Abstract
Huntington disease (HD) is a neurodegenerative disorder resulting from the expansion of a CAG trinucleotide repeat in the huntingtin (HTT) gene. Worldwide prevalence varies geographically with the highest figures reported in populations of European ancestry. HD in South Africa has been reported in Caucasian, black and mixed subpopulations, with similar estimated prevalence in the Caucasian and mixed groups and a lower estimate in the black subpopulation. Recent studies have associated specific HTT haplotypes with HD in distinct populations. Expanded HD alleles in Europe occur predominantly on haplogroup A (specifically high-risk variants A1/A2), whereas in East Asian populations, HD alleles are associated with haplogroup C. Whether specific HTT haplotypes associate with HD in black Africans and how these compare with haplotypes found in European and East Asian populations remains unknown. The current study genotyped the HTT region in unaffected individuals and HD patients from each of the South African subpopulations, and haplotypes were constructed. CAG repeat sizes were determined and phased to haplotype. Results indicate that HD alleles from Caucasian and mixed patients are predominantly associated with haplogroup A, signifying a similar European origin for HD. However, in black patients, HD occurs predominantly on haplogroup B, suggesting several distinct origins of the mutation in South Africa. The absence of high-risk variants (A1/A2) in the black subpopulation may also explain the reported low prevalence of HD. Identification of haplotypes associated with HD-expanded alleles is particularly relevant to the development of population-specific therapeutic targets for selective suppression of the expanded HTT transcript.
Keywords: Huntington disease, South Africa, haplotypes, haplogroups, prevalence, CAG expansion
Introduction
The neurodegenerative disorder Huntington disease (HD) results from the expansion of a CAG trinucleotide repeat in the huntingtin gene (HTT). The prevalence of HD varies geographically with the highest rates reported in European populations at approximately 5–7 affected individuals per 100 000 and significantly lower rates in Asian and African populations.1, 2 The average CAG-tract size in unaffected individuals varies between 17–20 CAG repeats across populations, whereas a CAG tract of 36 or more repeats is within the affected range.1, 3 Despite extensive characterisation of the main genetic defect underlying HD, which is identical across populations, differences in worldwide prevalence are not fully understood.
The age of onset of disease is inversely related to the size of the CAG tract, with longer tracts correlating to earlier onset.4 The CAG repeat exhibits instability and is prone to expansion particularly when transmitted on paternal alleles. Expansion of HD alleles thus tends to result in earlier disease onset in subsequent generations, known as anticipation. Intermediate alleles (IAs), also known as large normal alleles, are between 27 and 35 CAG repeats and may give rise to de novo expansions on transmission.4, 5, 6, 7, 8 Individuals with IAs will not develop HD, however, their offspring are at risk of inheriting a CAG tract that has expanded into the pathogenic range. Although the size of the CAG tract contributes to instability and is associated with age of onset, there is significant phenotypic variation among individuals with identical repeat lengths.1, 9
Various other factors, including non-CAG genetic modifiers, have been suggested to have a role in expansion and the age of onset of disease symptoms.5, 7, 10 Several studies have attempted to make use of genome-wide analyses in order to identify genetic modifiers and better understand HD aetiology and pathogenesis.11, 12, 13 A number of processes were implicated in early disease pathogenesis; however, specific genetic modifiers are still under investigation.14 Support for the hypothesis that genetic background may modulate the CAG mutation rate has recently been shown at a population level.15
In a 2011 study by Warby et al.15, differences in HD prevalence rates reported in European and Asian populations were associated with different haplotype distributions of disease alleles. The genetic diversity in the HTT gene was delineated using a subset of tagging SNPs (tSNPs) associated with disease-causing expanded alleles in the European population.16 In particular, haplogroup A variants A1 and A2 were strongly associated with HD alleles in individuals of European ancestry, whereas variants A4 and A5 were nearly absent from expanded alleles. The presence of high-risk variants A1 and A2 in the general population led the authors to suggest a step-wise model for the occurrence of de novo mutations, with expansions arising from a pool of alleles in the general population that were predisposed to expansion.16 These high-risk variants were absent from an East Asian cohort, consistent with the hypothesis that HD prevalence could be explained by geographical differences in HTT haplotypes.15
No such studies have yet been performed in South Africa where the prevalence of HD has been reported to differ significantly between different population groups.17 The largest proportion of the South African population consists of black, Caucasian (or white) and mixed (or coloured) peoples (www.statssa.gov.za). For purposes of this manuscript, these groups are referred to as subpopulations and are defined as outlined. The term ‘mixed' is used to describe a dynamic, historically distinct group with significant genetic contributions from Khoisan- and Bantu-speaking peoples, in addition to smaller contributions from Europe and Asia. Black refers to African Bantu-speaking peoples, whereas Caucasian refers to descendants of European settlers.
