Abstract
Williams–Beuren syndrome (WBS) is a neurodevelopmental disorder with multi-systemic manifestations, caused by a heterozygous segmental deletion of 1.55–1.83 Mb at chromosomal band 7q11.23. The deletion can include the NCF1 gene that encodes the p47phox protein, a component of the leukocyte NADPH oxidase enzyme, which is essential for the defense against microbial pathogens. It has been postulated that WBS patients with two functional NCF1 genes are more susceptible to occurrence of hypertension than WBS patients with only one functional NCF1 gene. We now describe two extremely rare WBS patients without any functional NCF1 gene, because of a mutation in NCF1 on the allele not carrying the NCF1-removing WBS deletion. These two patients suffer from chronic granulomatous disease with increased microbial infections in addition to WBS. Interestingly, one of these patients did suffer from hypertension, indicating that other factors than NADPH oxidase in vascular tissue may be involved in causing hypertension.
Keywords: Williams–Beuren syndrome, chronic granulomatous disease, NADPH oxidase, p47phox, ROS production, Nox
Introduction
Williams–Beuren syndrome (WBS; MIM #194050) is a neurodevelopmental disorder (frequency 1/20 000 births) with multi-system manifestations, including supravalvular aortic stenosis, hypercalcemia in infancy, mild-to-moderate mental retardation and characteristic craniofacial features. WBS is caused by a heterozygous segmental deletion of about 1.5–1.8 Mb at chromosomal band 7q11.23, which includes ELN (coding for elastin (MIM *130160)) located in the center of the deleted region, and about 20 additional genes.1, 2, 3 Hemizygosity at ELN is known to be the cause of the vascular stenoses in many of WBS patients that predispose to hypertension.1 The microdeletion arise through non-allelic homologous recombination between low-copy repeats (LCRs) flanking this region.4 However, implication of genes other than ELN in the deletion or flanking regions on the pathogenesis of this disease is still uncertain.
The NCF1 gene maps to the flanking region of the microdeletions at 7q11.23 and is present in multiple copies;5 the functional NCF1 gene (MIM #608512) is present in the telomeric repeat unit, whereas two nearly identical NCF1 pseudogenes presumably arisen by gene duplication appear at both centromeric and telomeric loci. The NCF1 gene encodes p47phox, one of the essential NADPH oxidase components. This enzyme catalyzes the formation of reactive oxygen species responsible for the microbicidal activity of phagocytes and has an essential role in host defense. The most common autosomal recessive (AR) form of chronic granulomatous disease (CGD; OMIM #233700) is caused by a 2-bp GT deletion at the beginning of exon 2 in NCF1 (about 60% of AR CGD cases).5, 6, 7 This mutation leads to a frameshift and a premature stop codon and consequently to loss of p47phox protein expression. The clinical consequence of this mutation is severe, with life-threatening, recurrent infections in childhood.8 However, p47phox is also expressed in non-phagocytic cells and may be involved in many other diseases, such as cardiovascular disorders.9 Depending on the LCR in which the recombination event leading to the WBS deletion occurs, the NCF1 gene may or may not be deleted in WBS individuals.1, 3 Hemizygosity at the NCF1 locus may protect against hypertension in WBS3 but as such has no effect on the susceptibility for microbial infections, because heterozygotes for AR 470CGD caused by NCF1 mutations are asymptomatic.
In extremely rare inherited situations, the WBS microdeletion is combined with a mutation in NCF1 on the other allele, thus leading to CGD if the WBS deletion includes NCF1. To our knowledge, only two cases of WBS associated with CGD have been reported in the literature, but in both reports the genetic defects were not fully characterized.10, 11
Here we report the complete phenotypic and genotypic characterization of two cases of WBS associated with AR 470CGD. Unlike in other previously reported cases of WBS, in which the size of the microdeletion at 7q11.23 was approximately determined by fluorescence in situ hybridization (FISH), a genome-wide array-comparative genomic hybridization study was used to detect the size of the deletion in both patients. We also identified the carrier status of AR 470CGD in the fathers of both patients. Despite common clinical characteristics of WBS found in both patients, there were some differences especially in the chronology and the severity of the infectious and inflammatory symptoms, as well as in the presence of hypertension.
