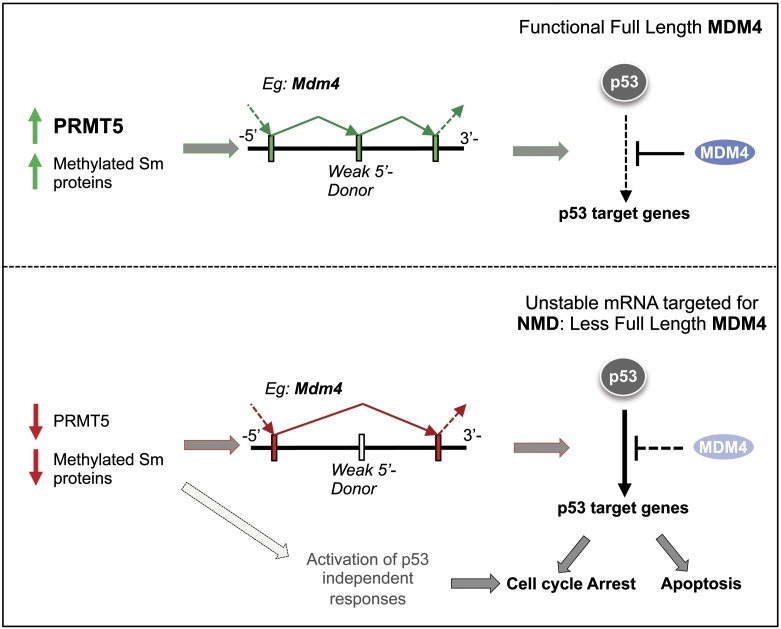
Mdm4 pre-mRNA senses defects in the spliceosomal machinery in cancer lines. Schematic model of the data presented in the study: Upon PRMT5 deletion (or reduction), we observed a loss of symmetric arginine dimethylation at key components of the splicing machinery (SmB/B′, SmD1, SmD3, and possibly others). This leads to aberrant snRNP maturation. The consequence is the activation of a sensing mechanism, which is linked to alternative splicing of key mRNAs (mainly RIs and SEs). As an example, we show Mdm4, which induces a potent p53 transcriptional activation. (Bottom) Other alternative splicing events might be equally important and will ultimately result in a p53-independent cell cycle arrest.
Discussion
Role of PRMT5 in splicing
The initial aim of our project was to dissect the role of PRMT5 in mouse CNS development. We showed by using a conditional knockout mouse model and a combination of in vitro and in vivo approaches that PRMT5 is an essential regulator of splicing in mammals. The first key finding is that these defects in splicing lead to activation of the p53 pathway in the CNS and, in general, in mammalian cells, as opposed to what was observed in other organisms. The role of PRMT5 in vivo has indeed been studied in Planaria, Drosophila, and plants. However, the lack of PRMT5 and, subsequently, symmetrically dimethylated Sm proteins results in distinct phenotypes in different systems. In Planaria, PRMT5 depletion results in defects in homeostasis, growth, and regeneration (Rouhana et al. 2012). In flies, it leads to problems in germ cell specification and circadian rhythm (Gonsalvez et al. 2006; Sanchez et al. 2010), while in plants, it leads to defects in flowering and circadian rhythm (Deng et al. 2010; Sanchez et al. 2010).
PRMT5 is not the only type II symmetric arginine methyltransferase in higher eukaryotes. PRMT7 is an essential gene in Drosophila (unlike PRMT5) (Gonsalvez et al. 2008), while in HeLa cells, it has been shown to methylate Sm proteins and have nonredundant functions in cytoplasmic snRNP biogenesis (Gonsalvez et al. 2007). Our data suggest that, at least in development, PRMT7 is not compensating for the absence of PRMT5, but to what extent, if any, PRMT7 plays a role in mammalian splicing in vivo remains to be addressed.
