Abstract
Background:
The microRNA, miR-34c, is a well-established regulator of tumour suppression. It is downregulated in most forms of cancers and inhibits malignant growth by repressing genes involved in processes such as proliferation, anti-apoptosis, stemness, and migration. We have previously reported downregulation and tumour suppressive properties for miR-34c in prostate cancer (PCa).
Methods:
In this study, we set out to further characterize the mechanisms by which miR-34c deregulation contributes to PCa progression. The genes regulated by miR-34c in the PCa cell line PC3 were identified by microarray analyses and were found to be enriched in cell death, cell cycle, cellular growth, and cellular movement pathways. One of the identified targets was MET, a receptor tyrosine kinase activated by hepatocyte growth factor, that is crucial for metastatic progression.
Results:
We confirmed the inhibitory effect of miR-34c on both MET transcript and protein levels. The binding of miR-34c to two binding sites in the 3′-UTR of MET was validated using luciferase reporter assays and target site blockers. The effect of this regulation on the miR-34c inhibition of the migratory phenotype was also confirmed. In addition, a significant inverse correlation between miR-34c expression levels and MET immunostaining was found in PCa patients.
Conclusion:
These findings provide a novel molecular mechanism of MET regulation in PCa and contribute to the increasing evidence that miR-34c has a key tumour suppressive role in PCa.
Keywords: microRNA, miR-34c, MET, prostate cancer, androgen receptor, c-met
Prostate cancer (PCa) is the most common malignancy in men and the leading cause of cancer-related death for European men (Jemal et al, 2011). While confined to the prostate gland, the tumour is curable by either prostatectomy or radiation therapy. As the tumour progresses, it develops the abilities to invade surrounding tissues and metastasize. The effect of androgens through the androgen receptor (AR) is important for the growth and survival of prostate cells (Balk, 2002; Gelmann, 2002), but also represents an important oncogenic pathway for PCa progression. Androgen deprivation therapy, either chemical or surgical castration, is the standard treatment for advanced PCa. This treatment option results in initial regression of cancer growth in almost all patients. However, the majority of the tumours will eventually assume growth in a castration-resistant form (CRPC), for which there is currently no effective cure. The progression to the castration-resistant state is dependent on AR signalling, involving, for example, increased expression of the AR, increased local production of androgens, or increased activity of the AR (Feldman and Feldman, 2001). CRPC progression is associated with increased incidence of metastases, the major cause of cancer-related deaths in these men. The met proto-oncogene (hepatocyte growth factor receptor), abbreviated MET, is known to promote motility and invasive capability of tumour cells (Birchmeier et al, 2003; Trusolino et al, 2010). MET is a member of the receptor tyrosine kinase family and has been shown to be upregulated in CRPC and in metastases (Humphrey et al, 1995; Knudsen et al, 2002). The level of MET is suggested to be increased by, for example, gene amplification, hypoxia via HIF1α, but repressed by androgens through the AR (Verras et al, 2007). The microRNA, miR-34c, has also been shown to negatively modulate MET expression in cell lines derived from, forexample, lung, colon and gastric tumours (Migliore et al, 2008).
The tumour suppressor miR-34c has been shown to be downregulated in a number of different malignancies, such as neuroblastoma (Cole et al, 2008), breast (Yu et al, 2012), lung (Liang, 2008; Liu et al, 2009) and colorectal cancer (Toyota et al, 2008). Downregulation of miR-34c has been suggested to occur due to, for example, loss of heterozygosity within the 11q23 region (Dahiya et al, 1997; Ellsworth et al, 2008), epigenetic events such as CpG island methylation (Toyota et al, 2008; Corney et al, 2010; Vogt et al, 2011) or lost transcriptional induction of p53 (Bommer et al, 2007; Corney et al, 2007; He et al, 2007), as a consequence of p53 often being mutated in advanced cancer (Lane and Levine, 2010). This miRNA is involved in the regulation of proliferation, apoptosis, stem cell renewal, migration, senescence, and chemotherapy resistance (Bommer et al, 2007; Corney et al, 2007; Migliore et al, 2008; Catuogno et al, 2012). Our team has previously shown that miR-34c is decreased in PCa, inversely correlates to the grade of the tumour, occurrence of metastases, and shortened survival, and that reconstituted levels of miR-34c induce changes in proliferation, apoptosis, migration, invasion (Hagman et al, 2010), and AR levels (Ostling et al, 2011). In this article, we continue to characterise the targets and mechanisms of action of miR-34c in PCa. By ectopic expression of miR-34c in vitro, followed by gene expression array, we found MET to be a target of miR-34c in prostate cancer cells. This finding was confirmed on both mRNA and protein levels, and reporter gene luciferase assays verified direct binding of miR-34c to two specific binding sites in the 3′-UTR of MET. Further, we found MET expression to inversely correlate to miR-34c levels in a small but well-documented PCa cohort. Our working hypothesis is that miR-34c has a role in the initiation, progression, metastases, and transition to castration resistance of PCa, and that some of these effects are mediated by targeting MET.