Materials and methods
Case reports of patient 1 and patient 2
Blood samples were obtained from the CGD patients and their family members with appropriate institutional informed consent.
Patient 1
Patient 1 is a French young adult man born in 1989. At the age of 16 years, he was first referred to our infectious diseases department for treatment of multiple liver abscesses. He was known to have a WBS. He showed characteristic clinical features: broad forehead with bitemporal narrowing, flat midface, long philtrum, thick lips and periorbital fullness with hyperacusis, joint hyperlaxity and global cognitive impairment. FISH analysis revealed a hemizygous deletion at the chromosomal location 7q11.23. At 8 years old, he showed a supravalvular aortic stenosis that was corrected by percutaneous endoluminal dilatation. For the 2 weeks preceding the hospitalization the patient suffered from fever, sweats and weight loss. Hepatic abscesses were confirmed by CT scan. A partial portal venous thrombosis was present. Examination of the liver biopsy samples showed inflammatory granuloma with epithelioid cells, suggesting an intracellular bacterial infection. Bacterial and fungal cultures were negative. Serum antibodies (IgG and IgM) were positive for Coxiella burnetii and IgG for Bartonella henselae. A combination of clindamycin plus doxycycline was given for 3 months, with clinical improvement. Six months later, the hepatic CT scan showed tiny scars without abscesses. Despite the lack of microbiological documentation, the diagnosis of inflammatory granulomatous hepatitis because of C. burnettii was retained and the patient was cured by long duration antibiotics. During the following 3 years patient 1 was well. Thereafter, he was readmitted for similar symptoms and multiple hepatic abscesses. A liver biopsy showed microabscesses surrounding inflammatory granulomata, similar to those of the previous biopsy. Systemic inflammatory reaction was present. CGD was suspected and confirmed by the absence of reactive oxygen species formation by his activated granulocytes. A combination of antibiotics (clindamycin plus ciprofloxacin) associated with antifungal therapy (caspofungin) and gamma interferon was started. During the first weeks of this treatment, clinical status and hepatic abscesses worsened. Improvement occurred when white cell transfusions were given, which were well tolerated without adverse effects. Now patient 1 is well and he only receives prophylactic treatment with itraconazole and co-trimoxazole.
Patient 2
A 3-year-old boy born in the Netherlands in 2004 with a previous diagnosis of WBS was referred for the evaluation of spiking fever and a granulomatous lesion in the liver. His clinical symptoms included a typical facial appearance, pulmonary valve stenosis, nephrocalcinosis and hypercalciuria. The diagnosis of WBS had been confirmed by FISH (RP11-105614), showing a 7q11.23 deletion. The patient suffered from hypertension for which he received hydrochlorothiazide. The medical history further included an S. aureus neck abscess at age 6 months, which was treated with drainage and antibiotic therapy, recurrent bacterial periungual infections and unexplained feeding difficulties for which a percutaneous gastric catheter was placed. The liver granulomata had been biopsied, and were negative for infectious causes. Based on the above-mentioned symptoms, CGD was suspected, which was confirmed by the absence of NADPH oxidase activity in activated granulocytes. Immunosuppressive therapy (prednisone) was started, in combination with broad spectrum antibiotics. Fever disappeared and inflammatory parameters normalized. However, the liver granulomata persisted, and prednisone could not be tapered without recurrence of fever and inflammatory parameters. The patient subsequently underwent hematopoietic stem cell transplantation with 6/6 matched unrelated stem cells from cord blood after myeloablative conditioning with busulfan–fludarabine–anti-thymocyte globulin. Graft versus host disease prophylaxis consisted of prednisone and cyclosporine-A (according to European Bone Marrow Transplantation guidelines). This procedure was only complicated by auto-immune thrombocytopenia (responsive to low-dose steroids). The patient is now 2 years post transplant, off immunosuppressive agents, with 100% donor signal in peripheral blood mononuclear cells, normal granulocyte function and resolution of the liver granulomata. The stem cell transplantation did so far not change any behavioral aspects of WBS. The patient is still taking enalapril and ramipril for his hypertension.
