A binary switch between telomerase-extendible and telomerase-nonextendible states determines telomere length homeostasis. Here, Qiao and coworkers address how shelterin complex component Tpz1 regulates telomere length in fission yeast. Separation-of-function mutant analyses indicate that Tpz1-mediated linkage within the shelterin complex defines the telomerase-nonextendible state. Interestingly, the authors show that Tpz1 also plays a role in the activation of telomeres to the extendible state via its interaction with shelterin component Ccq1. Thus, this study suggests that Tpz1 coordinates both positive and negative regulators of telomere length homeostasis.
Keywords: nucleoprotein complex, shelterin, telomerase, telomere length homeostasis, telomeres
Abstract
Telomeres are nucleoprotein complexes comprising telomeric DNA repeats bound by the multiprotein shelterin complex. A dynamic binary switch between telomerase-extendible and telomerase-nonextendible telomeric states determines telomere length homeostasis. However, the molecular nature of the nonextendible state is largely unknown. Here, we show that, in fission yeast, Tpz1 (the ortholog of human TPP1)-mediated complete linkage within the shelterin complex, bridging telomeric dsDNA to ssDNA, controls the telomerase-nonextendible state. Disruption of this linkage leads to unregulated telomere elongation while still retaining the shelterin components on telomeres. Therefore, the linkage within the shelterin components, rather than the individual shelterin components per se, defines the telomerase-nonextendible state. Furthermore, epistasis analyses reveal that Tpz1 also participates in the activation of telomeres to the extendible state via its interaction with Ccq1. Our results suggest critical regulatory roles of Tpz1 in the telomere binary switch.
Telomeres are DNA–protein complexes that protect the ends of eukaryotic chromosome ends from degradation and prevent their recognition as DNA damage sites (Palm and de Lange 2008; de Lange 2009). Telomere integrity is essential for cell survival and proliferation (Artandi and Cooper 2009; Jain and Cooper 2010), and, accordingly, dysfunctional telomeres can initiate genomic instability, cellular senescence, and organismal aging. The telomeric DNA consists of short tandem DNA repeats, which are G-rich in one strand (called the G strand) and C-rich in the complementary stand (called the C strand). The G strand extends beyond the C strand and forms a single-stranded G overhang. The 3′ end of the G overhang acts as the substrate for telomerase—a reverse transcriptase (Lingner et al. 1997; Nakamura et al. 1997) with its intrinsic RNA containing the template—to extend the telomeric DNA (Autexier and Lue 2006; Collins 2006). The basic structure and function of telomeres are conserved among eukaryotes; this conserved telomere protein complex that interacts with specific telomere sequences and caps chromosome ends is called shelterin (de Lange 2005). Identification of fission yeast (Schizosaccharomyces pombe) Pot1, the telomere ssDNA-binding protein, immediately allowed the discovery of human POT1 (Baumann and Cech 2001). A protein interaction partner of human POT1, TPP1, was subsequently identified (Houghtaling et al. 2004; Liu et al. 2004a; Ye et al. 2004). The POT1/TPP1 complex binds single-stranded telomere DNA with higher affinity than POT1 itself, forming an ortholog of the ciliate TEBP-α/TEBP-β complex—the archetypal telomere ssDNA overhang-binding module (Horvath et al. 1998; Wang et al. 2007). Thanks to the recent development of the powerful proteomic techniques, a nearly complete suite of telomere-localized proteins has been identified in human and fission yeast (Liu et al. 2004b; de Lange 2005; Miyoshi et al. 2008). Similar to the human telomeres, the fission yeast telomere dsDNA repeats are covered directly by sequence-specific dsDNA-binding protein Taz1 (TRF1 and TRF2 in humans) (Cooper et al. 1997). A “protein bridge,” consisting of Rap1, Poz1, and Tpz1 (human TPP1 ortholog), connects the dsDNA and ssDNA regions of the telomere through their direct protein–protein interactions with Taz1 and Pot1, respectively (Miyoshi et al. 2008).
In telomerase-positive cells, such as human embryonic stem cells (Gunes and Rudolph 2013), adult germline cells (Tan et al. 2012), most cancer cells (Shay and Wright 1996, 2010), and single-celled eukaryotes (ciliated protozoa and yeasts) (Greider and Blackburn 1985; Cohn and Blackburn 1995), telomeres are not maintained at a defined length but instead within a rather species-specific range; this telomere length homeostasis is proposed to be mediated through dynamic switching of the chromosome ends between two states: telomerase-extendible and telomerase-nonextendible states (Teixeira et al. 2004). The accessibility of the telomere substrate to telomerase is the key difference distinguishing the two states. The shelterin complex is believed to regulate telomere accessibility and thus control the telomere length homeostasis (Smogorzewska and de Lange 2004). The shelterin complex is a group of interlinked telomere proteins; deleting any member of this group alters telomere length homeostasis. Most if not all telomere proteins are multifunctional; their many functions enable telomere regulation and telomere protection. In the mammalian systems, shelterin components are recruited to telomeres through the dsDNA binders TRF1/TRF2, and interactions between shelterin components are essential for their localizations to the telomeres (Loayza and de Lange 2003; Liu et al. 2004a; Ye et al. 2004; Takai et al. 2011; Sfeir and de Lange 2012). This interdependent telomere association behavior of the mammalian shelterin components hampers a precise understanding of how each individual component and the linkage between them separately contribute to telomerase regulation. S. pombe has a similar shelterin complex (Miyoshi et al. 2008); however, the recruitments of its shelterin components to telomeres are not solely dependent on dsDNA-binding protein Taz1. The Pot1 complex is found to associate with telomeres in a Taz1- or Rap1-independent manner (Miyoshi et al. 2008). This nifty feature of S. pombe shelterin makes it an ideal system to investigate how telomere length homeostasis is achieved through interactions among members of the telomere protein complex without the complications of their dissociations from the telomere due to the loss of interactions.
Among S. pombe telomere proteins, Tpz1 physically lies at the interface of telomeric dsDNA- and ssDNA-binding proteins and is functionally positioned between the positive and negative regulators of telomere elongation (Fig. 1A). In addition to its interactions with Poz1 and Pot1, Tpz1 is also associated with Ccq1—a telomerase recruiter and checkpoint response inhibitor (Flory et al. 2004; Miyoshi et al. 2008; Tomita and Cooper 2008; Jain et al. 2010; Moser et al. 2011; Webb and Zakian 2012; Yamazaki et al. 2012; Nandakumar and Cech 2013). The unique position of Tpz1 in the shelterin complex signifies its architectural role in shelterin complex assembly and implies its coordination roles in communicating the telomeric dsDNA length and structural information to the 3′ end of the G overhang—the ultimate destination of telomerase. Thus, studying Tpz1 could help us understand the molecular mechanism by which the shelterin complex regulates telomeres in the nonextendible state and how it is switched to the telomerase-extendible state—two central questions that await answers. However, previous studies showed that most tpz1Δ cells are inviable. Surviving haploid cells completely lose the telomeric signal and form self-circularized chromosomes (Miyoshi et al. 2008), indicating the critical role of Tpz1 in chromosome end protection. The dominant telomere deprotection consequence of tpz1+ deletion completely masks other important functions of Tpz1 in telomere length homeostasis, making it impossible to study Tpz1's role in telomerase regulation using tpz1Δ cells. Clearly, separation-of-function mutants of Tpz1 are necessary to comprehensively understand the multifunctionality of Tpz1 in telomerase regulation and telomere protection. Recent studies using the separation-of-function mutations on the TEL patch of human TTP1 successfully uncovered its roles in recruiting telomerase to telomeres and promoting telomerase processivity (Nandakumar et al. 2012; Sexton et al. 2012; Zhong et al. 2012). However, the important function of Tpz1 in regulating different telomeric states via its interactions with other shelterin components is still not clear.