Materials and methods
Cell culture and transfection
The prostate cancer cell lines were obtained from American Type Culture Collection (DU145, PC3, 22Rv1 and LNCaP clone FGC; Manassas, VA, USA) and European Collection of Cell Cultures (PNT2 and VcaP; Public Health England, Salisbury, UK). The cells were cultured according to the manufacturer's recommendations. Cells were transiently transfected with miRIDIAN microRNA Mimic (80 nM probe, Dharmacon, Lafayette, CO, USA) or miRCURY LNA knockdown probes (200 nM probe, Exiqon, Vedbaek, Denmark) using an Oligofectamin reagent (Invitrogen, Carlsbad, CA, USA). Control experiments were performed in parallel, transfecting cells with miRIDIAN microRNA Mimic Negative Control (Dharmacon) or scramble-mimics (Exiqon), respectively. The MET pRRL.2 lentiviral vector was a kind gift from professor Trusolino (University of Turin). It contains the coding region of MET and the first part of the 3′-UTR (1-188 nt). We co-transfected cells with miR-34c mimics and 1 μg vector using Lipofectamin 2000 (Invitrogen).
RNA extraction and qRT-PCR
PC3 cells were transiently transfected with miR-34c or scramble mimics, and total RNA was extracted with Trizol, according to the manufacturer's recommendations (Invitrogen). RNA was also extracted from the different cell lines as described previously (Hagman et al, 2010). The RNA was reverse transcribed using the RevertAid H minus First Strand cDNA Synthesis Kit (Fermentas, Thermo Scientific, Wilmington, DE, USA). QRT–PCR was performed with MET specific primers (Applied Biosystems, Foster City, CA, USA, cat. #4331182) and the TaqMan master mix (Applied Biosystems, cat. #4369016). HPRT and PGK1 were used as endogenous controls.
Microarray hybridization and data analysis
Single-stranded cDNA was generated using the GeneChip Whole Transcript (WT) cDNA Synthesis and Amplification Kit (Affymetrix Inc, Santa Clara, CA, USA) using 100 ng of total RNA. Amplified cDNA was fragmented and labelled using the GeneChip WT Terminal Labelling Kit (Affymetrix). Subsequently, the biotinylated cDNA was hybridized to the GeneChip Human Gene 1.0 ST Arrays (Affymetrix). The arrays were washed and stained on a GeneChip Fluidics Station 450 (Affymetrix) according to the manufacturer's recommendations. Arrays were scanned using the GeneChip Scanner 3000, and image analysis was performed using GeneChip Operating Software (GCOS, Affymetrix), and in Genepix 4.0 (Axon Instruments, Inverurie, Scotland). The data were normalized, background corrected, and summarised using the Robust Multichip Average (RMA) algorithm implemented in the Expression Console version 1.1.2 software (Affymetrix). The data were analysed using SAM analysis to identify significantly differentially expressed genes between the groups. The list of differentially expressed genes with q-values over zero and lower expression in the test groups than in the controls were analysed using Ingenuity Pathway Analyses (Ingenuity, Mountain View, CA, USA). A score was computed for each network according to the fit of the original set of significant genes. This score reflects the negative logarithm of the P-value, which indicated the likelihood of the focus genes in a network being enriched in the data set by chance.
Western blot
Cells were lysed with M-PER (Pierce, Rockford, IL, USA) supplemented with HaltT protease inhibitor cocktail EDTA-free (Pierce) and 13.4 mM EDTA. Protein concentration was measured, and equal amount of proteins were separated on a NuPAGE (Invitrogen) and transferred onto an Immobilon PVDF membrane (Millipore Corporation, Bedford, MA, USA). The membrane was incubated with antibodies directed against MET (sc-10, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), GAPDH 1:20000 (a-GAPDH, MAB374, Chemicon, CA, USA), and α-actinin (sc-17829, Santa Cruz Biotechnology). Signals from HRP-coupled polyclonal secondary antibodies (mouse, Dako, Glostrup, Denmark) were generated by ECL plus (GE Healthcare, Waukesha, WI, USA) and recorded using a CCD camera (LAS-3000, Fujifilm, Tokyo, Japan). Band intensities on western blot were quantified and normalized to the band intensities of GAPDH, using the ImageJ software (Madison, WI, USA).
Luciferase assay
The full MET 3′-UTR was amplified by nested PCR using PC3 cDNA as template and cloned into miREPORT (Ambion, Austin, TX, USA). For primer sequences see Table 1. In silico analyses using TargetScan 5.2 identified two miR-34c binding sites in the 3′-UTR of MET, at position 51–57 and position 2162–2168. We used QuickChange mutagenesis to mutate these sites (Stratagene, Santa Clara, CA, USA). For primer sequences see Table 1. DNA sequencing of the vectors were performed using the Big Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems). Cells were co-transfected with mimics or LNA inhibitors together with the miREPORT vector containing MET 3′-UTR with or without mutations. After 24 or 48 h, the cells were lysed and the luciferase signal was detected using a dual luciferase reporter assay, according to manufacturer's recommendations (Promega, Madison, WI, USA). A co-transfected Renilla vector was used for normalisation (Ambion).