Splicing disorders are the cause of several neurodegenerative diseases (Dredge et al. 2001; Tollervey et al. 2011), such as SMA, in which the expression of the SMN1 gene is perturbed. Interestingly, also in the case of SMA, there is apoptosis of motor neurons. Although SMA is often referred to as a motor neuron disease, recent evidence suggests that the splicing defects are present in multiple organs (Zhang et al. 2008), and this can lead to disease-relevant phenotypes in cells other than motor neurons (Hayhurst et al. 2012). Surprisingly, neurospheres derived from the Smn−/−;SMN2 mice, which represent a severe model of human SMA, did not differ in number when compared with wild type (Shafey et al. 2008). Our data place PRMT5 upstream of SMN in the maturation cycle of Sm proteins in vivo. This discrepancy can be reconciled by two considerations. First, we are looking at a full deletion of PRMT5, as opposed to a hypomorphic SMA model. Indeed, the full SMN knockout has an early embryonic phenotype similar to the PRMT5 full knockout (Hsieh-Li et al. 2000; Tee et al. 2010). Second, PRMT5 might control splicing more broadly than simply through regulating the maturation of snRNPs. In this respect, the splicing regulator CA150 (Cheng et al. 2007) has been identified as a PRMT5 target, and this could be the case for other splicing proteins (Boisvert et al. 2003; Ong et al. 2004).
Role of PRMT5 in stem cell biology
A previous study has reported the constitutive knockout of PRMT5 using a gene trap model (Tee et al. 2010). Phenotypically, lack of PRMT5 leads to derepression of differentiation genes in mouse embryonic stem cells (mESCs) due, at least in part, to the lack of symmetric Arg3 methylation on histone H2A (H2AR3me2s). However, because of the early embryonic lethality, the investigators were not able to perform large-scale molecular experiments, and thus whether PRMT5 plays any role in controlling splicing in mESCs remains to be explored. It is of note that PRMT5 has been used to improve induced pluripotent stem cell (iPSC) derivation in combination with klf4 and Oct3/4 (Nagamatsu et al. 2011). Reducing p53 activity is known to be very important to enhance iPSC derivation (Hong et al. 2009; Kawamura et al. 2009). We thus believe that our data, which link PRMT5 methyltransferase activity to the regulation of MDM4 abundance by controlling its alternative splicing, will provide new insights into this expanding field of research.
Role of PRMT5 in regulating cell cycle progression and cell death
We described here how cells can sense general defects in the core splicing machinery (such as the one caused by PRMT5 depletion) by regulating the alternative splicing of a key p53 activator such as Mdm4. This alternative splicing event reduces the full-length MDM4 protein and gives rise to the unstable MDM4S product (Lenos and Jochemsen 2011), thus activating the p53 transcriptional program. Our findings describe for the first time the link between the methylosome (Friesen et al. 2001b), the core splicing machinery, alternative splicing, and activation of a p53 response in mammalian development.
What we uncovered here is indeed a much broader picture of how cells can activate the alternative splicing of sensor mRNAs (e.g., Mdm4) at key exons, characterized by weak 5′ donor sites. This occurs upon perturbation of the general splicing machinery—whether because of PRMT5 deletion or chemical inhibition (Fig. 5D)—to arrest growth and/or induce apoptosis. Besides Mdm4, there are other mRNAs that can potentially play a similar role. To mention a few, the SE event observed in the mRNA of 5′ cap-binding protein eIF4E, which is a rate-limiting component in the translation process, could affect genes involved in apoptosis and cell cycle arrest (Supplemental Fig. S4D; Mamane et al. 2004), while the RI events observed in Dvl1 might lead to the inactivation of the Wnt/Dvl1/β-catenin signaling pathway, which is known to support NPCs' growth and self-renewal potential (Supplemental Fig. S4E; Faigle and Song 2013).
We described here how the complete depletion of PRMT5 in mouse cells leads to a minor induction of γH2AX but a strong activation of p53 target genes such as p21 even when compared with etoposide. This is in contrast to what has been observed previously using siRNA/shRNA strategies to reduce the levels of PRMT5 in human cancer lines (Jansson et al. 2008; Scoumanne et al. 2009). This discrepancy could be due to differences between mouse and human cells and between primary cells and cancer cells and the fact that PRMT5 levels have to fall below a certain threshold in order to exhibit an activation of the p53 pathway. The latter concept has already been observed for other splicing regulators such as SMN (Zhang et al. 2008).
Both the concept that perturbation of the core splicing machinery can lead to regulation of alternative splicing (Saltzman et al. 2011) and the concept that apoptosis is regulated by alternative splicing (Schwerk and Schulze-Osthoff 2005; Moore et al. 2010) have been described previously. The fundamental advance that we describe here is that the two pathways are directly linked because Mdm4, the target of alternative splicing, unlike Bcl2-like factors, caspases, death receptors, and proapoptotic ligands, is as a direct upstream regulator of p53.