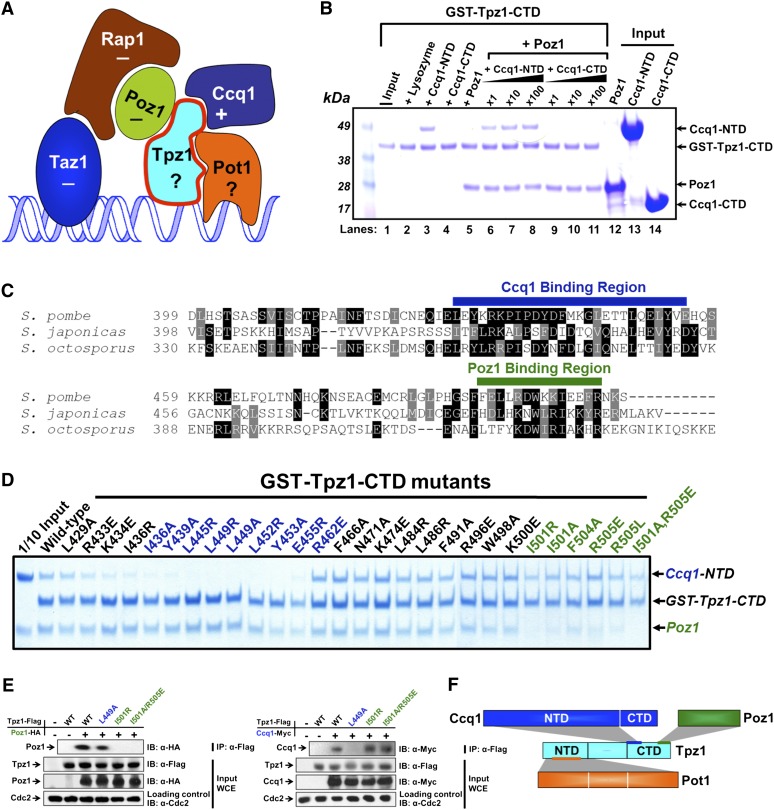
Figure 1.
Tpz1 interacts with Poz1 and Ccq1 simultaneously using two different patches. (A) Fission yeast shelterin complex. It is composed of telomeric sequence-specific dsDNA- and ssDNA-binding proteins Taz1 and Pot1, respectively, accompanied by their protein interaction partners, Rap1, Poz1, and Tpz1, forming a bridge between Taz1 and Pot1. Ccq1 was recently found to recruit telomerase to telomeres through Est1. If the deletion of a telomere protein causes telomere elongation, this protein is regarded as a negative regulator of telomere length and is therefore labeled “−”; otherwise, it is labeled “+”. For clarity, the stoichiometry of each individual component is not indicated in the figure; only one copy of each component is shown. (B) In vitro GST pull-down assays examining Tpz1–Poz1 and Tpz1–Ccq1 binary and Tpz1–Poz1–Ccq1 ternary interactions. Tpz1-CTD binds to Ccq1-NTD and Poz1 individually (lanes 3,5, respectively) and also at the same time (lanes 6–8). Competition experiments shown in lanes 7 and 8 demonstrate that even when the concentration of Ccq1-NTD is 100 times higher than that of Poz1, Tpz-CTD and Poz1 interaction remains stable. GST-Tpz1-CTD input (shown in lane 1) is one-tenth of the total protein used in the binding assays. Inputs shown in lanes 12–14 are one-fourth of the total Poz1, Ccq1-NTD, and Ccq1-CTD used in the binding assays. (C) Sequence alignment of Tpz1 from three different fission yeasts showing two conserved patches. Highly conserved residues are highlighted in black, and similar residues are highlighted in gray. (D) In vitro GST pull-down assays testing the binding of Tpz1 mutants to Poz1 and Ccq1-NTD. Two groups of mutants were identified that are defective in binding to Ccq1 (colored in blue) and Poz1 (colored in green), respectively. (E) Tpz1-L449A and Tpz1-I501R disrupt Tpz1–Ccq1 and Tpz1–Poz1 interactions, respectively, as evaluated by coimmunoprecipitation assays. Cdc2 is shown as the loading control. (Input) One-thirtieth of input WCE (whole-cell extract). (F) Schematic representation of Tpz1's interactions with Ccq1, Poz1, and Pot1. Tpz1 interacts simultaneously with Ccq1 and Poz1 via its CTD and with Pot1 via its NTD.
Here, we biochemically identified Tpz1 mutants that can individually but specifically disrupt its interactions with Poz1, Ccq1, or Pot1. Using these separation-of-function mutants of Tpz1, we found that the complete linkage between telomere dsDNA- and ssDNA-binding proteins within the shelterin complex is required for defining the telomerase-nonextendible state of telomeres. Disruption of the linkage on either the dsDNA binder or the ssDNA binder side of Tpz1 causes unregulated elongation of telomeres without delocalizing shelterin components from telomeres. Moreover, epistasis analyses of functional roles of Tpz1-centered interactions indicate that Ccq1 may activate the telomerase-nonextendible state of telomeres through its interaction with Tpz1, acting upstream of telomerase recruitment. Our results suggest that Tpz1 acts as the coordinator of positive and negative regulators of telomere length homeostasis and is also a critical regulatory target for controlling the competence of telomeres for elongation.
Results
Tpz1 interacts with Poz1 and Ccq1 simultaneously using two different but adjacent patches
We set out to mechanistically dissect the shelterin complex in S. pombe by focusing on Tpz1 due to its physical connections to multiple other shelterin components. Previous studies using yeast two-hybrid assays demonstrated that both Poz1 and Ccq1 bind to the Tpz1 C-terminal domain (Tpz1-CTD), a domain of ∼100 residues (Miyoshi et al. 2008). However, the investigation was unable to determine whether the two binding events happen simultaneously on Tpz1-CTD due to the limitations of the yeast two-hybrid assay. Using Escherichia coli-expressed, recombinant Tpz1-CTD (residues 406–508), Poz1, the Ccq1 N-terminal domain (Ccq1-NTD; residues 2–439), and Ccq1-CTD (residues 544–716), we performed GST pull-down assays to distinguish whether Poz1 and Ccq1 bind to Tpz1 simultaneously (as drawn in Fig. 1A) or their interactions with Tpz1 are mutually exclusive. As shown in Figure 1B, we found that GST-Tpz1-CTD interacted with Poz1 and Ccq1-NTD but not with Ccq1-CTD. Moreover, increasing the concentration of Ccq1-NTD to 100 times higher than that of Poz1 does not disassociate Poz1 from Tpz1-CTD. This experiment demonstrates that both Poz1 and Ccq1 can bind to Tpz1-CTD at the same time.
Simultaneous binding of Ccq1-NTD and Poz1 to Tpz1-CTD implies that there are two different surfaces on the Tpz1-CTD that mediate its interactions with Ccq1-NTD and Poz1, respectively. We then aimed to determine whether Tpz1–Poz1 interaction and Tpz1–Ccq1 interaction could be biochemically separated by distinct Tpz1 mutations. We hypothesized that functionally important residues should be evolutionarily conserved and thus represent leading candidates for residues to be targeted by mutagenesis. Sequence alignment of Tpz1 from S. pombe and two other fission yeast relatives, Schizosaccharomyces japonicus and Schizosaccharomyces octosporus (Rhind et al. 2011), reveals two clusters of relatively conserved patches (labeled with green and blue bars above the sequences in Fig. 1C). We hypothesized that these two patches correspond to the binding sites for Ccq1 and Poz1, respectively. Therefore, we made GST-Tpz1-CTD mutants and tested their binding ability to Poz1 and Ccq1-NTD. As predicted, mutations on one conserved patch (labeled in blue in Fig. 1C) disrupted Tpz1–Ccq1 binding while retaining Tpz1–Poz1 interaction, with the converse result for mutations on the other patch (labeled in green in Fig. 1C). As controls, we introduced mutations in the nonconserved residues between the two patches. These mutations disrupted neither Tpz1–Ccq1 nor Tpz1–Poz1 interaction. Given that the Tpz1 mutants on each conserved patch retain their binding to at least one protein, it is unlikely that the mutations cause global unfolding of Tpz1-CTD. Furthermore, using coimmunoprecipitation, we confirmed that mutations that abrogate Tpz1-CTD and Poz1 or Tpz1-CTD and Ccq1 interaction in vitro also disrupted the interactions between the full-length Tpz1 and Poz1 or between the full-length Tpz1 and Ccq1, respectively (Fig. 1E). Therefore, we identified two adjacent patches merely ∼50 residues away from each other on Tpz1-CTD that mediate its binding to Poz1 and Ccq1, respectively (as shown in Fig. 1F). Having obtained and validated Tpz1 separation-of-function mutations, we next used these mutants for functional dissection of the roles of Tpz1 in telomere maintenance in vivo.