Similar prevalence estimates have been reported for the Caucasian and mixed subpopulations (2.22 and 2.17 per 100 000, respectively), whereas the prevalence in black South Africans was estimated at 0.01 per 100 000.17 The genetic background underlying HD in these subpopulations is unknown. Furthermore the association of specific variants with HD CAG-expanded alleles is essential for the development of population-specific allele-silencing techniques. This study therefore undertook an investigation of HTT haplotypes in individuals from the different subpopulations in South Africa.
Materials and methods
Cohort
Genetic diagnostic testing for HD has been available in South Africa since the mid 1980s and is currently offered in the public domain by the National Health Laboratory Service (NHLS) in Cape Town (Western Cape province) and Johannesburg (Gauteng province). Blood and DNA samples were originally collected from HD patients and their families for diagnostic testing and stored by the Divisions of Human Genetics at both the NHLS/University of the Witwatersrand (Wits), Johannesburg and at the NHLS/University of Cape Town (UCT), Cape Town. Patient information was recorded on databases at the two centres. Selection criteria for this study included known subpopulation (indicated by the individual) and disease status (determined by sizing the CAG repeat). Unrelated general population control samples consisted of specific unaffected HD family members as well as permanently de-identified archived South African control cohorts stored at the two centres (NHLS/UCT, NHLS/Wits) and in the HD BioBank at the University of British Columbia (UBC). The study was approved by the Human Research Ethics Committees of the different centres and renewed annually (UCT: HREC REF: 450/2010; Wits: M110443/M10745; and UBC: UBC-CREB H05-70532 and H06-70467).
CAG and CCG repeat sizing
CAG and CCG repeat sizes were determined as previously described3, 18 using fluorescently labelled primers flanking the CAG (HD344F, 5′-HEX-CCTTCGAGTCCCTCAAGTCCTTC-3′ and HD450R, 5′-GGCGGCGGTGGCGGCTGTTG-3′) and CCG (HD419F, 5′-AGCAGCAGCAGCAACAGCC-3′ and HD482R, 5′-6FAM-GGCTGAGGAAGCTGAGGAG-3′) repeats. A third PCR encompassing both CAG and CCG sequences (HD344F, 5′-HEX-CCTTCGAGTCCCTCAAGTCCTTC-3′ and HD482R, 5′-GGCTGAGGAAGCTGAGGAG-3′) was used to phase CAG and CCG sizes from the first two assays. Sizing was performed relative to a control panel of sequenced CAG and CCG repeat lengths. A total of 660 alleles representing the general population of South Africa and 128 expanded alleles from presumed unrelated HD patients were sized.
SNP genotyping
Genotyping was performed at 96 SNP positions across the HTT gene using a customised Illumina GoldenGate assay on the BeadArray platform (Illumina, San Diego, CA, USA), as previously described.16 Raw fluorescence output was analysed using Illumina GenomeStudio software (Illumina) to assign SNP genotype calls to each individual. For a small number of SNPs, automated clusters were manually adjusted to improve call accuracy. Of the 96 SNPs, 3 demonstrated no clear clustering and were excluded from downstream haplotype construction.