Table 1. Sequences of primers for cloning MET 3′-UTR.
| Outer forward( Fw) |
5′-GTACTATGTCAAAGCAACAGT-3′ |
| Outer reverse (Rv) |
5′-CATCACTTTACTTTAATTGCAT-3′ |
| Inner Fw |
5′-CTCTCACTAGTCAAAGCAACAGTCCACACT-3′ |
| Inner Rv |
5′-CTCTCAAGCTTGCATGATTTATCAGAACAACT-3′ |
| Fw_mut1 |
5′-TGTCCAATGGTTTTTTAAATGCCTGACCTTTAAAAGGCCAT-3′ |
| Rv_mut1 |
5′-ATGGCCTTTTAAAGGTCAGGCATTTAAAAAACCATTGGACA-3′ |
| Fw_mut2 |
5′-TTAGATACTTGTAAATGCCTATACCTGCAGCTGAACTGAAT-3′ |
| Rv_mut2 | 5′-ATTCAGTTCAGCTGCAGGTATAGGCATTTACAAGTATCTAA-3′ |
Sequences of target site blockers to block the miR-34c-predicted sites in the MET 3′-UTR. Primers for PCR amplification of MET 3′-UTR and the primers used to mutate the miR-34c binding sites.
miR-34c and MET expression levels in PCa
The prostate cohort of 47 patients with PCa and the RNA extraction protocol have been described previously (Hagman et al, 2010). The miRNA levels were quantified using the TaqMan MicroRNA Assay (Applied Biosystems) according to the manufacturer's protocol with a few modifications described previously (Hagman et al, 2010). Adjacent formalin-fixed paraffin-embedded prostate tissue section slides were used for the miR-34c quantification and the detection of MET. The percentage of cancer cells in the samples (5–90%) was determined by a pathologist, more detailed information regarding the clinical parameters of each individual can be found in Hagman et al (Hagman et al, 2013). The immunohistochemical detection of MET was done as described in Östling et al (Ostling et al, 2011). MET (ab51067, Abcam, Cambridge, UK) was used as a primary antibody. The stained sections were scored independently, based on the MET staining intensity, by one pathologist and two other researchers. Overall staining intensity was scored (0=no, 1=weak, 2=distinct). For each patient, the MET intensity was scored in the malignant epithelium and benign epithelium.
Migration assay
The migration capacity of the cells was studied using migration inserts and plates (BD Biosciences, cat # 353093 and 353502) according to the manufacturer's instructions. Briefly, 3 days after transfection of DU145 cells with miR-34c or scrambled mimics and MET expression vector, cells were seeded in serum-free medium onto the membrane of the upper chamber. Cell media with 10% FBS was added to the lower chamber. The cells that had passed through the membrane after 17 h were fixed in paraformaldehyde and stained with crystal violet. The number of cells was counted manually in three independent experiments.
Statistical analysis
For the results from the luciferase assay, western blot and qRT–PCR on MET expression in cell lines, we used Student's t-test, where P<0.05 was considered significant. GraphPad Prism version 5 was used for analyses on patient material (GraphPad Software, La Jolla, CA, USA).
Results
Identification of pathways affected by miR-34c in prostate cancer
To identify mRNAs negatively affected by miR-34c in PCa, mRNA microarray analysis was performed with androgen-independent PC3 cells transfected with miR-34c or a scramble mimics. The targets identified are provided in Supplementary Table 1 (cutoff>1.3-fold changed). The data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002), accession number GSE41322.
By ingenuity pathway analysis, we investigated which pathways that are enriched for molecules deregulated by miR-34 c. Among the top biofunctional pathways are cell death (P=2.5 · 10−7), cell cycle (P=1.2 · 10−5), cellular growth (P=1.6 · 10−5), and cellular movement (P=3.2 · 10−5), see Table 2. Among the canonical pathways affected by miR-34c are NF-κB and hepatocyte growth factor (HGF) signalling, see Table 3. Among the downregulated genes after miR-34c expression are genes important in cancer, for example, CDC25A, E2F5, NOTCH2, RRAS, JAG1, MET, and PDGFRA (target list is found in Supplementary Table 1). All the specified targets contain miR-34c binding sites predicted by at least three target prediction programs (Miranda (Enright et al, 2003), miRtarget2 (Wang and El Naqa, 2008), Pictar (Krek et al, 2005), Pita (Kertesz et al, 2007), RNAhybrid (Rehmsmeier et al, 2004), and TargetScan (Lewis et al, 2005)). We chose to focus our efforts on MET, found to be 1.4-fold downregulated in the transcriptome analyses. The rationale behind this was that HGF signalling was one of the highest ranking canonical pathways, cellular movement was one of the top biofunctional pathways, MET has been suggested to have a key role in PCa progression affecting migration and invasion (features previously described by us that miR-34c induces in PCa cells (Hagman et al, 2010)) and MET has been shown to be a target of miR-34 family members in other cancer cell lines (Migliore et al, 2008).