Disruption of Tpz1–Poz1 interaction results in dramatically elongated telomeres
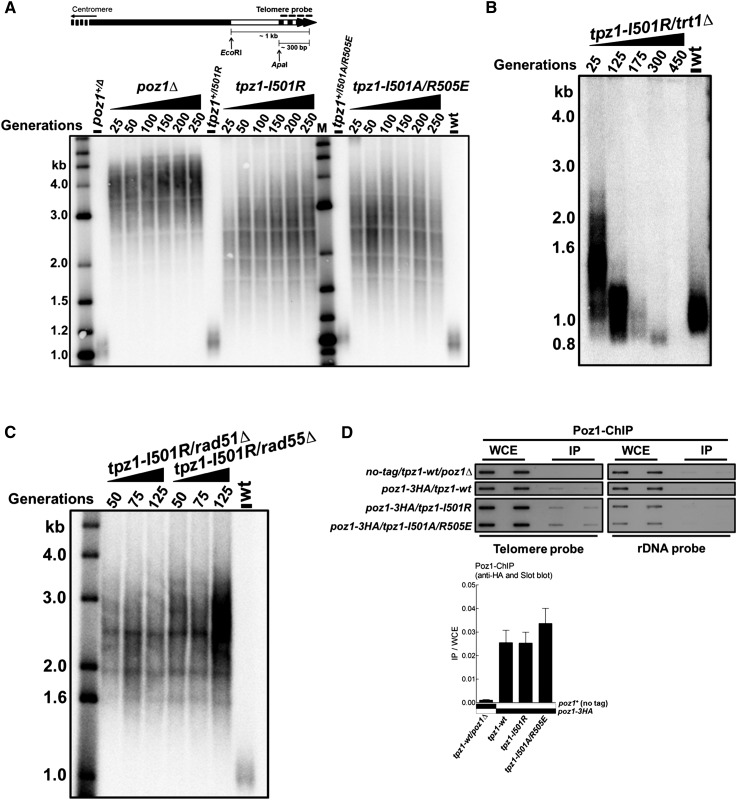
To elucidate the functional roles of Tpz1–Poz1 interaction in telomere maintenance, we made two S. pombe strains with tpz1 mutants encoding proteins defective in Poz1 binding: tpz1-I501R and tpz1-I501A/R505E. Both mutants had dramatically elongated telomeres, although not to the extent of telomeres in the poz1Δ cells (Fig. 2A; Miyoshi et al. 2008). This experiment indicates that the direct interaction of Poz1 with the telomeric ssDNA-binding complex Tpz1–Pot1 is required for the negative regulatory function of Poz1. To further investigate whether the negative regulatory role of Tpz1–Poz1 interaction in telomere length is imposed on telomerase, we deleted trt1+ in the tpz1-I501R mutant to make tpz1-I501R/trt1Δ and test whether telomere elongation resulting from loss of Tpz1–Poz1 interaction is telomerase-dependent. The classic “ever-shorter telomere” phenotype appears in the tpz1-I501R/trt1Δ double-mutant cells, indicating that telomerase is downstream from the defective Tpz1 function in tpz1-I501R (Fig. 2B). Consistent with the above, the telomere lengths of the tpz1-I501R/rad51Δ or tpz1-I501R/rad55Δ double mutant were not shortened upon deleting rad51+ or rad55+, which are required for homologous recombination (HR)-dependent telomere elongation (Fig. 2C). In addition, chromatin immunoprecipitation (ChIP) assays demonstrated that in both tpz1-I501R and tpz1-I501A/R505E mutant cells, Poz1 remains associated with telomeres (Fig. 2D), possibly via its interaction with Rap1. This result further indicates that it is the interaction between Tpz1 and Poz1, not Poz1 per se, that leads to the negative regulation of telomerase. Taken together, we conclude that the interaction between Tpz1 and Poz1 negatively regulates telomerase because disruption of this interaction results in telomerase-dependent, dramatically elongated telomeres. It is worth noting that the ∼1.5-kb more telomere elongation in poz1Δ cells compared with tpz1-I501R or tpz1-I501A/R505E cells suggests that Poz1 may have some additional roles in negatively regulating telomerase via a pathway that is separable from Poz1–Tpz1 interaction.
Figure 2.
Tpz1–Poz1 interaction negatively regulates telomerase. (A, top) Diagram of the telomere region showing relative positions of restriction enzyme sites and the telomere DNA probe for Southern blots. (Bottom) Southern blot analysis of EcoRI-digested genomic DNA using the telomere DNA probe for the indicated tpz1-I501R and tpz1-I501A/R505E mutant strains from successive restreaks on agar plates. Disruption of Tpz1–Poz1 interaction causes elongated telomeres that are ∼1.5 kb shorter than poz1Δ. In the telomere length analysis Southern blots presented in this study, either the 2-log DNA ladder from New England Biolabs (for A and Fig. 3D) or the 1-kb plus marker from Invitrogen (the rest of the telomere blots) was used and is denoted as “M.” Wild-type cells are denoted as “wt” in the blot. (B) Telomere maintenance in tpz1-I501R cells is telomerase-dependent. (C) Telomere maintenance in tpz1-I501R cells is not HR-dependent. (D) Recruitment of Poz1 to telomeres in tpz1-wt, tpz1-I501R, and tpz1-I501A/R505E cells was monitored by ChIP assays. Telomere association of Poz1 in each genetic background was monitored by slot blot. After hybridization with the telomere probe, the same membrane was stripped and then hybridized with the rDNA probe. Telomeric enrichment of Poz1 was expressed as immunoprecipitate (IP)/whole-cell extract (WCE) from the telomere DNA probe. Error bars in the quantitation of the slot blot analysis represent standard deviations of two individual repeats.
Disruption of Tpz1–Pot1 interaction also elongates telomeres
In fission yeast, the telomeric dsDNA is linked to a single-stranded G overhang through a protein bridge composed of Taz1, Rap1, Poz1, Tpz1, and Pot1, as shown in Figure 1A. Deletion of Taz1, Rap1, or Poz1 causes telomere elongation (Cooper et al. 1997; Miyoshi et al. 2008), while deletion of either Tpz1 or Pot1 leads to telomere deprotection and shortening and the subsequent formation of circularized chromosomes (Baumann and Cech 2001; Miyoshi et al. 2008; Pitt and Cooper 2010). This strong phenotype masks the functional roles of Tpz1 and Pot1 in telomerase regulation. Our finding shows that just like deletion of taz1+, rap1+, or poz1+, disruption of Tpz1–Poz1 interaction also disrupts negative regulation of telomerase and leads to elongated telomeres. What these alterations share in common is that they all break the complete linkage between the telomeric dsDNA and ssDNA mediated by the shelterin components. It is reasonable to hypothesize that if any linkage point between the telomeric dsDNA- and ssDNA-binding proteins is removed or broken, negative regulation of telomerase will be abrogated, thereby keeping telomeres in a telomerase-extendible state constitutively. The full connection between telomeric dsDNA-binding protein and ssDNA-binding protein may define the telomerase-nonextendible state of the telomere. This model predicts that disruption of Tpz1–Pot1 interaction will also lead to elongated telomeres in a telomerase-dependent manner, just as disruption of Tpz1–Poz1 interaction does. To test this, we set out to identify a specific mutant of Tpz1 that disrupts its interaction with Pot1, as we did for Poz1 and Ccq1 above.
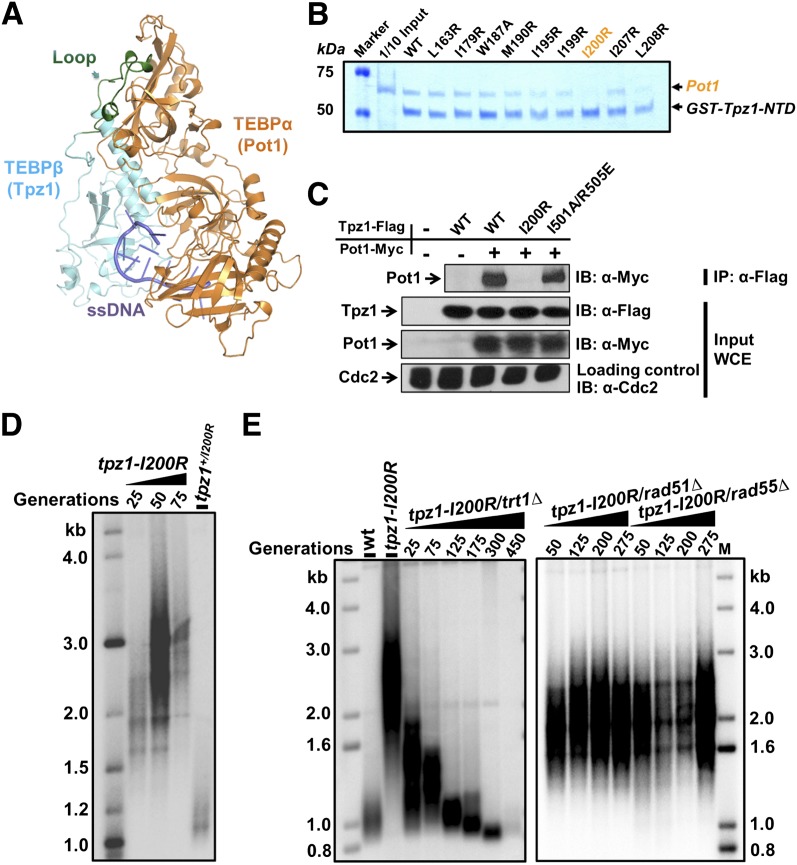
To this end, we resorted to the crystal structure of TEBP-α/β, the Oxytricha nova homolog of the S. pombe Pot1/Tpz1 complex (Horvath et al. 1998). Through a secondary structure prediction of Tpz1 and its homology with the TEBP-α/β structure (as shown in Supplemental Fig. S1), we identified a loop region in the Tpz1-NTD (residues 1–234) (Nandakumar and Cech 2012) as a strong candidate to mediate Tpz1–Pot1 interaction (colored green in Fig. 3A). Within that loop of Tpz1, we introduced point mutations individually to residues that are conserved among fission yeasts. The purified recombinant GST-Tpz1-NTD mutants, produced in E. coli, were then subjected to GST pull-down assays to evaluate their binding ability to Pot1. As shown in Figure 3B, among nine Tpz1-NTD mutants, Tpz1-I200R evidently abolished the in vitro association between Tpz1 and Pot1.