Phasing and haplogroup assignment
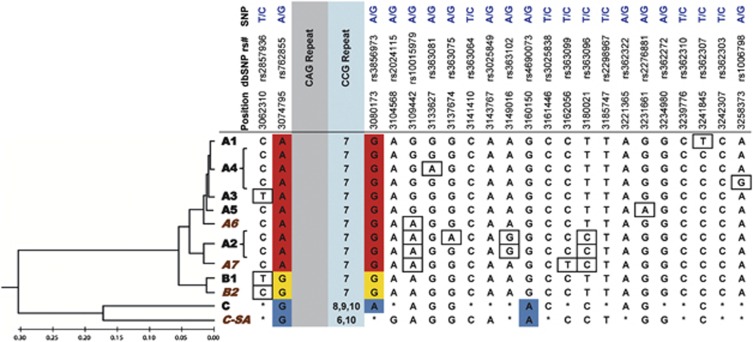
Using PHASE (v2.1),19 SNP genotypes were assigned to complete haplotypes based on a Bayesian inference model. Where possible, haplotypes were phased to an individual CAG-tract size based on segregation of alleles within familial trios (i.e., parents and offspring or sibships). In the absence of familial phasing, haplotypes were phased to CAG size where possible by CCG repeat size associations with specific haplotypes (Figure 3). To control against bias created by duplications of alleles in multiple family members, haplotype frequencies were calculated from total numbers of presumed unrelated chromosomes with >35 CAG repeats (N=72) and <35 CAG repeats (N=311). HD pedigree sizes ranged between 2 and 5 generations, with the average being 3.4 generations. ‘Unrelated' here refers to alleles that, to the best of our knowledge, do not share identical haplotypes because of familial relationships. Haplogroups were assigned based on 21 previously described tSNPs derived from a European population.15 Previously unknown variants were defined in part by the addition of tSNP rs10015979, reflecting a linkage disequilibrium block not captured by the initial 21 tSNP panel (Figure 3). African variants A6 and A7 were added to haplogroup A, whereas haplogroups B and C were sub-divided into B1 and B2, and C and C-South Africa (C-SA), respectively. Note that C-SA is a novel variant encompassing haplotypes that were not previously identified in European or Asian populations, whereas C comprises haplotypes that could be similarly clustered under haplogroup C.
Statistical analysis
Statistical analysis was performed using GraphPad Prism or ‘R' software (www.r-project.org). Statistical significance is reported by P-values and indicated with non-significant (n.s.) at P≥0.05. Mean CAG-repeat sizes were compared between subpopulations by one-way ANOVA, using Tukey's test for post-hoc analysis. Odds ratios were calculated to examine association of haplotype variants with expanded HD alleles in each subpopulation, and Fisher's exact test was used to ascertain significance.
Results
CAG repeat distributions differ significantly across South African subpopulations
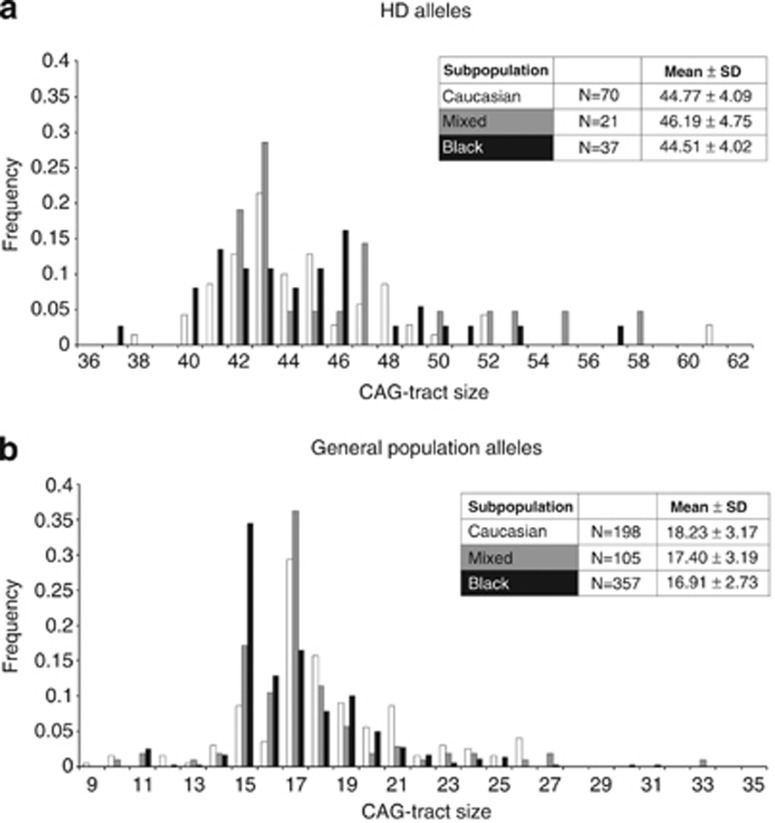
CAG-tract sizes were determined for a total of 660 general population alleles and 128 CAG-expanded HD alleles from the different subpopulations (Figure 1). There were no significant differences seen in this cohort between the mean CAG-tract sizes of expanded HD alleles from the different subpopulations (Figure 1a). In the general population, the mean CAG in black South Africans (16.91±2.73) is significantly lower than the mean CAG in the Caucasian group (18.23±3.17; P=3.673E-06 one-way ANOVA with Tukey post-hoc). Mean CAG size in the mixed subpopulation is intermediate between blacks and Caucasians (17.40±3.19), but does not significantly differ from either the Caucasian or the black subpopulations signifying admixture in the mixed group (95% CI black: 16.63–17.19 versus 95% CI Caucasian: 17.78–18.67 and 95% CI mixed: 16.78–18.02). The most frequent allele in both the Caucasian and mixed subpopulations has 17 repeats, whereas in the black subpopulation, the most frequent allele has 15 repeats (Figure 1b).