Table 2. Top biofunctional pathways affected by miR-34c.
| Category | Function annotation | P-value |
|---|---|---|
| Cell death |
Cell death of cell lines |
2.5E-07 |
| Cell death |
Cell death of tumor cell lines |
8.8E-07 |
| Cell death |
Apoptosis of cell lines |
2.3E-06 |
| Cell death |
Apoptosis of tumor cell lines |
4.8E-06 |
| Cell death |
Apoptosis of eukaryotic cells |
6.5E-06 |
| Cell death |
Cell death of eukaryotic cells |
6.8E-06 |
| Cell death |
Apoptosis |
1.1E-05 |
| Cell cycle |
Cell division process |
1.2E-05 |
| Cellular growth and proliferation |
Proliferation of tumor cells |
1.6E-05 |
| Tumor morphology |
Proliferation of tumor cells |
1.6E-05 |
| Cancer |
Ovarian cancer |
1.8E-05 |
| Reproductive system disease |
Ovarian cancer |
1.8E-05 |
| Cell death |
Cell death |
2.2E-05 |
| Cellular development |
Angiogenesis of cells |
2.8E-05 |
| Organismal development |
Angiogenesis of cells |
2.8E-05 |
| Cardiovascular system development/function |
Angiogenesis of cells |
2.8E-05 |
| Cell-to-cell signaling and interaction |
Formation of gap junctions |
3.0E-05 |
| Cellular assembly and organization |
Formation of gap junctions |
3.0E-05 |
| Cell death |
Cell death of breast cancer cell lines |
3.1E-05 |
| Cellular movement |
Invasion of cell lines |
3.2E-05 |
| Infection mechanism |
Replication of Herpesviridae |
3.7E-05 |
| Cellular growth and proliferation |
Proliferation of cancer cells |
4.5E-05 |
| Tumor morphology | Proliferation of cancer cells | 4.5E-05 |
Table 3. Top canonical pathways affected by miR-34c.
| Ingenuity canonical pathways | P-value |
|---|---|
| Antigen presentation pathway |
2.5E-05 |
| Interferon signalling |
9.6E-05 |
| Prolactin signalling |
5.3E-04 |
| Erythropoietin signalling |
9.6E-04 |
| Hepatic fibrosis / hepatic stellate cell activation |
0.0013 |
| p70S6K signaling |
0.0020 |
| Role of pattern recognition receptors in recognition of bacteria/viruses |
0.0026 |
| Arginine and proline metabolism |
0.0028 |
| Growth hormone signalling |
0.0030 |
| NF-κB signaling |
0.0052 |
| Sphingolipid metabolism |
0.0072 |
| HGF signalling |
0.0087 |
| Pentose phosphate pathway | 0.0091 |
miR-34c affects MET expression
To validate that miR-34c modulates the levels of MET in PCa cells, we transfected PC3 cells with miR-34c or scramble mimics. As shown in Figure 1a, the mRNA level of MET was dramatically affected by miR-34c expression (a 97% decrease, P=0.003). The amount of MET protein was also significantly reduced when ectopically expressing miR-34c, (88% decrease for the precursor of 170 kDa, P=0.012; and 94% decrease for the β-subunit of 145 kDa, P=0.0002) in PC3 cells, see Figure 1B. The effect on MET protein levels was also confirmed in PNT2 cells (39% decrease, P=0.028) and DU145 cells (73% decrease, P=0.038), see Figure 1b. This indicates that miR-34c affects MET expression at the transcriptional level by, for example, cleavage of the mRNA or by destabilizing the mRNA. This effect is carried on to the protein level, but miR-34c might also accomplish additional blocking of the translation.
Figure 1.
(A) MET transcript levels, as measured by qRT–PCR, after ectopic expression of miR-34c mimic in PC3. Mean of quadruplicate is shown. HPRT and PGK1 were used as endogenous controls. (B) Western blot on the protein levels of MET after ectopically expressing miR-34c in PC3, PNT2, and DU145 cells. Both the 170 kDa precursor and the 140 kDa mature form of Met are detected. GAPDH and α-actinin were used as loading controls.