Figure 3.
Tpz1–Pot1 interaction negatively regulates telomerase. (A) Structural representation of the O. nova TEBP-α/TEBP-β complex, an ortholog of the Pot1/Tpz1 complex. The loop region of TEBP-β that mediates protein–protein interaction between TEBP-α and TEBP-β is indicated by an arrow and colored green. (B) In vitro GST pull-down assays examining the binding of Tpz1-NTD mutants to Pot1. (C) Tpz1-I200R disrupts Tpz1–Pot1 interaction, as evaluated by coimmunoprecipitation assays. Cdc2 was shown as the loading control. (Input) One-thirtieth of input WCE (whole-cell extract). (D) Telomeres elongate in the tpz1-I200R mutant, as shown by Southern blot analysis. (E) Telomere maintenance in the tpz1-I200R mutant is telomerase-dependent but not HR-dependent.
Next, we confirmed that Tpz1-I200R mutation, which abrogates Tpz1-NTD and Pot1 interaction in vitro, also disrupts full-length Tpz1–Pot1 interaction in coimmunoprecipitation assays (Fig. 3C). Consistent with our model, telomeres in tpz1-I200R mutant cells are also elongated (Fig. 3D), reminiscent of the tpz-I501R mutant in which Tpz1–Poz1 interaction is disrupted. In addition, just like we observed in the tpz-I501R mutant, deletion of telomerase resulted in an “ever-shorter telomere” phenotype in the tpz1-I200R/trt1Δ double mutant, whereas both the tpz1-I200R/rad51Δ and tpz1-I200R/rad55Δ double mutants maintain elongated telomeres (Fig. 3E), indicating that telomere elongation in the tpz1-I200R mutant is telomerase-dependent but not HR-dependent.
Both Tpz1–Poz1 and Tpz1–Pot1 interactions function in the same pathway as Rap1 and Poz1 in negatively regulating telomerase
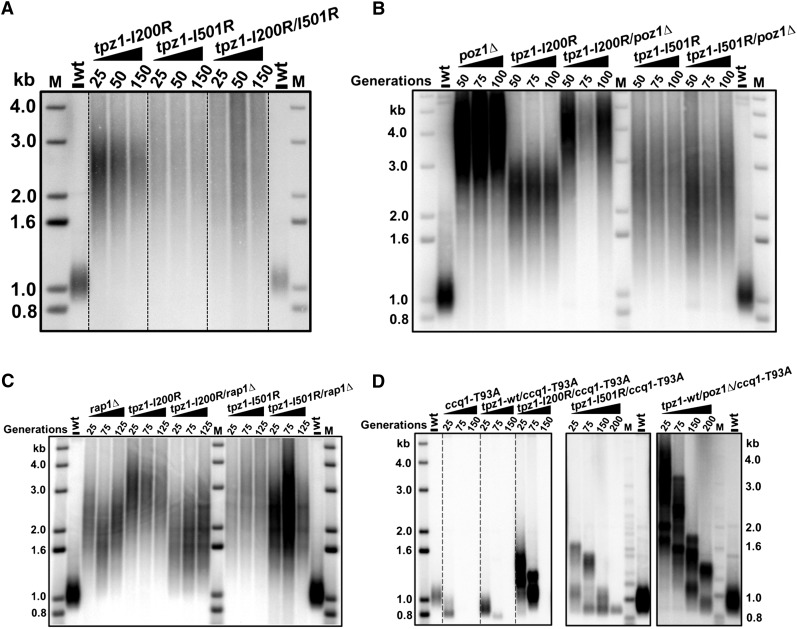
We further asked whether Tpz1–Pot1 interaction functions in the same pathway as Tpz1–Poz1 in telomerase regulation. To address this, we constructed a tpz1-I200R/I501R double-mutant strain and found that its telomeres were also elongated (Fig. 4A), similar to the single-mutant tpz1-I200R or tpz1-I501R but not additionally, indicating epistasis between the mutants. Hence, Tpz1–Pot1 interaction negatively regulates telomerase-mediated telomere elongation in the same pathway as Tpz1–Poz1 interaction. In the S. pombe shelterin complex, Tpz1, Poz1, and Rap1 connect the telomeric dsDNA-binding protein Taz1 to the G-overhang-binding protein Pot1. Deletion of rap1+ or poz1+causes telomerase-dependent telomere elongation, much like the phenotypes observed for Tpz1–Poz1 or Tpz1–Pot1 interaction-defective mutants. To determine whether the “connector” proteins Rap1 and Poz1 also act in the same pathway as Tpz1–Poz1 and Tpz1–Pot1 interactions to negatively regulate telomerase, we employed epistasis analysis and generated a set of double mutants in which rap1+ or poz1+ was individually deleted in tpz1-I200R and tpz1-I501R mutant strains. As shown in Figure 4, B and C, deletion of either rap1+ or poz1+ in both Tpz1–Pot1 interaction-defective (tpz1-I200R) and Tpz1–Poz1 interaction-defective (tpz1-I501R) mutant cells still produced elongated telomeres in the double-mutant cells; moreover, none of the double mutants had telomeres longer than those of the single mutants, suggesting that Rap1 and Poz1 act through Tpz1 and then Pot1 to prevent telomerase from elongating telomeres. Thus, we conclude that breaking any linkage within the shelterin complex connecting telomeric dsDNA and ssDNA leads to loss of negative regulation and therefore to elongated telomeres. Recent work showed that telomerase recruitment can be mediated by the interaction between telomerase subunit Est1 and Ccq1 when Thr93 of Ccq1 is phosphorylated by Tel1ATM and/or Rad3ATR. A nonphosphorylable mutant of Ccq1, Ccq1-T93A, cannot bind to Est1 and therefore fails to promote telomerase recruitment (Moser et al. 2011; Webb and Zakian 2012; Yamazaki et al. 2012). Taking advantage of this discovery, we constructed tpz1-I200R/ccq1-T93A and tpz1-I501R/ccq1-T93A double mutants and found that both double-mutant cells showed progressive telomere loss (Fig. 4D), suggesting that Tpz1-mediated negative telomerase regulation functions upstream of telomerase recruitment.
Figure 4.
Tpz1–Poz1 and Tpz1–Pot1 interactions act in the same pathway as Rap1 and Poz1 to negatively regulate telomerase. (A) tpz1-I200R/I501R cells have elongated telomeres, similar to those of tpz1-I501R cells. (B) Double-mutant strains tpz1-I200R/poz1Δ and tpz1-I501R/poz1Δ both have telomere length similar to the rap1Δ single-mutant strain. (C) Double-mutant strains tpz1-I200R/rap1Δ and tpz1-I501R/rap1Δ both have telomere length similar to the poz1Δ single-mutant strain. (D) Double-mutant strains tpz1-I200R/ccq1-T93A, tpz1-I501R/ccq1-T93A, and poz1Δ/ccq1-T93A show progressive telomere shortening.
Taken together, our finding implies that the complete linkage between the double-stranded and single-stranded telomeric DNA may define the telomerase-nonextendible telomeric state in which the telomere is a “dead” substrate for telomerase to elongate and has to be activated before telomerase can extend it. This mechanism provides a key molecular element about how the negative regulatory information is delivered from the telomere dsDNA side to 3′ of the G overhang—where telomerase works.