Cell preparations
Human neutrophils and mononuclear cells (lymphocytes plus monocytes) were isolated from 5–15 ml of citrated blood from patients, their parents and healthy volunteers as described.12 Lymphocytes purified by Ficoll–Hypaque density gradient centrifugation were infected with the B95-8 strain of EBV and cultured, as previously described.13
Measurement of NADPH oxidase activity in neutrophils
Measurement of H2O2 production by NADPH oxidase was performed by resorufin (Amplex Red, Life Technologies SAS, Saint-Aubin, France) fluorescence kinetics.14 Human neutrophils (5 × 104 cells per well in a 96-well plate) in PBS containing 0.9 mM CaCl2, 0.5 mM MgCl2, 20 mM glucose, 50 μM resorufin and 10 U/ml horse radish peroxidase were stimulated at 37 °C with 10 ng/ml phorbol myristate acetate, 1 mg/ml opsonized zymosan15 or 1.25 μM platelet-activating factor+formyl methionyl leucyl phenylalanine. The increase in fluorescence caused by resorufin oxidation was recorded at 590 nm every 30 s for 15 min in a fluorometer (Twinkle LB 970 Berthold SA, Thoiry, France) connected to a computer. Fluorescence values at 10–20 min after activation were registered and compared with a calibration curve made with increasing amounts of H2O2. The results were expressed as nmoles of H2O2 produced per minute per 106 neutrophils. The absence of NADPH oxidase activity in the patients' neutrophils was confirmed by the NBT reduction test (data not shown).16
SDS-PAGE and immunoblotting
Solubilized proteins were run on SDS-PAGE,17 electrotransferred to nitrocellulose18 and immunodetected by rabbit polyclonal antipeptide antibodies directed against p47phox.19 P67phox, p22phox and Nox2 were immunodetected by rabbit polyclonal antipeptide antibodies,19 and mouse monoclonal antibodies 449 and 48, respectively. Proteins were detected by the ECL Advance Western Blotting Detection Kit (GE Healthcare, formerly Amersham Biosciences, Piscataway, NJ, USA) or by infrared-based fluorometric detection by the Odyssey system (LI-COR Biosciences, Cambridge, UK).
Gene-scan method for determination of the ratio of NCF1 and ΨNCF1 genes
Genomic DNA was purified with a purification kit (ref A1120 Wizard genomic DNA, Promega Biosciences Inc., San Luis Obispo, CA, USA). Fragments of genomic DNA from AR470CGD patients and their relatives were amplified by PCR with labeled primers that anneal to regions in NCF1 as well as to regions in the two pseudo-NCF1 genes (ΨNCF1) around the GTGT sequence at the start of exon 2. The ratio between the number of NCF1 and ΨNCF1 genes was determined.20
Fluorescence in situ hybridization
FISH analysis was performed on metaphase chromosomes with the LSI WBS (elastin gene) region probe (orange, Abbott Molecular, Rungis, France). A 7q31 telomeric probe (green, Abbott Molecular) was used as a control. FISH probes were denatured for 10 min at 73 °C and slides were incubated for 20 h at 37 °C. The post-hybridization washes were performed according to the manufacturer's instructions (Vysis, Abbott Molecular). Slides were counterstained with DAPI/Vectashield (Vector Laboratories, Peterborough, UK). FISH signals were captured with a fluorescence microscope (M1, Carl Zeiss SAS, Le Pecq, France) equipped with appropriate filters, a charge-coupled device camera, and the FISH imaging software (Isis, Metasystems, Altlussheim, Germany). Twenty metaphases were analyzed per slide.