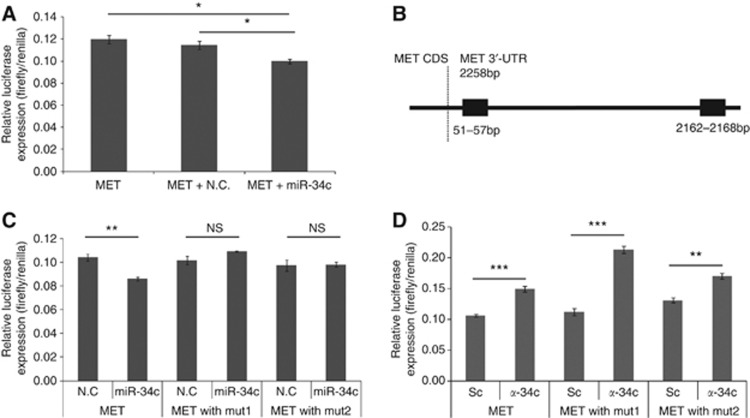
To test if miR-34c directly binds the 3′-UTR of MET mRNA, we performed luciferase reporter assays in PC3 cells. Co-transfection of MET 3′-UTR reporter vector and miR-34c mimics resulted in a decreased luciferase signal (compared with scramble mimics, P=0.006), confirming that binding of miR-34c to the 3′-UTR of MET have a direct inhibitory effect, see Figure 2A. To locate the specific binding site, an in silico prediction was performed, and two miR-34c binding sites were identified in the MET 3′-UTR, se Figure 2B. These sites were confirmed in vitro: the repression of MET upon ectopic expression of miR-34c in PC3 cells (P=0.044) was abolished if either of the two predicted binding sites was mutated, as measured using luciferase reporter assays with the 3′-UTR of MET, see Figure 2C, indicating that both sites are needed for the repression of MET to occur. The reverse, blocking of endogenous miR-34c with a LNA-antisense probes, resulted in increased luciferase signal (P=0.001), see Figure 2d, implying that MET is repressed to a certain degree by endogenous miR-34c. But, when plasmids containing MET 3′-UTR with either of the two miR-34c binding sites mutated were co-transfected with antisense miR-34c, this resulted in an increase in luciferase signal for both mutated binding sites, indicative of miR-34c being able to act through either of two sites, see Figure 2D. This experiment, however, suggests that the second binding site is the primary binding site, as the ratio of miR-34c and negative control for ‘mut1' that only has an intact second target site is higher than for ‘mut2' (P=0.019).
Figure 2.
(A) PC3 cells were transiently co-transfected with MET 3′-UTR and miR-34c or scramble mimics. (B) A schematic picture of the two predicted miR-34c binding site in the 3′-UTR of MET. (C) PC3 cells were transiently co-transfected with MET 3′-UTR and miR-34c mimic or negative control. Mutating either of the miR-34c binding sites abolished the effect. (D) PC3 cells were transiently co-transfected with MET 3′-UTR and antisense LNA miR-34c or scramble antisense control. Mutating either of the miR-34c binding sites did not abolish the effect. Luciferase activity was measured after 24 h and normalised to the co-transfected Renilla.
Effects on the phenotype
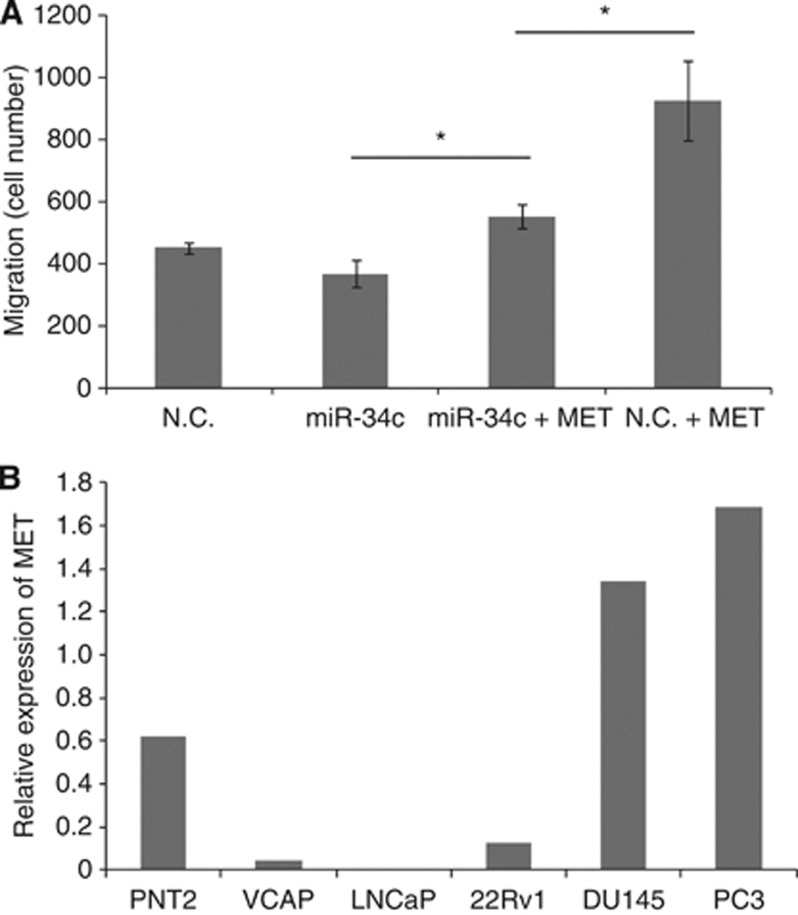
Next, we investigated if MET is the main target of miR-34c conveying the migratory phenotype. This was done by introducing miR-34c and investigating whether ectopic expression of MET could rescue the phenotype in DU145 cells. The MET construct contains the coding region and part of the 3′-UTR were the first miR-34c binding site is located, but not the second. Cells overexpressing miR-34c has a decreased migratory potential, but when exogenous MET is co-expressed with miR-34c the migratory potential is significantly increased (P=0.034), but still shows a significantly (P=0.05) lower migration compared with cells only overexpressing MET and no miR-34c, see Figure 3A. This experiment indicated that the decreased migratory potential of miR-34c-expressing cells can be counteracted by MET overexpression.