Loss of Ccq1–Tpz1 interaction causes telomere shortening and telomere maintenance via HR
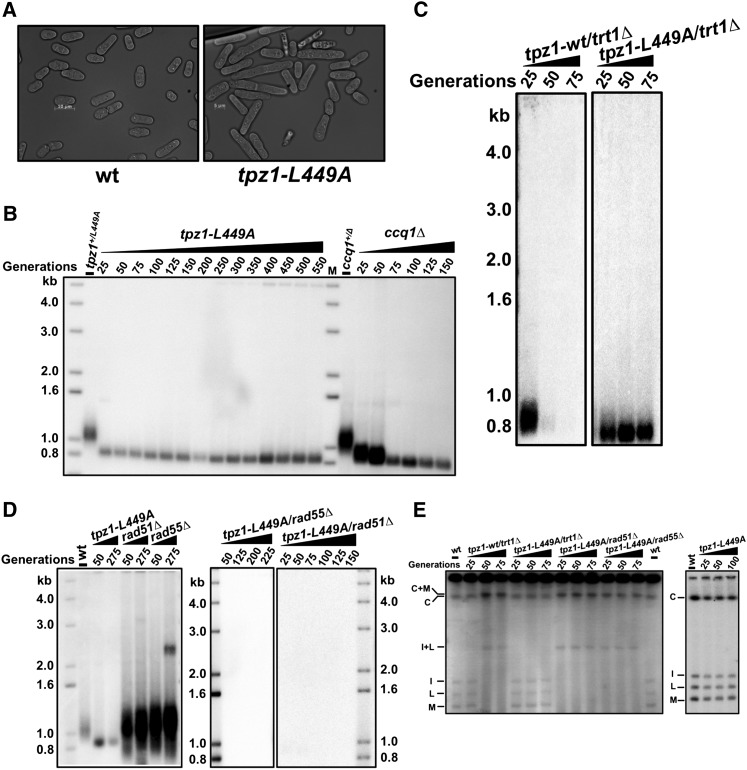
Ccq1, a more recently identified factor in fission yeast telomere maintenance, has been shown to be required for telomerase recruitment and inhibition of DNA damage-induced checkpoint activation at telomeres (Flory et al. 2004; Miyoshi et al. 2008; Tomita and Cooper 2008). Using our biochemically identified Ccq1–Tpz1 interaction-defective mutant (tpz1-L449A) in hand (Fig. 1D), we explored the functional significance of this interaction in telomere maintenance. We found that S. pombe cells containing the tpz1-L449A mutation elongated progressively with successive generations (Fig. 5A), a phenotype characteristic of DNA damage checkpoint activation. When tpz1-L449A cells reach 50 generations, ∼50% of the cells are twice as long as wild-type cells, reminiscent of ccq1Δ cells. This observation suggested that disruption of Ccq1–Tpz1 interaction triggers a checkpoint pathway, similar to the deletion of ccq1+. In addition, telomere length in tpz1-L449A cells, as shown in Figure 5B and Supplemental Figure S2A, is stable but ∼150 base pairs (bp) shorter than that in wild-type cells directly following sporulation from tpz1-L449A heterozygous diploid cells (tpz1-L449A/+). Similar to ccq1Δ cells (Tomita and Cooper 2008), tpz1-L449A cells appear to stably maintain short telomeres many more generations than the trt1Δ cells, in which telomeres shorten progressively. Furthermore, the telomere maintenance in tpz1-L449A cells is not dependent on telomerase (Fig. 5C); instead, it is achieved via a HR mechanism because deletion of either rad51+ or rad55+ immediately abrogates the stably maintained short telomeres in tpz1-L449A cells (Fig. 5D) and eventually generates survivals with circular chromosomes (Fig. 5E) that have lost ∼10-kb-long subtelomeric regions (Supplemental Fig. S2B). Since Rad55 is not required for telomere maintenance in ccq1Δ cells (Tomita and Cooper 2008) but is required for cells with defective Tpz1–Ccq1 interaction, Ccq1 might have additional roles in regulating HR-based telomere maintenance. In conclusion, shortened telomeres and loss of telomerase-mediated telomere elongation in Tpz1–Ccq1 interaction-defective mutant tpz1-L449A cells indicates that Ccq1 requires interaction with Tpz1 to carry out its functions as a positive regulator of telomerase.
Figure 5.
Loss of Ccq1–Tpz1 interaction causes telomere shortening and telomere maintenance via HR. (A) tpz1-L449A cells show elongated cell shape. (B) Telomeres in tpz1-L449A cells are ∼150 bp shorter than those of the wild-type cells, similar to those in ccq1Δ cells. (C) Telomere maintenance in tpz1-L449A cells is not dependent on telomerase. (D) Telomere maintenance in tpz1-L449A mutants is dependent on HR. (E) Deletion of either rad51+ or rad55+ in tpz1-L449A cells leads to chromosome circularization.
Loss of Ccq1–Tpz1 interaction does not affect the association of telomerase with telomeres or cause telomere deprotection
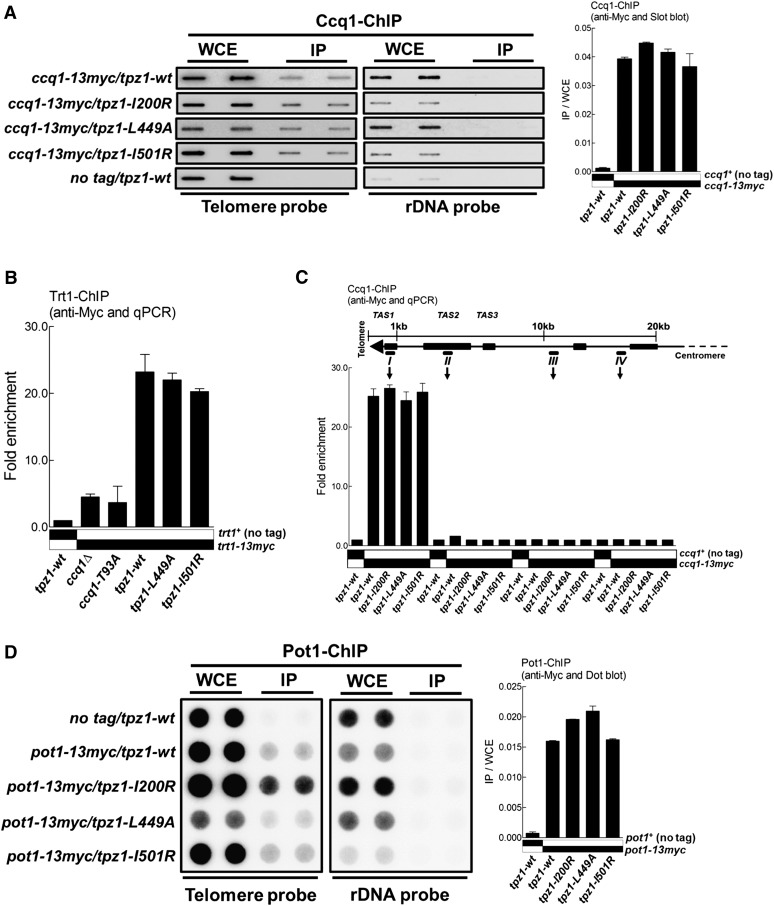
As mentioned above, recent studies demonstrated that Ccq1 is critical for telomerase recruitment via its interaction with Est1; this interaction is regulated by the phosphorylation of Ccq1-Thr 93 by Tel1ATM and/or Rad3ATR (Moser et al. 2011; Yamazaki et al. 2012). One possible outcome of disrupting Tpz1–Ccq1 interaction is that Ccq1 might not be able to localize to telomeres and thus fails to recruit Est1 and Trt1 to telomeres. To test this possibility, we performed ChIP assays to examine whether the associations of Ccq1 and/or Trt1 with telomeres were abolished in tpz1-L449A cells. To our surprise, we found that both Ccq1 and Trt1 bound to telomeres at levels similar to those in the wild-type strain, just as in the other two Tpz1 mutant strains (tpz1-I200R and tpz1-I501R) bearing elongated telomeres (Fig. 6A,B). In contrast, we determined from the same ChIP assay that Trt1 fails to localize to telomeres in ccq1Δ and ccq1-T93A cells (both are known to affect telomerase recruitment) (Fig. 6B). In addition, we found that, in all three Tpz1 interaction mutant strains, Ccq1 specifically bound to the telomere region or the first ∼300 bp of the subtelomeric region close to the telomere but not to the rest of the subtelomeric regions covering ∼20 kb from the chromosome end (Fig. 6C). This result indicated that telomere shortening in tpz1-L449A cells was not due to the failure to recruit telomerase to telomeres. This observation implies that Ccq1 and therefore telomerase may also be recruited to telomeres through another functional patch on Tpz1 or by other shelterin components.
Figure 6.