Array-comparative genomic hybridization (CGH-array)
Array-CGH was performed with an 180 000-oligonucleotide microarray (Human Genome CGH-Microarray Kit 180K, Agilent Technologies, Santa Clara, CA, USA). The average spacing of the probes was 13 kb. DNA from the patient was compared with DNA from two other patients with different diseases, according to the loop model.21 Genomic Workbench software, standard edition 6.5 (Agilent Technologies) was used to interpret the results with the following parameters: aberration algorithm ADM-2, threshold 6.0, fuzzy zero, centralization and moving average window 0.5 and 1 Mb. A copy number variation (CNV) was noted if at least three contiguous oligonucleotides showed an abnormal log2 ratio (>+0.5 or <−0.5 according to the Alexa 5 deviation, red curve) with a mirror image. The Database of Genomic Variants (http://projects.tcag.ca/variation/) was used to compare findings to previously reported studies. Coordinates of CNVs are based on the UCSC GRCh37/hg19 assembly.
Protein determination
The protein content was measured with the Bradford assay.22
Results
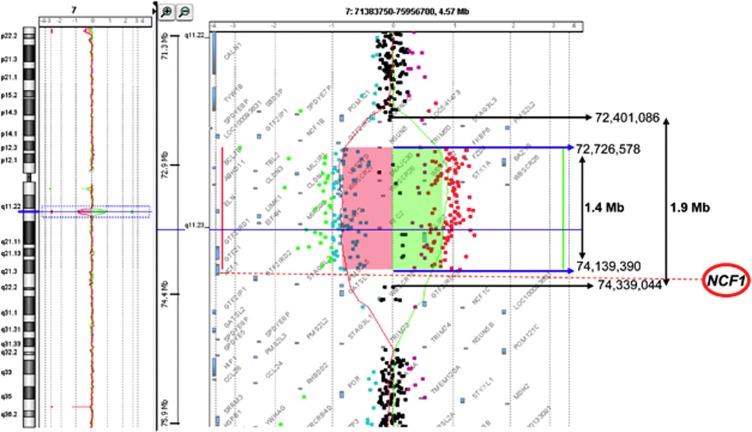
WBS diagnosis was established early in the childhood of both patients based on common clinical features, such as characteristic facial appearance, global cognitive impairment and cardiovascular diseases such as valvular aortic stenosis. However, only patient 2 suffered from hypertension, treated with hydrochlorothiazide. FISH analysis revealed a hemizygous deletion at the chromosomal location 7q11.23 (Figure 1). More precise information about the size of the deletion in chromosome band 7q11.23 of patients 1 and 2 was obtained with the array-CGH approach (Figure 2). The heterozygous deletion showed a similar extension in both patients. The minimum and maximum deletion size ranges from 1.4 (chr7:72 726 578–74 139 390, hg19) to 1.9 Mb (chr7:72 401 086–74 339 044, hg19), depending on the retained breakpoint and the oligonucleotide sequence position (Figure 2). The deletion comprises 24 genes including the ELN, LIMK1, STX1A, RFC2 and GFT2I genes. In both cases, the NCF1 gene is located between the last deleted and the first normal oligonucleotides. The 180 000 oligonucleotide microarray resolution level does not permit to determine more precisely the deletion breakpoint.
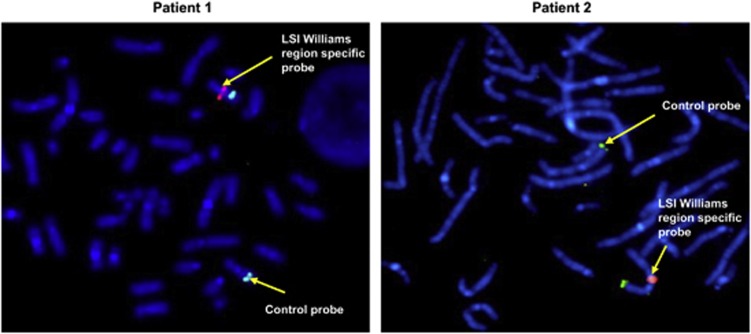
Figure 1.