Figure 3.
(A) Introduction of MET rescue the phenotype induced by miR-34c. The migration potential of DU145 cells after transfection with miR-34c or scrambled mimics, with or without co-transfecting with a MET expression vector. (B) MET transcript levels as measured by qRT–PCR in prostate cell lines. Mean of quadruplicate is shown. HPRT and PGK1 were used as endogenous controls.
MET expression in prostate cell lines and clinical samples
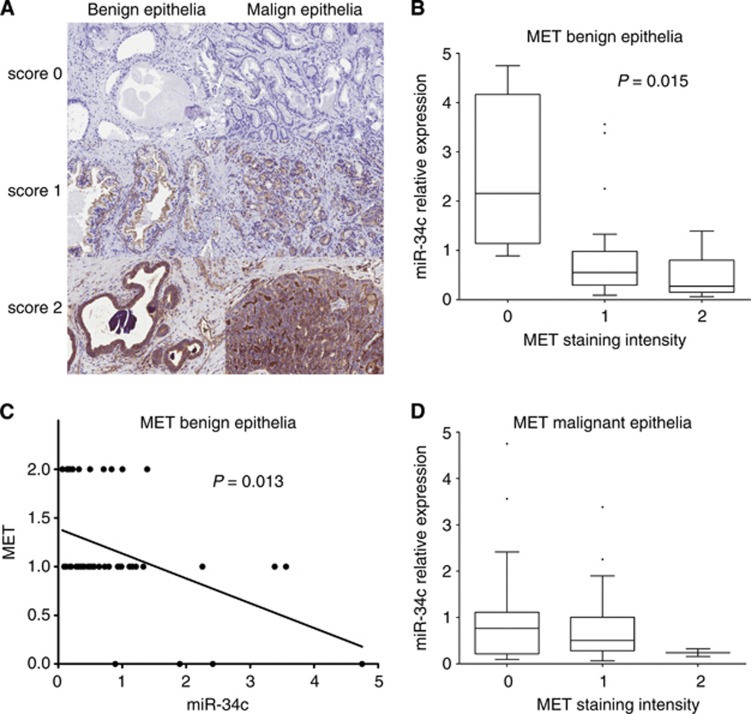
The level of MET in different prostate cell lines was determined by qRT–PCR. The MET expression is the highest in the highly metastatic and aggressive AR-negative cell lines PC3 and DU145, whereas the expression of MET was found to be the lowest in the AR-positive cell lines LNCaP and 22Rv1, see Figure 3B, in accordance with previous results (Humphrey et al, 1995; Knudsen et al, 2002; Verras et al, 2007), indicative of AR signalling repressing MET levels. We also analysed the expression of MET by immunohistochemical analyses using an antibody against MET in prostatic tissue sections from a PCa cohort (n=47). This resulted in a cytoplasmic or plasma membrane staining of epithelial cells, and no staining in stromal cells. The MET protein content in the benign and malignant epithelium in samples from each PCa patient was scored by overall intensity, see Figure 4A. We found a statistical significant increase of MET protein in patients with CRPC compared with untreated PCa patients or patients undergoing treatment (the benign epithelium P=0.0008, the malignant epithelium P=0.032). Reduced miR-34c expression has previously been linked to PCa aggressiveness and occurrence of metastases, whereas the opposite, increased expression, has been described for MET. Therefore, we analysed whether miR-34c levels determined by qRT–PCR were inversely correlated with MET levels in the PCa cohort. We found a significant inverse correlation between miR-34c expression levels and assessment of the MET immunostaining in the benign epithelium (P=0.0145, 1 way ANOVA; P=0.0126, Spearman), see Figure 4b and c. In the malignant epithelium there was no significant correlation, see Figure 4d.
Figure 4.
In vivo inverse correlation of miR-34c and MET levels in the prostate benign epithelium. (A) The MET protein level was determined by immunostaining and scored by overall intensity (score: 0=no staining; 1=weak; 2=distinct) in the prostatic tissue from 47 prostate cancer patients. (B, C) Significant inverse correlation was observed for miR-34c when comparing MET intensity in the benign epithelium with miR-34c detected with qRT-PCR on an adjacent slide. (D) No significant correlation was observed in malignant epithelium.