Loss of Ccq1–Tpz1 interaction does not affect the association of telomerase with telomeres or cause telomere deprotection. (A) Telomere association of Ccq1 in the indicated tpz1 mutants was assayed by ChIP assay. Immunoprecipitated DNA was applied to a slot blot for hybridization. After hybridization with the telomere probe, the same membrane was stripped and then hybridized with the rDNA probe. Error bars in the quantitation of the slot blot analysis represent standard deviations of two individual repeats. (B) ChIP analysis of Trt1. Telomere association of Trt1 in each background was monitored by quantitative real-time PCR with a primer pair against subtelomeric region I. The same PCR reaction using a primer pair against an fbp1 gene fragment was carried out as background control. Plots show mean values ± SD for two independent experiments. (C) Binding of Ccq1 to the telomeric and subtelomeric regions was evaluated by quantitative real-time PCR-ChIP analysis. (D) Telomere association of Pot1 in the indicated tpz1 mutant cells was monitored by a dot blot ChIP assay. Error bars in the quantitation of the dot blot analysis represent standard deviations of two individual repeats.
We next addressed whether disruption of Ccq1–Tpz1 interaction caused the dissociation of single-stranded telomere overhang-binding protein Pot1 from the telomeres, which would result in telomere deprotection and subsequent telomere shortening. As shown in Figure 6D, ChIP assays suggested that Pot1 interacted with telomeres of tpz1-L449A cells at a level similar to those in wild-type cells or Tpz1 mutants tpz1-I200R and tpz1-I501R. Furthermore, ChIP assays examining the telomeric association of Poz1 (Supplemental Fig. S3A) or Tpz1 (Supplemental Fig. S3B) showed that their presence at telomeres was not affected by tpz1-L449A mutation or the other two Tpz1 mutations (tpz1-I200R and tpz1-I501R) (Supplemental Fig. S3B,C). Altogether, we conclude that telomere integrity is not compromised in Ccq1–Tpz1 interaction-defective cells.
Ccq1 activates the telomerase-nonextendible state of telomeres via its interaction with Tpz1
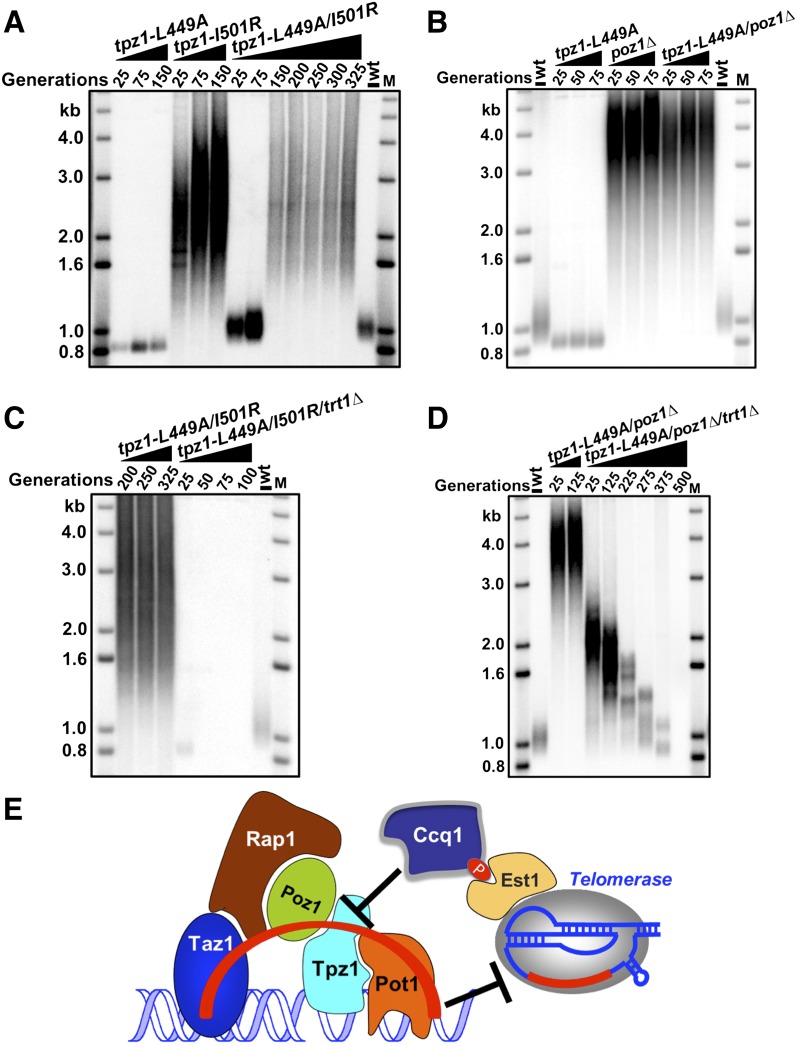
Since Ccq1–Tpz1 interaction does not affect telomerase recruitment to telomeres, we hypothesized that this interaction may stimulate telomerase action through the substrate side; namely, Ccq1 may interact with Tpz1 to induce a breakdown of Tpz1-mediated complete linkage between telomeric dsDNA and ssDNA, switching telomeres from a telomerase-nonextendible to a telomerase-extendible state. To investigate this possibility, we took advantage of a Tpz1 mutant strain characterized earlier in this study, tpz1-I501R, in which telomeres stay constitutively in the telomerase-extendible state due to the loss of complete linkage between the telomeric ssDNA and dsDNA by shelterin. If our hypothesis were correct, the tpz1-I501R mutant would bypass the requirement for the telomere activation step mediated by Ccq1–Tpz1 interaction. Indeed, tpz1-L449A/I501R double-mutant cells had telomeres elongated similarly to the tpz1-I501R single mutant (Fig. 7A). Moreover, we carried out the same experiment in the poz1Δ mutant background, which is similarly defective in negative regulation of telomere extension. poz1Δ/tpz1-L449A double-mutant cells showed telomere length identical to poz1Δ cells (Fig. 7B), further confirming bypass of Ccq1–Tpz1 interaction for telomere extension when telomeres are constitutively extendible. Not surprisingly, both the tpz1-I200R/L449A double-mutant cells and the tpz1-L449A/I501R/ poz1Δ triple-mutant cells also bypassed the necessity of Ccq1–Tpz1 interaction for telomerase-mediated telomere elongation (Supplemental Fig. S4A,B). However, tpz1-I501R cannot suppress ccq1Δ due to the lack of telomerase recruitment through Ccq1; instead, tpz1-I501R/ccq1Δ double-mutant cells appear to have the telomere deprotection phenotype (Supplemental Fig. S4C), similar to poz1Δ/ccq1Δ double-mutant cells (Miyoshi et al. 2008).
Figure 7.
Ccq1–Tpz1 interaction activates the telomerase-nonextendible state of telomeres. (A) tpz1-L449A/I501R cells have elongated telomeres, which are the same length as tpz1-I501R cells. (B) tpz1-L449A/poz1Δ cells have elongated telomeres, which are the same length as poz1Δ cells. (C,D) Telomere maintenance in the tpz1-L449A/I501R mutant (C) and tpz1-L449A/poz1Δ mutants (D) is telomerase-dependent. (E) A model for controlling telomerase-nonextendible telomeric state by the shelterin linkage and its switching to the extendible state by Ccq1–Tpz1 interaction.
Furthermore, we confirmed that in both tpz1-L449A/I501R and tpz1-L449A/poz1Δ cells, telomere elongation was mediated by telomerase because deletion of trt1+ in both strains led to telomere shortening (Fig. 7C,D). Thus, we conclude that Ccq1–Tpz1 interaction acts upstream of Tpz1–Poz1 interaction, Tpz1–Pot1 interaction, or likely all other negative regulators (such as Poz1) that retain telomeres in the nonextendible state. Therefore, Ccq1–Tpz1 interaction is necessary for the activation of telomeres to the extendible state for telomerase to elongate. Thus, as shown in Figure 7E, we propose that in addition to its telomerase recruitment role, Ccq1 is also a negative regulator of the shelterin-composed dsDNA–ssDNA negative regulatory bridge for telomerase-mediated telomere elongation; its interaction with Tpz1 is required to antagonize the negative force transmitted through Taz1–Rap1–Poz1–Tpz1–Pot1 (red curved line in Fig. 7E) on telomere extension. In other words, Ccq1 interacts with Tpz1 to induce the activation of the shelterin-controlled nonextendible state of telomeres, switching it to the extendible state.