Figure 1.
Distribution of CAG-tract sizes in the South African population. (a) CAG-tract sizes of 128 unrelated HD alleles. There are no significant differences in mean HD CAG size between the different subpopulations (P=0.308, one-way ANOVA).(b) CAG-tract sizes of 660 unrelated general population alleles. The mean CAG in black South Africans is significantly lower than that in the Caucasian subpopulation (P=3.673E-06, one-way ANOVA with Tukey post-hoc; 95% CI black: 16.63–17.19 versus 95% CI Caucasian: 17.78–18.67). Confidence intervals for mean CAG in the mixed subpopulation overlap Caucasian and black subpopulations (95% CI mixed: 16.78–18.02). The most frequent CAG-tract size is 17 repeats in the Caucasian and mixed subpopulations, but 15 repeats in the black subpopulation. Note that 15 repeats is the second most common CAG size in the mixed subpopulation.
Haplogroup variants are diverse and ethnically specific to South African subpopulations
Phased SNP genotypes for each individual were assigned to one of the three primary haplogroups based on previously published criteria.16 A total of 72 unrelated HD alleles and 311 unrelated general population alleles were phased and could be broadly categorised into A, B and C haplogroups. Five phased haplotypes were left unassigned as ‘other' (O).
HD haplogroups and variants
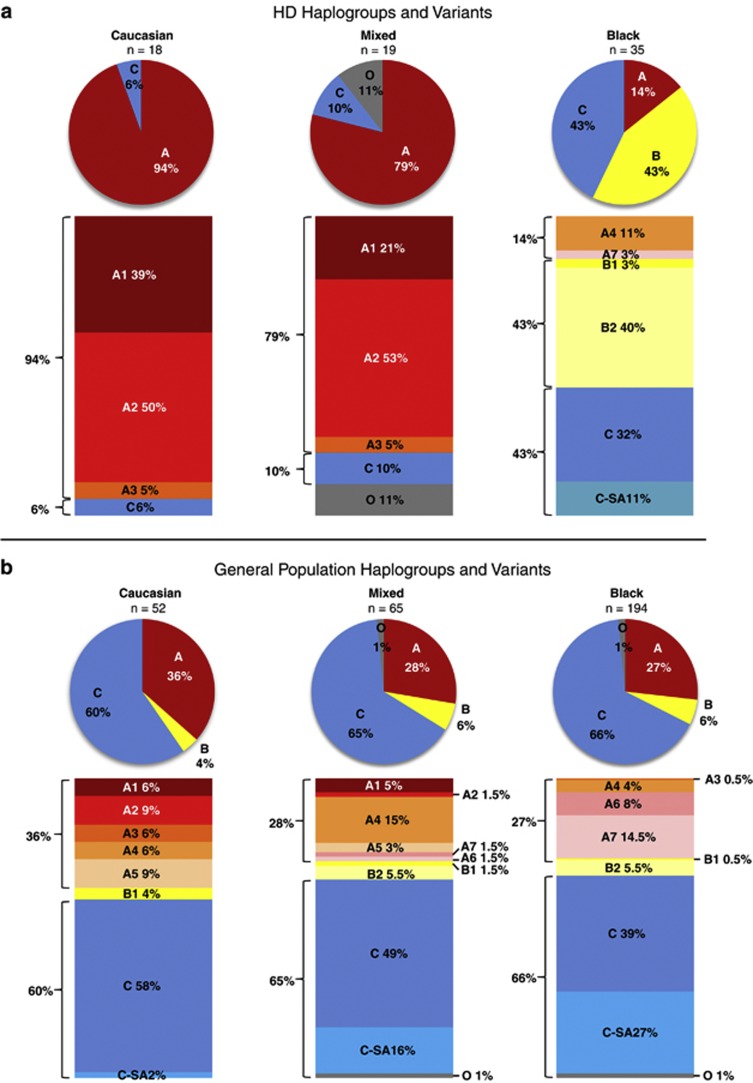
CAG-expanded HD alleles from the Caucasian subpopulation (N=18) were found almost exclusively on haplogroup A (94%), the largest proportion consisting of variants A1 (39%) and A2 (50%) (Figure 2a). In the mixed subpopulation (N=19), CAG-expanded HD alleles occurred predominantly on haplogroup A (79%) with a small proportion on haplogroup C (10%). Similar to the Caucasian subpopulation, the largest proportion of haplogroup A comprised variants A1 (21%) and A2 (53%) (Figure 2a). Apart from the unassigned alleles (N=2) in the mixed subpopulation, all the phased HD alleles in the Caucasian and mixed groups fit previously defined haplogroup variants.16 In stark contrast, CAG-expanded HD alleles in the black subpopulation (N=35) were found predominantly on haplogroup B (43%); with an equal