Discussion
In this study, we further investigate the mechanisms and implications of miR-34c deregulation in PCa. The analyses of the transcriptome after miR-34c ectopic expression showed that miR-34c targets major oncogenic pathways such as apoptosis, proliferation, and migration. These results agree with the effects of miR-34c on proliferation, apoptosis, migration, and invasion found in earlier studies, and with the correlation to clinical parameters such as aggressiveness, occurrence of metastases, and overall survival in PCa patients (Hagman et al, 2010). The analyses of the transcriptome were run in PC3 cells, as they have low levels of miR-34c and as ectopic expression of miR-34c in PC3 cells gives profound phenotypic changes such as increased apoptosis and decreased growth and migration. The effects that miR-34c conveyed on proliferation and apoptosis could be exerted through the identified targets E2F3 and Bcl-2, but the effect on migration has not yet been explained. It was thus interesting to find that HGF signalling was affected by miR-34c. HGF is the ligand of MET, and it is secreted by mesenchymal cells in the stroma to induce the activation of MET in epithelial cells (Birchmeier et al, 2003). MET has been described as a miR-34c target in other cellular settings (Migliore et al, 2008) and is known to promote motility and invasive capabilities of tumour cells.
To our knowledge, this is the first time miR-34c is shown to target MET in PCa cells. Our results clearly show that miR-34c targets MET in vitro, as both ectopic expression and blocking of miR-34c effect MET levels, and possibly in vivo as well. Our data further strongly suggest that the binding to MET in PCa cells is direct, as the effect can be counteracted by mutating either of the miR-34c predicted binding sites. There are three other miRNAs miR-34a, 449a and b that are predicted to bind to the same binding site according to the TargetScan. It is possible that miR-34c affects levels of these other miRNAs that subsequently targets MET; however, such feedback loops have not been reported and is unlikely to occur within the 24 h time frame used. Increased levels of MET as a consequence of alleviated miR-34c repression could explain the motile and invasive phenotype of miR-34c-depleted cells and the inverse correlation to metastases in patients those we previously reported (Hagman et al, 2010). By a gain-of-function experiment, we show that the migratory potential increased when overexpressing MET as could be expected. The migration induced by MET was significantly reduced by simultaneous ectopic expression of miR-34c, indicating that MET is an important target for the effect miR-34c has on migration.
It is likely that miR-34c and MET are connected by a positive feedback loop. MET triggers the activation of PI3K/AKT, which increases mTOR activation, which in turn inhibits p53 (by promoting the translation of Mdm2) (Moumen et al, 2007). As p53 activates the expression of miR-34c (He et al, 2007), the circle is closed by miR-34c targeting MET. This sheds light on the past work showing that MET expression is increased in cells devoid of p53 (Rong and Vande Woude, 1994). In the prostate cell line, the highest MET expression was seen in PC3 and DU145 that both contain mutated p53, corroborating this theory. It was recently reported that MET-dependent cell motility and invasion are controlled by p53, acting by two mechanisms: activation of miR-34 family genes (Migliore et al, 2008) and inhibition of Sp1 binding to the promoter of MET (Hwang et al, 2011).
To investigate the miR-34c/MET axis, we stained for MET expression in a cohort of 47 PCa patients, and found an inverse correlation to miR-34c in the benign parts of tissues, but in the malignant parts the trend seen was not statistically significant. The difference in the benign and malignant parts could be explained by the low level of miR-34c in the malignant parts and thus a reduced dynamic range or the higher frequency of p53 mutations and thus decreased positive feedback as described above. Another contributing factor might be target competition (Ebert et al, 2007; Poliseno et al, 2010; Salmena et al, 2011) between the hereby confirmed miR-34c target MET and previously confirmed AR (Ostling et al, 2011) in PCa cells. As the levels of AR transcripts are increased in malignant tissues (Hobisch et al, 1995; Ostling et al, 2011) and the miR-34c levels are decreased, the repressive effect miR-34c is having on MET might be relieved through targets competition. It has also been suggested that MET is repressed by androgens through AR (Verras et al, 2007). In the prostate cell lines, MET expression inversely correlates with active AR signalling; high MET expression in AR-negative cell lines DU145 and PC3, and lower expression in the AR-positive cell lines, in agreement with earlier observations (Humphrey et al, 1995; Knudsen et al, 2002). It has been suggested that AR, as well as p53, regulates the transcription of MET through the transcription factor Sp1 (Verras et al, 2007).
Currently, androgen deprivation therapy is used for advanced PCa, inducing apoptosis and repressing expression of growth promoting genes activated by the AR, with the result of a reduction in tumour size (Huggins and Hodges, 1972; Furuya and Isaacs, 1993). However, it is possible that androgen deprivation alleviates the repressive role of AR signalling in MET expression and hereby contributes to the metastatic potential of these cells. If this is the case, it is conceivable that androgen deprivation combined with therapeutic miR-34c replacement treatment can both delay the CRPC onset and keep MET levels low so that the metastatic potential of the PCa cells are diminished. In conclusion, the link between miR-34c and MET in prostate cancer opens up novel therapeutic opportunities especially for the advanced and currently incurable form of PCa and merits further investigations.