Splicing disorders have been estimated to occur in 50% of tumors (Ritchie et al. 2008; David and Manley 2010; Ward and Cooper 2010). These can contribute to tumor progression by giving rise to alternative isoforms of oncogenes or tumor suppressors. At the same time, while the perturbation of the splicing machinery was known to activate the p53 pathway, the underlying mechanisms by which this occurred were unknown (Allende-Vega et al. 2013). Our data uncover the mechanism of p53 activation by identifying Mdm4 as a key sensor mRNA. We believe these data to be extremely relevant for the entire p53 field. Mdm4 is indeed up-regulated in several p53wt tumors (Gembarska et al. 2012), and our findings provide new therapeutic avenues to alter its protein levels by affecting its splicing pattern.
In conclusion, this study expands our understanding of the complex network regulating correct splicing and cell fate decisions in mammalian development as well as in human cancer lines, providing new possibilities to target the arginine methyltransferase family to treat neurodegenerative diseases (Dredge et al. 2001; Tollervey et al. 2011) and cancer (Ritchie et al. 2008; David and Manley 2010; Ward and Cooper 2010).
Materials and methods
Mouse strains and genotyping
The PRMT5 knockout first mice were obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM; http://www.knockoutmouse.org). To generate the PRMT5 FLOX allele, the bgal-neomycin cassette was removed by crossing PRMT5 knockout first mice with β-actin-Flpe transgenic mice (Rodriguez et al. 2000) [strain name: B6.Cg-Tg(ACTFLPe) 9205Dym/J; stock no. 005703, The Jackson Laboratory]. These were then crossed to Nestin-CRE [B6.Cg-Tg(Nes-cre)1Kln/J; JAX Laboratory]. 4-OHT-inducible conditional knockouts were created by crossing PRMT5F/F mice with ER transgenic mice (Hameyer et al. 2007) in a mixed C57BL/6 × 129S1/SvlmJ background. Southern blotting was carried out according to a standard protocol (Southern 2006).
Immunohistochemistry (IHC) staining
Automated IHC staining and counterstaining was performed on the Leica Bond-Max autostainer. Detailed protocols are available in the Supplemental Material.
Cell culture
NPCs
Neurosphere cultures were established as previously described (Lim and Kaldis 2012). Briefly, E14.5 embryos were harvested, and cortices were carefully dissected in ice-cold PBS and incubated in trypsin (Invitrogen, 25300120) for 10 min at 37°C. The tissue was then mechanically dissociated, and single cells were cultured into complete NSC medium (DMEM, Life Technologies, 11965118 + 2% B-27, Life Technologies, 17504-044), 1% penicillin/streptomycin (Life Technologies, 15140122), 20 ng/mL recombinant human epidermal growth factor (EGF) (Peprotech, 100-15), and 20 ng/mL recombinant human fibroblast growth factor-basic (FGF-2) (Peprotech, 100-18B). PRMT5F/F ER day 4 neurospheres were treated with either 50 nM 4-OHT (Sigma, H7904) or the equivalent volume of EtOH for 24 h before splitting to induce PRMT5 knockout.
MEFs
Primary MEFs were prepared from E14.5 embryos as previously described (Xu 2005) and maintained in a humidified 5% CO2 atmosphere at 37°C in DMEM (Life Technologies, 11965118) supplemented with 10% fetal bovine serum (FBS) (Hyclone, SH30070.03) and 1% penicillin/streptomycin (Life Technologies, 15140122). To induce PRMT5 knockout, MEFs (passage 1) were grown to confluence in 15-cm dishes, and either 50 nM 4-OHT (H7904; Sigma) or the equivalent volume of EtOH was added 24 h before splitting.
Human cell lines
HEK293T, Phoenix-Eco, A549, U87, U2OS, and HCT116 were obtained from American Type Culture Collection (ATCC) and propagated according to ATCC data sheets.
Microarray analysis
The expression data from quadruplicate Illumina MouseRef-8 V2 microarrays were quantile-normalized, and only probes with an absolute fold change >1.5-fold and a Q-value <0.01 were labeled as significantly differentially expressed.