proportion on haplogroup C (43%) and only a small proportion on haplogroup A (14%) (Figure 2a). The most common haplogroup A variants (A1 and A2) in the Caucasian and mixed subpopulations were absent from the black subpopulation, and the small proportion of HD alleles on haplogroup A were found instead on variant A4 (11%) and novel variant A7 (3%) (Figure 2a). HD alleles in the black subpopulation occurred predominantly on novel variant B2 (40%). In addition within haplogroup C, 11% of the alleles occurred on novel variant C-SA (Figure 2a).
Figure 2.
Haplogroup variants on phased, unrelated alleles from the South African population. (a) HD haplogroups and variants (N=72). HD alleles in the Caucasian subpopulation occur almost exclusively on haplogroup A (94%), predominantly variants A1 and A2 (39% and 50%, respectively). In the mixed subpopulation, the largest proportion of HD alleles also occurs on haplogroup A (79%). Similar to the Caucasian subpopulation, variants A1 and A2 (21% and 53%, respectively) predominate. In the black subpopulation, haplogroup A accounts for only a small proportion of HD alleles (14%), with A1 and A2 absent, whereas the largest proportion occurs on novel variant B2 (40%). (b) General population haplogroups and variants (N=311). In all three subpopulations, the largest proportion of general population alleles occurs on haplogroup C. Haplogroup A in the Caucasian subpopulation accounts for 36% of general population alleles, with variants A1 and A2 present at 6% and 9%, respectively. In the mixed subpopulation 28% of general population alleles occur on haplogroup A, but variant A4 predominates (15%). Haplogroup A also occurs in the black subpopulation (27%) but with a markedly different distribution consisting of A4 and novel variants A6 and A7. A large proportion of all the general population haplotypes in the black subpopulation can be additionally grouped into South African group C-SA (27%), consisting of C haplotypes not previously identified in Caucasian or East Asian populations. Mixed general population haplotypes thus represent an admixture of variants observed in Caucasian and black subpopulations.