Acknowledgments
We thank Elise Nilsson for preparing prostatic tissue samples. The authors are grateful to Srinivas Veerla and the other staff at SCIBLU Genomics at Lund University, Sweden for carrying out the microarray experiments. We thank professor Livio Trusolino at the University of Turin for the vector containing MET. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n°201438, the Swedish Research council, Gunnar Nilssons Cancer foundation, Jeanssons foundation and Gyllenstiernska Krapperups foundation.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Balk SP.2002Androgen receptor as a target in androgen-independent prostate cancer Urology 60132–138.discussion 138-139. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, Macdougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Catuogno S, Cerchia L, Romano G, Pognonec P, Condorelli G, De Franciscis V. miR-34c may protect lung cancer cells from paclitaxel-induced apoptosis. Oncogene. 2012;32 (3:341–351. doi: 10.1038/onc.2012.51. [DOI] [PubMed] [Google Scholar]
- Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R, Mccarville J, Lee C, Hu W, Kaur G, Carroll P, Deng G. Deletion of chromosome 11p15, p12, q22, q23-24 loci in human prostate cancer. Int J Cancer. 1997;72:283–288. doi: 10.1002/(sici)1097-0215(19970717)72:2<283::aid-ijc14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth RE, Vertrees A, Love B, Hooke JA, Ellsworth DL, Shriver CD. Chromosomal alterations associated with the transition from in situ to invasive breast cancer. Ann Surg Oncol. 2008;15:2519–2525. doi: 10.1245/s10434-008-0051-7. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- Furuya Y, Isaacs JT. Differential gene regulation during programmed death (apoptosis) versus proliferation of prostatic glandular cells induced by androgen manipulation. Endocrinology. 1993;133:2660–2666. doi: 10.1210/endo.133.6.8243289. [DOI] [PubMed] [Google Scholar]
- Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- Hagman Z, Haflidadottir BS, Ceder JA, Larne O, Bjartell A, Lilja H, Edsjo A, Ceder Y. miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br J Cancer. 2013;108 (8:1668–1676. doi: 10.1038/bjc.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman Z, Larne O, Edsjo A, Bjartell A, Ehrnstrom RA, Ulmert D, Lilja H, Ceder Y. miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int J Cancer. 2010;127:2768–2776. doi: 10.1002/ijc.25269. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, De Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobisch A, Culig Z, Radmayr C, Bartsch G, Klocker H, Hittmair A. Distant metastases from prostatic carcinoma express androgen receptor protein. Cancer Res. 1995;55:3068–3072. [PubMed] [Google Scholar]
- Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232–240. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- Humphrey PA, Zhu X, Zarnegar R, Swanson PE, Ratliff TL, Vollmer RT, Day ML. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am J Pathol. 1995;147:386–396. [PMC free article] [PubMed] [Google Scholar]
- Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, Boccaccio C, Thorgeirsson SS, Comoglio PM, Hermeking H, Nikitin AY. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci USA. 2011;108:14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Gmyrek GA, Inra J, Scherr DS, Vaughan ED, Nanus DM, Kattan MW, Gerald WL, Vande Woude GF. High expression of the Met receptor in prostate cancer metastasis to bone. Urology. 2002;60:1113–1117. doi: 10.1016/s0090-4295(02)01954-4. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, Da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liang Y. An expression meta-analysis of predicted microRNA targets identifies a diagnostic signature for lung cancer. BMC Med Genomics. 2008;1:61. doi: 10.1186/1755-8794-1-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sempere LF, Galimberti F, Freemantle SJ, Black C, Dragnev KH, Ma Y, Fiering S, Memoli V, Li H, Direnzo J, Korc M, Cole CN, Bak M, Kauppinen S, Dmitrovsky E. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–1183. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134:1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- Ostling P, Leivonen SK, Aakula A, Kohonen P, Makela R, Hagman Z, Edsjo A, Kangaspeska S, Edgren H, Nicorici D, Bjartell A, Ceder Y, Perala M, Kallioniemi O. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011;71:1956–1967. doi: 10.1158/0008-5472.CAN-10-2421. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong S, Vande Woude GF. Autocrine mechanism for met proto-oncogene tumorigenicity. Cold Spring Harb Symp Quant Biol. 1994;59:629–636. doi: 10.1101/sqb.1994.059.01.072. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- Verras M, Lee J, Xue H, Li TH, Wang Y, Sun Z. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007;67:967–975. doi: 10.1158/0008-5472.CAN-06-3552. [DOI] [PubMed] [Google Scholar]
- Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X, Liu Y, He Y, Park EY, Zhang H, Lv X, Ma K, Su F, Park JH, Song E. MicroRNA 34c gene down-regulation via DNA methylation promotes self-renewal and epithelial-mesenchymal transition in breast tumor-initiating cells. J Biol Chem. 2012;287:465–473. doi: 10.1074/jbc.M111.280768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.