RNA-seq library preparation and splicing analysis
For RNA-seq library preparation, we followed the Illumina TruSeq RNA Sample Preparation kit version 2 manual. At least 70 million, 51-base-pair (bp)-long paired-end reads were mapped to the NCBI37/mm9 version of the mouse genome per replicate. Genes were labeled as significantly differentially expressed if the P-value as called by cuffdiff (Trapnell et al. 2012) was <0.05.
To determine differential splicing events, MATS 3.0.6 beta (Shen et al. 2012) was used for counting junction reads and reads falling into the tested region within ENSEMBL version 65 gene definitions. Matching embryos were analyzed individually, and only significant events occurring in at least two replicates were considered. Splicing events were labeled significant if the sum of the reads supporting a specific event exceeded 10 reads, the P-value was <0.05, and the minimum inclusion level difference as determined by MATS was >0.2. All sequencing and microarray data have been submitted to the Gene Expression Omnibus repository and are available under accession number GSE45285.
Functional annotation and network analysis
The functional annotation of the significant microarray and RNA-seq genes was performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang et al. 2009) using Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway (Kanehisa et al. 2012) representations. Network representations were generated using Cytoscape 3.0.1 (Shannon et al. 2003) with the plug-in ClueGO (Bindea et al. 2009). Only gene ontology terms with at least five members and a κ score >0.3 were used.
Polysome purification
Polysomes were isolated and separated as previously described (Zhang et al. 2012).
TAM injections
A single pulse of 2 mg of TAM (Sigma, T5648) plus 1 mg of progesterone (Sigma, P8783) in mineral oil (Sigma, M5904) was given to pregnant females at E10.5. Embryos were harvested and analyzed at E15.5 and E17.5.
Competing interest statement
The research of Z.Q.B. was funded by Lilly Research Laboratories, and he is an employee of Eli Lilly and Company.
Acknowledgments
We thank P.R. Kaldis, S.H. Lim, K. Diril, and U. Surana for sharing protocols and for helpful discussions. We thank L. Tora and P.R. Kaldis for critically reading the manuscript; K. Rogers, S. Rogers, and E.W. Sim for help with histopathology work; and M. Al-Haddawi for help with the pathology description of the PRMT5F/FNes brains. We thank P. Kaldis for sharing the Nestin-CRE and ER transgenic animals, V. Tergaonkar for sharing the p53-null mice, D. Lane and C.F. Cheok for the kind gift of Nutlin, Y. Minoru for the kind gift of Spliceostatin A, E. Makeyev for sharing the RFP-minigene backbone construct, and the BRC Shared facilities for technical support. We are grateful to X. Ruan and the GIS Genome Sequencing Team for help with the Solexa high-throughput sequencing, and the entire E.G. laboratory for critical discussion. This work was supported by an AGA-SINGA (Singapore Graduate Award) fellowship to M.B. and by IMCB, A-STAR. E.G. acknowledges support from JCO-ASTAR grants number 1134c001 and number 11/03/FG/07/04.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.219899.113.