Characterization of the 7q11.23 microdeletions from patients 1 and 2 by FISH. FISH studies were performed on EBV immortalized B lymphocytes from patient 1 and patient 2 as described in Materials and methods section. LSI William's syndrome (elastin gene) region probe (orange, Abbott Molecular) shows only one signal on the metaphase chromosomes. 7q31 Telomeric probe (green, Abbott Molecular) was used as control. Slides were counterstained with DAPI/Vectashield and FISH signals were captured with an equipped fluorescence microscope.
Figure 2.
CGH-array analysis of the microdeletion in chromosome 7 from patients 1 and 2. CGH-array profile of chromosome 7 shows the same heterozygous deletion in 7q11.23 detected in both patients, varying from 1.4 to 1.9 Mb, depending on the retained breakpoints. Genomic position of the normal centromeric (7:72 401 086) and the normal telomeric oligonucleotides (7:74 339 044) bordering the deletion are indicated with black arrows. Genomic position of the first centromeric deleted (7:72 726 578) and the last telomeric deleted oligonucleotides (7:74 139 390) are indicated with blue arrows. The NCF1 gene (red circle) is located between the last deleted and the first normal oligonucleotides.
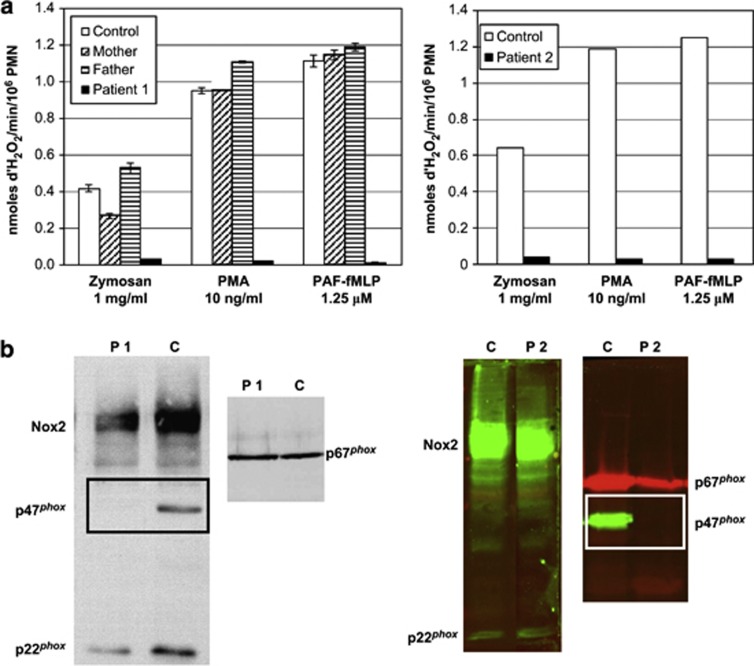
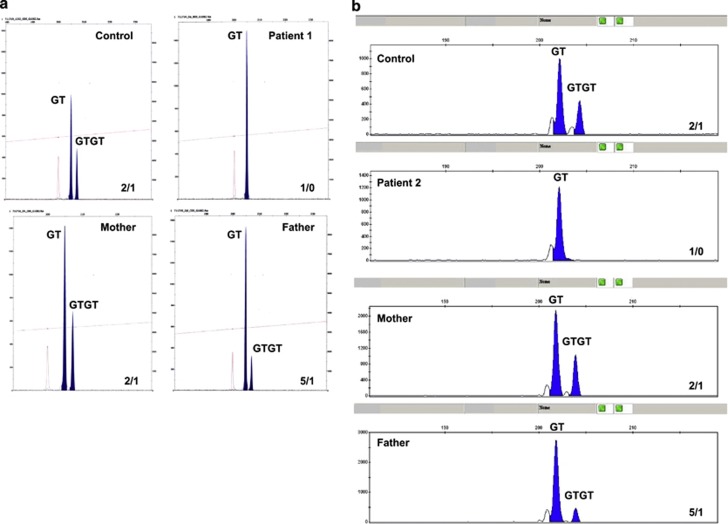
The clinical suspicion of CGD was raised because of liver abscesses in both patients. However, patient 1 was diagnosed with CGD at the age of 16, whereas patient 2 was diagnosed at 3 years of age. The test with resorufin (Amplex Red) demonstrated the absence of NADPH oxidase activity in the phagocytes in both patients. The activity was normal in the parents of patient 1, but we did not test this for the parents of patient 2 (Figure 3a). The absence of NADPH oxidase activity was confirmed by a negative NBT reduction test in the patients (data not shown). The absence of the p47phox protein observed by western blot analysis (Figure 3b) made clear that the hemizygous deletion of the WBS region includes the NCF1 gene and that it must be associated with a mutation in NCF1 in the second allele, leading to AR 470CGD in both patients. The AR 470CGD of the patients appeared to be due to the classical dinucleotide deletion (ΔGT) at a GTGT repeat at the beginning of exon 2 of the NCF1 gene, as shown by the gene-scan method (Figure 4). Detection of AR 470CGD carriers was also determined with the gene-scan method. The ratio of NCF1 pseudogene/NCF1 gene in both fathers of the patients was compatible with a carrier status of the GT deletion on one allele (Figures 4a and b).