General population haplogroups and variants
In the general population, the largest proportion of alleles occurred on haplogroup C in all three groups (Caucasian=60%, mixed=65% and black=66%) (Figure 2b). Furthermore, a small proportion of alleles from each subpopulation was found on haplogroup B (Caucasian=4%, mixed=7% and black=6%). In the Caucasian subpopulation, 36% of all general population alleles (N=52) were found on haplogroup A, of which variants A1 and A2 comprised 6% and 9%, respectively. In the mixed subpopulation (N=65), haplogroup A made up 28% of the alleles, with 5% and 1.5% on variants A1 and A2, respectively (Figure 2b). In the black subpopulation (N=194), variants A1 and A2 are absent from the general population. Variant B2, predominant in the CAG-expanded HD alleles from black patients (Figure 2a), accounts for a very small proportion (5.5%) in the general population (Figure 2b). It is noteworthy that overall, the largest proportion of alleles from the black subpopulation occurred on novel variants A6, A7, B2 and C-SA (totalling up to 55% of all black general population alleles). The mixed subpopulation encompasses the greatest diversity of haplotype variants, as expected, given that this group arose from the admixture of several ethnic groups. Nearly all defined variants, except for A3, are present in the mixed subpopulation (Figure 2b).
Novel variants
Genetic diversity in South Africa is reflected by the presence of haplogroup variants not previously identified in European or East Asian populations (Figure 3). Definitions of haplotypes that were previously determined are shown alongside novel variants. The variants are arranged by relationship in a neighbour-joining tree constructed from concatenated 93-SNP sequences. Concatenated tSNP sequences closely approximate this phylogeny (Figure 3). Table 1 provides haplotype counts for each subpopulation for both CAG-expanded HD alleles and alleles from the general population. In addition, the calculated odds ratios and significant differences in haplotype frequency between HD and general population alleles in each group (P-value<0.05, Fisher's exact test) are shown.
Figure 3.
Definitions of HTT haplogroup variants using 22 tSNPs spanning the gene region. Red, yellow and blue SNP genotypes near the CAG repeat define A, B and C haplogroups, respectively. Boxed SNP genotypes represent variant-specific defining SNPs within haplogroups. Asterisks (*) represent variable SNP alleles within haplogroups C and C-SA. Haplogroup variants are grouped by 93-SNP sequence similarity in a neighbour-joining tree, with branch distances drawn to a constant rate of evolution in units of base substitutions per site. Tree was constructed using the Maximum Composite Likelihood method in MEGA5. Haplogroup variants in brown italics indicate those unique to the South African population.
Table 1. HD and general population haplotype counts for all populations, with calculated odds ratios and Fisher's exact test significance values.
| |
Caucasian |
Mixed |
Black |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HD | GP | OR | P-value | HD | GP | OR | P-value | HD | GP | OR | P-value | |
| A1 | 39% (7) | 6% (3) | 10.39 | 0.0019 | 21% (4) | 5% (3) | 5.51 | 0.0431 | — (0) | — (0) | Not present | |
| A2 | 50% (9) | 9% (5) | 9.4 | 0.0007 | 53% (10) | 1.5% (1) | 71.11 | 3.27E-07 | — (0) | — (0) | Not present | |
| A3 | 5% (1) | 6% (3) | 0.96 | 1.0000 | 5% (1) | — (0) | n/a | 0.2262 | — (0) | 0.5% (1) | 0.00 | 1.0000 |
| A4 | — (0) | 6% (3) | 0.00 | 0.5640 | — (0) | 15% (10) | 0.00 | 0.1071 | 11% (4) | 4% (8) | 3.00 | 0.0919 |
| A5 | — (0) | 9% (5) | 0.00 | 0.3180 | — (0) | 3% (2) | 0.00 | 1.0000 | — (0) | — (0) | Not present | |
| A6 | — (0) | — (0) | Not present | — (0) | 1.5% (1) | 0.00 | 1.0000 | — (0) | 8% (15) | 0.00 | 0.1357 | |
| A7 | — (0) | — (0) | Not present | — (0) | 1.5% (1) | 0.00 | 1.0000 | 3% (1) | 14.5% (28) | 0.17 | 0.0923 | |
| B1 | — (0) | 4% (2) | 0.00 | 1.0000 | — (0) | 1.5% (1) | 0.00 | 1.0000 | 3% (1) | 0.5% (1) | 5.58 | 0.2829 |
| B2 | — (0) | — (0) | Not present | — (0) | 5% (3) | 0.00 | 1.0000 | 40.0% (14) | 5% (10) | 12.27 | 2.06E-07 | |
| C | 6% (1) | 58% (30) | 0.04 | 8.09E-05 | 10% (2) | 49% (32) | 0.12 | 0.0029 | 32% (11) | 39% (75) | 0.73 | 0.4543 |
| C-SA | — (0) | 2% (1) | 0.00 | 1.0000 | — (0) | 16% (10) | 0.00 | 0.1071 | 11% (4) | 27% (53) | 0.34 | 0.0552 |
| O | — (0) | — (0) | Not present | 11% (2) | 1% (1) | 7.53 | 0.1268 | — (0) | 1.5% (3) | 0.00 | 1.0000 | |
Abbreviations: C-SA, C-South Africa; GP, general population alleles; HD, expanded HD alleles; n/a, not applicable; O, other; OR, odds ratio.