Discussion
The equilibrium between telomerase-extendible and telomerase-nonextendible states is regulated by telomere length; short telomeres are elongated by telomerase more frequently than long telomeres (Teixeira et al. 2004). This equilibrium contributes to telomere length homeostasis, which in turn defines the telomere length in a species-specific range. Understanding the molecular nature of these two telomeric states depends on elucidating the functional roles of each individual molecular interaction among the telomere proteins. As telomere proteins are all interconnected with more than one interacting partner, separation-of-function mutants of them, mostly identified genetically (such as cdc13-1, cdc13-2, etc.), have been instrumental in revealing their multifunctionality in telomere maintenance (Evans and Lundblad 1999; Qi and Zakian 2000; Chandra et al. 2001; Pennock et al. 2001). However, in this “genetics first” approach, the biochemical properties of some of the mutants are difficult to clarify. In this study, we focused on fission yeast Tpz1, which physically lies in the interface of telomeric ssDNA- and dsDNA-binding proteins and is functionally positioned between the positive and negative regulators of the telomere elongation. It is almost impossible to study Tpz1's role in telomerase regulation because tpz1+ deletion immediately leads to circular chromosomes, presumably due to the loss of the telomere protection function of Tpz1. To overcome this complication, we biochemically identified Tpz1 mutants, which can individually but specifically disrupt its interaction with Pot1, Poz1, or Ccq1 while maintaining critical telomere protection functions. Coupled with epistasis analyses, these Tpz1 separation-of-function mutants allowed us to further dissect the multifaceted roles of Tpz1 in regulating telomerase-extendible and telomerase-nonextendible states.
A model for the nonextendible telomeric state
Shelterin is believed to have an inhibitory effect on telomerase (Bianchi and Shore 2008). Earlier studies, primarily using genetic deletions in the yeast systems and RNAi knockdown in the human system, have revealed that most of the shelterin components act as negative regulators of telomerase, which includes TRF1 and TRF2 (homologs of Taz1), RAP1, and TIN2 (homolog of Poz1). Telomere length phenotypes of POT1/TPP1 knockdown in human cells are controversial (Colgin et al. 2003; Veldman et al. 2004), and deletion of pot1+ or tpz1+ in fission yeast leads to telomere deprotection. These observations support the proposal that inhibition of telomerase through integration of telomere length information is transduced from the dsDNA-binding TRF1 complex, including TRF2, TIN2, and TPP1, to the telomere terminus through recruiting POT1 to the very end, thereby controlling telomere accessibility to telomerase (Marcand et al. 1997; Loayza and de Lange 2003; Barrientos et al. 2008; Kendellen et al. 2009). Alternatively, one or more shelterin components could also directly act on telomerase to enforce their negative roles. Deletion or knockdown of whole components cannot distinguish between these two possibilities. Here, by examining the telomere length of our Tpz1 separation-of-function mutants, in which only one residue of a protein is altered, likely preserving all the other functions, we can unambiguously evaluate the contributions of specific protein–protein interactions to telomerase regulation and avoid the complications from simultaneously losing other functional interactions mediated by the same protein. Our data together with previous work (Cooper et al. 1997; Miyoshi et al. 2008; Chen et al. 2011) demonstrate that the complete linkage between the double-stranded telomeric DNA and single-stranded G overhang through shelterin, but not individual shelterin components per se, controls telomeres in the telomerase-nonextendible state (Fig. 7E). The importance of the linkage in controlling telomeres in the nonextendible state is evident in both tpz1-I200R and tpz1-I501R mutants, in which shelterin components are disconnected due to the loss of Tpz1–Pot1 and Tpz1–Poz1 interactions, respectively, but are still associated with telomeres (shown by the ChIP assays in Figs. 2D, 6D; Supplemental Fig. S3). In particular, we show through the tpz1-I200R mutant that fission yeast Pot1 itself cannot inhibit telomerase even if it is associated with telomeres. The complete linkage from Pot1 to Taz1 is an essential element to prevent telomerase from elongating telomeres. Structurally, either the T-loop or G-overhang fold-back model can render telomerase-nonextendible telomeres because in both structures the very 3′ end of the G overhang is sequestered from being accessible to the telomerase. Importantly, the linkage between the dsDNA and ssDNA telomere binders is a shared key requirement for both structures. The longer the telomere, the more telomere dsDNA-binding proteins will be on the telomere and therefore more likely to form the linkage with the ssDNA-binding protein via the bridging proteins. Thus, our findings help explain why long telomeres tend to fall in the telomerase-nonextendible state.
The physical association of telomerase with telomeres is necessary but not sufficient to elongate telomeres, as we observed in the Tpz1–Ccq1 interaction-defective tpz1-L449A cells, in which Trt1 still binds to the telomere (Fig. 6B), but new telomere addition by telomerase does not occur (Fig. 5B) unless the negative regulation of telomeres resulting from the shelterin linkage is removed. Telomeres in the nonextendible state are basically “dead” substrates for telomerase and need to be activated. It is not hard to envision that the model that we propose for fission yeasts is applicable to the human system as well, given the conservation of the functional counterparts and pairwise interactions in both systems. In human cancer cells, telomerase is suggested to be prepositioned on telomeres. Under telomere steady-state maintenance conditions with normal telomere length, most telomeres are elongated only one round by telomerase every cell cycle; however, after telomeres are artificially shortened, the extendible telomeric state (also called open conformation) lasts longer, and therefore more telomerase molecules were observed to carry out additional rounds of telomere extensions in order to rapidly elongate telomeres to be above the critical length to avoid the activation of DNA damage signaling (Zhao et al. 2009, 2011).
Activation of the telomere substrate acts upstream of the telomerase recruitment
To be elongated by telomerase, the very 3′ end of the telomere has to become accessible. How does the telomere switch from the telomerase-nonextendible state to the telomerase-extendible state? Our in-depth epistasis analyses of Tpz1 mutants provide a clue. Without Ccq1–Tpz1 interaction (in the tpz1-L449A background), telomerase can still localize to telomeres but cannot elongate them; however, when poz1+ is deleted or Tpz1–Poz1 interaction is disrupted in this background, telomeres can be elongated again (Fig. 7A,B). This result indicates that Ccq1–Tpz1 interaction acts upstream of the shelterin linkage, the negative force keeping telomeres in the telomerase-nonextendible state. In other words, Ccq1 interacts with Tpz1 to activate the telomerase-nonextendible state. Ccq1 is therefore an inhibitor of nonextendible telomeres (net activation of extension) along with its role as a telomerase recruiter. In addition to the telomerase activation step (Taggart et al. 2002), our observation suggests that telomeres also need to be activated to make the elongation happen. This finding opens a brand new route to explore the biochemical mechanisms whereby the nonextendible telomeres are activated and become optimal substrates for telomerase. One possibility is that Ccq1–Tpz1 interaction may induce post-translational modifications of Tpz1 (such as phosphorylation by a kinase). The modified Tpz1 then loses its interaction with either Poz1 or Pot1, switching the telomere to the telomerase-extendible state.
In summary, our study reveals a key mechanistic aspect of the telomerase-nonextendible telomeric state and provides the first genetic evidence of how it is activated to the extendible state. Telomerase enzyme inhibitors have been developed as promising anti-cancer drugs (Harley 2008). Our mechanistic understanding of the nature of the telomerase-nonextendible state makes the telomere, the substrate, also “druggable.” Locking telomeres in the nonextendible state represents a possible new therapeutic approach in addition to inhibiting the enzyme.
Materials and methods
Yeast strains, gene tagging, and mutagenesis
Fission yeast strains used in this study are listed in Supplemental Table S1. Single-mutant strains were constructed by one-step gene replacement of the entire ORF with the selectable marker. Double-mutant and triple-mutant strains were produced by mating, sporulation, dissection, and selection followed by PCR verification of genotype. Genes were fused to specific epitope tags at the C terminus by HR; the pFA6a plasmid modules were used as a template for the PCR reaction (Bahler et al. 1998; Sato et al. 2005). Point mutations were made by mutagenesis PCR using the high-fidelity polymerase Pfu. All mutations were confirmed by DNA sequencing (Eton).
Protein expression and purification
The constructed plasmids were transformed into Rosetta-BL21(DE3) cells; protein expressions were induced by adding IPTG to a final concentration of 0.3–0.4 mM for 4–5 h at 30°C or 0.1–0.2 mM IPTG overnight at 16°C. Cells were harvested by centrifugation at 5000 rpm, and pellets were resuspended in lysis buffer (25 mM Tris-HCl at pH 8.0, 350 mM NaCl, 5 mM β-mercaptoethanol, 2 mM PMSF). Cells were disrupted by sonication, and the supernatant was incubated with equilibrated Ni-NTA (Qiagen) resin for 1 h. After centrifugation at 2000 rpm for 2 min, the resin was washed twice with B350 wash buffer (25 mM Tris-HCl at pH 8.0, 350 mm NaCl, 15 mM imidazole, 2 mM β-mercaptoethanol), and the protein was stepwise-eluted with elution buffer containing up to 300 mM imidazole.
GST pull-down assay
Fifteen microliters of 1 mg/mL GST fusion protein was incubated with 20 μL of glutathione sepharose beads for 1 h at 4°C. After incubation, the beads were washed twice with 800 μL of GST pull-down buffer (50 mM Tris-HCl at pH 8.0, 200 mM NaCl, 10 mM β-ME, 0.05% Tween-20). The bound proteins were then incubated with 20 μL of target protein (1 mg/mL) for 1 h at 4°C with gentle rocking. After washing three times with 800 μL of GST pull-down buffer, the supernatants were removed by centrifugation at 3,000 rpm for 30 sec and then boiled for 5 min in 15 μL of 2× SDS loading buffer. Eluted proteins were resolved by 10% SDS-PAGE and then visualized by Coomassie blue staining.