Figure 3.
Phenotypic profiles of neutrophils from patients 1 and 2. (a) The absence of NADPH oxidase activity was evidenced in the neutrophils of patients 1 and 2 by resorufin (Amplex Red) fluorescence kinetics as described in Materials and methods section. Parents of patient 1 exhibited normal activity as it is usually the case in AR CGD. (b) The absence of oxidase activity was confirmed by the absence of immunodetection of p47phox expression in both patients, whereas the other oxidase components were present. The experiment was performed on 1% Triton X100 soluble extract of neutrophils subjected to SDS–PAGE and blotted onto nitrocellulose sheet. Proteins were detected by the ECL Advance Western Blotting Detection Kit (GE Healthcare, formerly Amersham Biosciences) for patient 1 and by infrared-based fluorometric detection by the Odyssey system (LI-COR Biosciences, Ltd,) for patient 2.
Figure 4.
Gene-scan analysis of NCF1 and NCF1 pseudogenes (ψNCF1) regions around the beginning of exon2 for patients 1 and 2. Fragments of genomic DNA from AR470CGD patient 1 (a) and patient 2 (b) and their parents were amplified with primers that anneal with regions in NCF1 as well as in ψNCF1 regions around the GTGT sequence at the start of exon 2 as described in Materials and methods section. The mixture of NCF1 and ψNCF1 PCR products was analyzed in a sequencer (Applied Biosystems, Foster City, CA, USA) to determine the ratio between the number of NCF1 and ψNCF1 genes. Both patients show only ψNCF1 PCR product; both fathers show a ψNCF1:NCF1 product ratio of 5; both mothers and both controls show a ψNCF1:NCF1 product ratio of 2.
Discussion
The array-CGH analysis performed in our patients highlights the fact that the NCF1 gene is not covered by any of the 180 000 oligonucleotides spotted on the microarray in spite of its involvement in a human inherited disease. No NCF1-specific probes are currently available on microarray used routinely in most French medical genetics laboratories. This can probably be explained by the fact that NCF1 has two nearly identical (>99.5%) pseudogenes (NCF1B and NCF1C). Therefore, in our patients, the distal breakpoint of the deletion could not be determined more precisely by array-CGH. However, the clinical features, the biochemical analysis and the AR inheritance of CGD, led us to conclude that the deletion encompasses totally or partially the NCF1 gene.