Number of alleles for each haplotype is indicated in brackets. P-value <0.05 represents significant difference in haplotype count between HD and control distributions within a subpopulation. Given significance, odds ratio indicates whether a given haplotype is more likely on HD alleles than control alleles (OR >1), equally likely (OR=1) or more likely on control alleles (OR <1). Bold text represents haplotype variants which significantly differ in frequency between HD and general population alleles in a given subpopulation.
HTT alleles are associated with different haplogroup variants in South African subpopulations
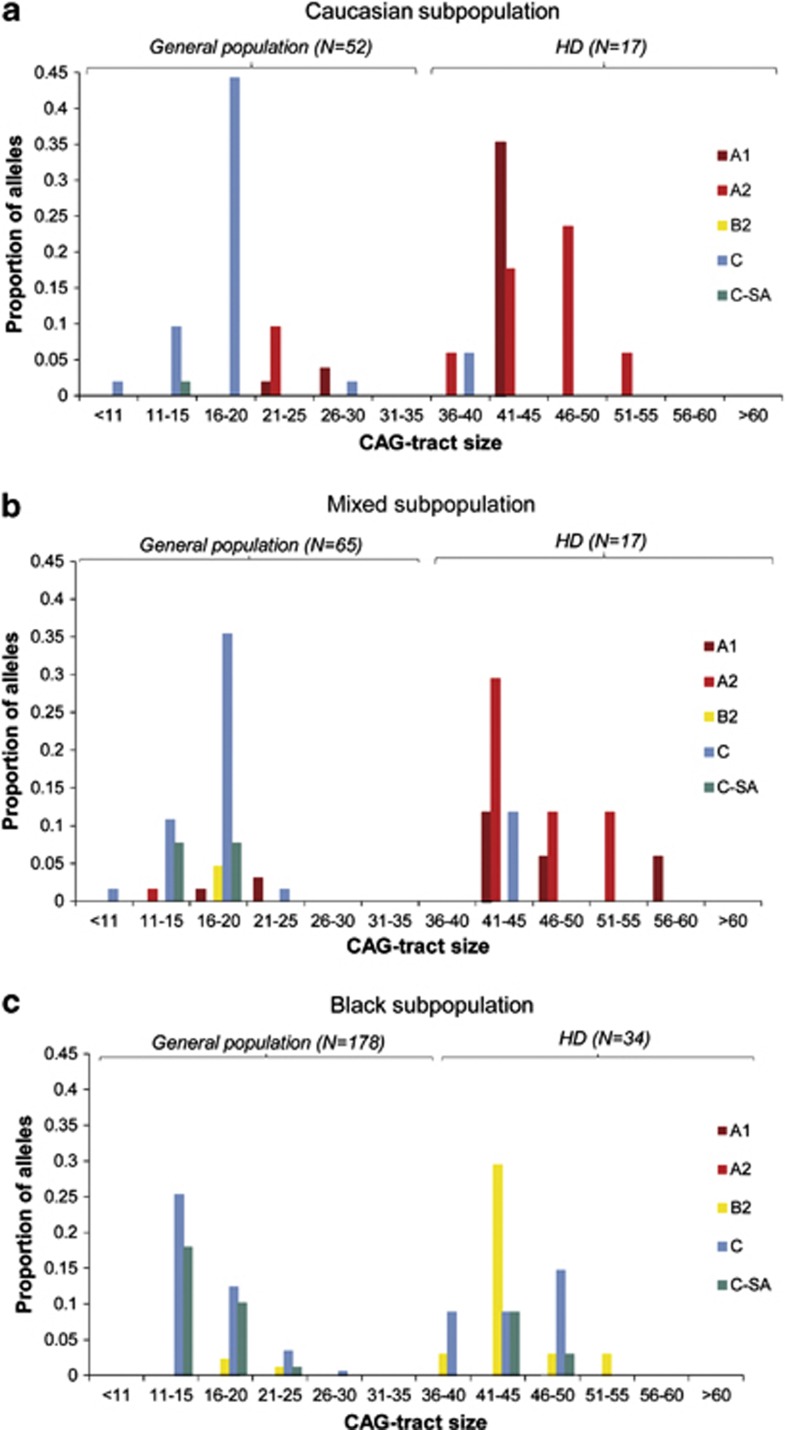
The South African population showed significant differences in the distribution of CAG-tract sizes across haplogroup variants (Figure 4). In the Caucasian subpopulation, variants A1 and A2 were associated with CAG-expanded HD alleles (CAG>35) (OR 10.39, P=0.0019 and OR 9.4, P=0.0007, respectively, Fisher's exact test) (Figure 4a, Table 1). Furthermore, HD alleles were unlikely to occur on haplogroup C (OR 0.04, P=8.09E-05, Fisher's exact test). Similarly, in the mixed subpopulation, variants A1 and A2 were associated with HD alleles (OR 5.51, P=0.0431 and OR 71.11, P=3.27E-07, respectively, Fisher's exact test) (Figure 4b, Table 1). In comparison with alleles from the general population, HD alleles were also less likely to occur on haplogroup C in this subpopulation (OR 0.12, P=0.0029, Fisher's exact test). The black subpopulation is markedly different, with HD alleles associated with the novel variant B2 (OR 12.27, P=2.06E-07, Fisher's exact test) (Figure 4c, Table 1). In addition, haplogroup C variants do not differ significantly between HD and general population alleles in the black subpopulation (Table 1).
Figure 4.
CAG-tract distributions of haplogroup variants. Only haplogroups with differences between HD and general population alleles are shown. (a) Caucasian subpopulation. Variants A1 and A2 are associated with HD alleles (CAG>35) (OR 10.39, P=0.0019 and OR 9.3, P=0.0007, respectively, Fisher's exact test). Haplogroup C is largely absent from HD alleles (OR 0.04, P=8.09E-05, Fisher's exact test). (b) Mixed subpopulation. Similar to the white subpopulation, variants