Coimmunoprecipitation
Frozen S. pombe cells cells were cryogenically disrupted using CryoMill (Retsch) and then resuspended in ice-cold lysis buffer (50 mM Tris-HCl at pH 7.5, 200 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, Complete proteinase inhibitor [Roche], 1 mM DTT, 2 mM PMSF, 2 mM benzamidine, 1 mM Na3VO4, 1 mM NaF). Extracts were clarified, and a final concentration of extracts was adjusted to 10 mg/mL. Anti-Flag M2 affinity gel (from Sigma) was equilibrated and washed twice with the same lysis buffer. Immunoprecipitations were performed for 4 h at 4°C and washed; proteins were eluted from the beads by incubating for 10 min at room temperature with 30 μL of 0.1 M glycine solution (pH 2.0) followed by the addition of 2 μL of 1 M Tris-HCl (pH 8.0). Eluted proteins were resolved by 10% SDS-PAGE and then subjected to Western blotting. Western blot analysis was performed using monoclonal anti-Flag (M2-F1804, from Sigma), monoclonal anti-Myc (9E10, from Covance), anti-Cdc2 (y100.4, from Abcam), or monoclonal anti-HA (3F10, from Roche). Whole-cell extracts were prepared using either trichloracetic acid (TCA) or urea lysis buffer with protease and phosphatase inhibitors.
Pulsed-field gel electrophoresis and Southern blotting for telomere length analysis
S. pombe cells grown in 2 mL of YEAU medium were used to extract chromosomal DNA, which was then digested by NotI. The digested DNA in plugs was subjected to pulsed-field gel electrophoresis as described (Moser et al. 2011). For telomere length analysis by Southern blotting, EcoRI-digested genomic DNA from 2 mL of YEAU S. pombe culture was separated on 1% agarose gel and probed with a telomeric DNA probe as previously described (Moser et al. 2011).
ChIP
S. pombe cells were grown at 32°C in YEAU to OD600 0.5–0.6, shifted for 1 h to 20°C prior to 20-min fixation with an 11% formaldehyde solution (11% formaldehyde, 100 mM NaCl, 1 mM EDTA at pH 8.0, 0.5 mM EGTA, 50 mM Tris-HCl at pH 8.0). After addition of 125 mM glycine and incubation for 5 min at 20°C, cells were chilled on ice, washed with ice-cold 1× TBS, and resuspended in 400 μL of lysis buffer (50 mM Hepes at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Trition X-100, 0.1% sodium deoxycholate, Complete proteinase inhibitor [Roche], 1 mM PMSF, 1 mM benzamidine, 1 mM Na3VO4, 1 mM NaF). Crude extracts were prepared by four pulses (60 sec) of bead-beating in FastPrep MP with Cryo-adaptor until 90% of cells were broken. Extracts were sonicated three times for 30 sec in 18 cycles using a Bioruptor until chromatin was sheared to an average size of less than ∼300 bp and subsequently cleared of insoluble cell debris by centrifugation at 15,000 rpm for 10 min. Ten microliters of the whole-cell extract was saved as an input control. Immunoprecipitation was performed for 2 h with antibody-conjugated beads (anti-Flag M2 affinity gel from Sigma; agarose-conjugated HA-probe F-7 or c-Myc 9E10 from Santa Cruz Biotechnology was used according to the tag on the protein). Precipitates were washed twice with 800 μL of lysis buffer, 800 μL of lysis buffer plus salt (lysis buffer with 500 mM NaCl), 800 μL of wash buffer, and 800 μL of 1× TE buffer, respectively. After the addition of 100 μL of 10% Chelex100 resin into the input controls and precipitates, respectively, those samples were boiled for 15 min at 100°C and then cooled at room temperature. Each sample was incubated with 2 μL of proteinase K (10 mg/mL) for 30 min at 55°C with gentle shaking. After denaturing with 0.4 M NaOH, ChIP and input samples were then transferred to a Hybond-XL membrane by using a slot or dot blot module. The membrane was hybridized with a probe specific for the telomeric sequence and then reprobed with rDNA after stripping. The hybridization signals were quantified using ImageQuant software. In addition, quantitative real-time PCR was used to analyze the same ChIP and input samples. Fold enrichment values were calculated based on ΔCt between ChIP and input samples after performing independent duplicate SYBR Green-based real-time PCR (Bio-Rad) using primer pairs of subtelomere and an fbp1 (fructose-1,6-bisphosphatase) gene fragment (as the background control); the values were expressed as immunoprecipitate/whole-cell extract (subtelomere) divided by immunoprecipitate/whole-cell extract (fbp1+) (Dehe et al. 2012).
Amplification of subtelomeric regions
Genomic DNAs were analyzed using primers selected to amplify specific subtelomeric regions by PCR (Moser et al. 2011). PCR products were loaded onto 2% agarose gels. Agarose gels stained with ethidium bromide were visualized using a Bio-Rad imaging system.
Acknowledgments
We thank Toru Nakamura, Fuyuki Ishikawa, Julie Cooper, Virginia Zakian, and Takashi Toda for providing plasmids and strains; Toru Nakamura for sharing detailed protocols of ChIP assay and pulsed-field gel electrophoresis; Wei Yao and Songtao Jia for sharing yeast protocols; and Craig Kaplan, Peter Kaiser, Songtao Jia, Jayakrishnan Nandakumar, Suzanne Sandmeyer, and Dorothy Shippen for comments on the manuscript and helpful discussions. The work in the laboratory of F.Q. is supported by a Basil O'Connor Starter Scholar Research Award from March of Dimes, a Beginning Grant-in-Aid from American Heart Association, and National Institutes of Health grant R01 GM098943.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.219485.113.
References
- Artandi SE, Cooper JP 2009. Reverse transcribing the code for chromosome stability. Mol Cell 36: 715–719 [DOI] [PubMed] [Google Scholar]
- Autexier C, Lue NF 2006. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem 75: 493–517 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge KT, Counter CM 2008. Distinct functions of POT1 at telomeres. Mol Cell Biol 28: 5251–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D 2008. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell 31: 153–165 [DOI] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH 1995. Telomerase in yeast. Science 269: 396–400 [DOI] [PubMed] [Google Scholar]
- Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR 2003. Human POT1 facilitates telomere elongation by telomerase. Curr Biol 13: 942–946 [DOI] [PubMed] [Google Scholar]
- Collins K 2006. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747 [DOI] [PubMed] [Google Scholar]
- Dehe PM, Rog O, Ferreira MG, Greenwood J, Cooper JP 2012. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol Cell 46: 797–808 [DOI] [PubMed] [Google Scholar]
- de Lange T 2005. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- de Lange T 2009. How telomeres solve the end-protection problem. Science 326: 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Flory MR, Carson AR, Muller EG, Aebersold R 2004. An SMC-domain protein in fission yeast links telomeres to the meiotic centrosome. Mol Cell 16: 619–630 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Gunes C, Rudolph KL 2013. The role of telomeres in stem cells and cancer. Cell 152: 390–393 [DOI] [PubMed] [Google Scholar]
- Harley CB 2008. Telomerase and cancer therapeutics. Nat Rev Cancer 8: 167–179 [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95: 963–974 [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Jain D, Cooper JP 2010. Telomeric strategies: Means to an end. Annu Rev Genet 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227 [DOI] [PubMed] [Google Scholar]
- Kendellen MF, Barrientos KS, Counter CM 2009. POT1 association with TRF2 regulates telomere length. Mol Cell Biol 29: 5611–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z 2004a. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z 2004b. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR 2012. DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res 40: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR 2013. Finding the end: Recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Pitt CW, Cooper JP 2010. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res 38: 6968–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22: 583–591 [DOI] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem 287: 34455–34464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T 2012. Removal of shelterin reveals the telomere end-protection problem. Science 336: 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE 1996. Telomerase activity in human cancer. Curr Opin Oncol 8: 66–71 [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE 2010. Telomeres and telomerase in normal and cancer stem cells. FEBS Lett 584: 3819–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T 2004. Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TC, Rahman R, Jaber-Hijazi F, Felix DA, Chen C, Louis EJ, Aboobaker A 2012. Telomere maintenance and telomerase activity are differentially regulated in asexual and sexual worms. Proc Natl Acad Sci 109: 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman T, Etheridge KT, Counter CM 2004. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol 14: 2264–2270 [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase–telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T 2004. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]