WBS has prevalence in newborns between 1/7500 and 1/20 000.3 Carriers of AR 470CGD have an estimated prevalence of 1/500, because AR 470CGD patients make up about 25% of all CGD patients, and CGD in total occurs in about 1/250 000 newborns.7, 8 Therefore, the combination of WBS and AR 470CGD may be expected in about 1/15 000 × 1/500 × 1/2=1/15 000 000 newborns. Indeed, this combination is an extremely rare event, with only two reports in the literature. Kabuki et al10 determined WBS syndrome in a 2-month-old boy by FISH analysis and CGD by NADPH oxidase activity, western blotting and DNA analysis. However, this article is in Japanese, with only an abstract in English. Gilbert-Barness et al11 described a 20-year-old female patient diagnosed with WBS by FISH analysis, suffering from abdominal pain, diarrhea and a dry, hacking cough. Colon biopsies showed focal active colitis and granulomata; lung biopsy showed granulomatous inflammatory changes with fungal hyphae identified by microbiological culture as aspergillus infection. From these last observations, the conclusion was drawn that the patient had CGD in addition to WBS. Extensive laboratory studies did not mention hypertension.
This last observation is of special interest, because about half of the WBS patients described suffer from hypertension as demonstrated.3 This is supposed to be due to haploinsufficiency at the elastin gene, in the middle of all WBS deletions, leading to structural changes in vessel walls mediated by endothelial cell proliferation and smooth-muscle reorganization, resulting in decreased vascular lumen. This in turn may lead to angiotensin elevation and hypertension. Del Campo et al3 observed that WBS patients with two functional NCF1 genes (NCF1 not included in the WBS deletion) cluster in the group of patients with hypertension, whereas WBS patients with one functional NCF1 gene (NCF1 included in the WBS deletion) are overrepresented in the group with normal blood pressure. These authors explain their results by an activating action of angiotensin II on an NADPH oxidase present in vascular tissue, of which p47phox may be a component.9 The resulting increase in superoxide production and concomitant decrease in nitric oxide levels (through dissipation by superoxide and production of tissue-damaging peroxynitrite) will result in hypertension. This process may be limited by the expression of p47phox. This concept was recently confirmed in a mouse model.23
One would expect, therefore, that in WBS patients without any p47phox expression, such as the two patients described in this study, hypertension would not occur. However, one of our two patients did suffer from hypertension, for which he is treated since its diagnosis by hydrochlorothiazide, enalapril and ramipril. Therefore, we must assume that this angiotensin-mediated increase in vascular NADPH oxidase activity cannot be the only or decisive cause of hypertension. That was already indicated by the fact that in Del Campo's study3 also some WBS patients with only one functional NCF1 gene did suffer from hypertension. Clearly, further studies are needed for unraveling the exact mechanism of hypertension induction in WBS. Genetic comparison between the two patients described in this study may be helpful in this respect.
Acknowledgments
MJS is grateful for support from the University Joseph Fourier, Faculty of Medicine; the Ministry of Education and Research, MENRT; the Regional Clinical Research Department, DRCI, Grenoble University Hospital, the CGD Research Trust grant award reference J4G/09/09. DR is co-recipient of an E-Rare research grant on CGD from the European Union.
The authors declare no conflict of interest.