References
- Akhtar RS, Geng Y, Klocke BJ, Latham CB, Villunger A, Michalak EM, Strasser A, Carroll SL, Roth KA 2006. BH3-only proapoptotic Bcl-2 family members Noxa and Puma mediate neural precursor cell death. J Neurosci 26: 7257–7264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende-Vega N, Dayal S, Agarwala U, Sparks A, Bourdon JC, Saville MK 2013. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 32: 1–14 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J 2009. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Richard S 2003. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics 2: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Cheng D, Cote J, Shaaban S, Bedford MT 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25: 71–83 [DOI] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE 2004. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW 2011. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol 21: 904–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL 2010. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev 24: 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, et al. 2010. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, Cookson MR 2013. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci 16: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumont G, Martoriati A, Beekman C, Bogaerts S, Mee PJ, Bureau F, Colombo E, Alcalay M, Bellefroid E, Marchesi F, et al. 2005. G1 checkpoint failure and increased tumor susceptibility in mice lacking the novel p53 target Ptprv. EMBO J 24: 3093–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Polydorides AD, Darnell RB 2001. The splice of life: Alternative splicing and neurological disease. Nat Rev Neurosci 2: 43–50 [DOI] [PubMed] [Google Scholar]
- Faigle R, Song H 2013. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim Biophys Acta 1830: 2435–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci 103: 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M, Asang C, Kammler S, Konermann C, Krummheuer J, Hipp M, Meyer I, Gierling W, Theiss S, Preuss T, et al. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res 31: 6963–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G 2001a. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 7: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G 2001b. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem 277: 8243–8247 [DOI] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, et al. 2012. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG 2006. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG 2007. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 178: 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Praveen K, Hicks AJ, Tian L, Matera AG 2008. Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster. RNA 14: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Muller U 2001. β1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31: 367–379 [DOI] [PubMed] [Google Scholar]
- Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, et al. 2007. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics 31: 32–41 [DOI] [PubMed] [Google Scholar]
- Hayhurst M, Wagner AK, Cerletti M, Wagers AJ, Rubin LL 2012. A cell-autonomous defect in skeletal muscle satellite cells expressing low levels of survival of motor neuron protein. Dev Biol 368: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H 2000. A mouse model for spinal muscular atrophy. Nat Genet 24: 66–70 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB 2008. Arginine methylation regulates the p53 response. Nat Cell Biol 10: 1431–1439 [DOI] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S 2011. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci 36: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA 1996. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature 384: 368–372 [DOI] [PubMed] [Google Scholar]
- Lane DP 1992. Cancer. p53, guardian of the genome. Nature 358: 15–16 [DOI] [PubMed] [Google Scholar]
- Lenos K, Jochemsen AG 2011. Functions of MDMX in the modulation of the p53-response. J Biomed Biotechnol 2011: 876173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Kaldis P 2012. Loss of Cdk2 and Cdk4 induces a switch from proliferation to differentiation in neural stem cells. Stem Cells 30: 1509–1520 [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N 2004. eIF4E—from translation to transformation. Oncogene 23: 3172–3179 [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 11: 1990–1994 [DOI] [PubMed] [Google Scholar]
- Migliori V, Phalke S, Bezzi M, Guccione E 2010. Arginine/lysine-methyl/methyl switches: Biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics 2: 119–137 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell-cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, et al. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem 279: 24246–24254 [DOI] [PubMed] [Google Scholar]
- Nagamatsu G, Kosaka T, Kawasumi M, Kinoshita T, Takubo K, Akiyama H, Sudo T, Kobayashi T, Oya M, Suda T 2011. A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J Biol Chem 286: 10641–10648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mittler G, Mann M 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods 1: 119–126 [DOI] [PubMed] [Google Scholar]
- Rallapalli R, Strachan G, Tuan RS, Hall DJ 2003. Identification of a domain within MDMX-S that is responsible for its high affinity interaction with p53 and high-level expression in mammalian cells. J Cell Biochem 89: 563–575 [DOI] [PubMed] [Google Scholar]
- Ritchie W, Granjeaud S, Puthier D, Gautheret D 2008. Entropy measures quantify global splicing disorders in cancer. PLoS Comput Biol 4: e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140 [DOI] [PubMed] [Google Scholar]
- Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA 2012. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 25: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. 2010. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 19: 1–13 [DOI] [PubMed] [Google Scholar]
- Scoumanne A, Zhang J, Chen X 2009. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res 37: 4965–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafey D, MacKenzie AE, Kothary R 2008. Neurodevelopmental abnormalities in neurosphere-derived neural stem cells from SMN-depleted mice. J Neurosci Res 86: 2839–2847 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P 1987. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y 2012. MATS: A Bayesian framework for flexible detection of differential alternative splicing from RNA-seq data. Nucleic Acids Res 40: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E 2006. Southern blotting. Nat Protoc 1: 518–525 [DOI] [PubMed] [Google Scholar]
- Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA 2010. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev 24: 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Wang Z, Hortobagyi T, Witten JT, Zarnack K, Kayikci M, Clark TA, Schweitzer AC, Rot G, Curk T, et al. 2011. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21: 1572–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L 2012. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA 2010. The pathobiology of splicing. J Pathol 220: 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. 2013. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103: 3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J 2005. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol 70: 28.1.1–28.1.8 [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhao T, Ang HS, Chong P, Saiki R, Igarashi K, Yang H, Vardy LA 2012. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev 26: 461–473 [DOI] [PMC free article] [PubMed] [Google Scholar]