References
- Francke U. Williams-Beuren syndrome: genes and mechanisms. Hum Mol Genet. 1999;8:1947–1954. doi: 10.1093/hmg/8.10.1947. [DOI] [PubMed] [Google Scholar]
- Perez Jurado AL. Williams-Beuren syndrome: a model of recurrent genomic mutation. Horm Res. 2003;59 (Suppl 1:106–113. doi: 10.1159/000067836. [DOI] [PubMed] [Google Scholar]
- Del Campo M, Antonell A, Magano LF, et al. Hemizygosity at the NCF1 gene in patients with Williams-Beuren syndrome decreases their risk of hypertension. Am J Hum Genet. 2006;78:533–542. doi: 10.1086/501073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer A, Dutly F, Balmer D, et al. High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum Mol Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- Gorlach A, Lee PL, Roesler J, et al. A p47-phox pseudogene carries the most common mutation causing p47-phox- deficient chronic granulomatous disease. J Clin Invest. 1997;100:1907–1918. doi: 10.1172/JCI119721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimir CM, Bu-Ghanim HN, Rodaway AR, Bentley DL, Rowe P, Segal AW. Autosomal recessive chronic granulomatous disease caused by deletion at a dinucleotide repeat. Proc Natl Acad Sci USA. 1991;88:2753–2757. doi: 10.1073/pnas.88.7.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos D, Kuhns DB, Maddalena A, et al. Hematologically important mutations: The autosomal recessive forms of chronic granulomatous disease (second update) Blood Cells Mol Dis. 2010;44:291–299. doi: 10.1016/j.bcmd.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg JM, van Koppen E, Ahlin A, et al. Chronic granulomatous disease: the European experience. PLoS One. 2009;4:e5234. doi: 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassegue B, San Martin A, Griendling KK. Biochemistry, physiology, and pathophysiology of NADPH oxidases in the cardiovascular system. Circ Res. 2012;110:1364–1390. doi: 10.1161/CIRCRESAHA.111.243972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuki T, Kawai T, Kin Y, et al. A case of Williams syndrome with p47-phox-deficient chronic granulomatous disease. Nihon Rinsho Meneki Gakkai Kaishi. 2003;26:299–303. doi: 10.2177/jsci.26.299. [DOI] [PubMed] [Google Scholar]
- Gilbert-Barness E, Fox T, Morrow G, Luquette M, Pomerance HH. Williams syndrome associated with Crohn disease, multiple infections, and chronic granulomatous disease. Fetal Pediatr Pathol. 2004;23:29–37. doi: 10.1080/15227950490423016. [DOI] [PubMed] [Google Scholar]
- Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest. 1968;9 (Suppl:77–89. [PubMed] [Google Scholar]
- Cohen-Tanugi L, Morel F, Pilloud-Dagher MC, et al. Activation of O2(-)-generating oxidase in an heterologous cell-free system derived from Epstein-Barr-virus-transformed human B lymphocytes and bovine neutrophils. Application to the study of defects in cytosolic factors in chronic granulomatous disease. Eur J Biochem. 1991;202:649–655. doi: 10.1111/j.1432-1033.1991.tb16419.x. [DOI] [PubMed] [Google Scholar]
- Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253:62–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Suzuki K, Nakaji S, Sugawara K. Analysis and assessment of the capacity of neutrophils to produce reactive oxygen species in a 96-well microplate format using lucigenin- and luminol-dependent chemiluminescence. J Immunol Methods. 1997;210:1–10. doi: 10.1016/s0022-1759(97)00159-2. [DOI] [PubMed] [Google Scholar]
- Stasia MJ, Brion JP, Boutonnat J, Morel F. Severe clinical forms of cytochrome b-negative chronic granulomatous disease (X91-) in 3 brothers with a point mutation in the promoter region of CYBB. J Infect Dis. 2003;188:1593–1604. doi: 10.1086/379035. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnaud S, Paclet MH, El Benna J, Pocidalo MA, Morel F. Complementation of NADPH oxidase in p67-phox-deficient CGD patients p67-phox/p40-phox interaction. Eur J Biochem. 2000;267:1059–1067. doi: 10.1046/j.1432-1327.2000.01097.x. [DOI] [PubMed] [Google Scholar]
- Dekker J, de Boer M, Roos D. Gene-scan method for the recognition of carriers and patients with p47(phox)-deficient autosomal recessive chronic granulomatous disease. Exp Hematol. 2001;29:1319–1325. doi: 10.1016/s0301-472x(01)00731-7. [DOI] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Segura-Puimedon M, Terrado V, et al. Reduction of NADPH-oxidase activity ameliorates the cardiovascular phenotype in a mouse model of Williams-Beuren Syndrome. PLoS Genet. 2009;8:e1002458. doi: 10.1371/journal.pgen.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]