A1 and A2 are associated with HD alleles (OR 5.51, P=0.0431 and OR 71.11, P=3.27E-07, respectively). Haplogroup C is uncommon on HD alleles versus those in the mixed general population (OR 0.12, P=0.0029, Fisher's exact test). (c) Black subpopulation. In contrast to Caucasian and mixed groups, HD alleles are associated with the novel variant B2 (OR 12.27, P=2.06E-07, Fisher's exact test). Novel South African C group, C-SA is uncommon on HD alleles versus the general population, although this difference does not reach significance (OR 0.34, P=0.0552, Fisher's exact test). Notably, other haplogroup C variants do not significantly differ between HD and general population alleles in the black subpopulation (OR 0.73, P=0.4543).
Discussion
In this study of HD alleles in the South African population, we report an association of the expanded CAG tract with specific haplotypes distinct in each subpopulation. The results may help to explain the differences in reported prevalence between the diverse groups that comprise the HD population in South Africa. A similar principle has been shown in European and East Asian populations.15 The South African population comprises largely Caucasian, ‘mixed' and black groups. The Caucasian subpopulation had its origins in European settlers from several countries (Holland, Germany, England and France) who arrived in South Africa around the 17th century; with on-going contributions from European migration.17, 20 The black subpopulation as used here refers to individuals belonging to different tribes of Bantu-speaking peoples from sub-Saharan Africa who settled in South Africa over 400 years ago. The term ‘mixed' refers to a dynamic multi-cultural group that arose from the admixture of European settlers, the Khoisan peoples and slaves from West Africa and East India.17, 20 The individuals included in this retrospective study self-identified with one of these three subpopulations.
HD has been reported in all three groups described here, albeit at very different rates; approximately 20–30 times higher in the Caucasian and mixed subpopulations than in black South Africans.17, 21, 22 It is important to note, however, that these figures are very likely underestimates as they are based on clinical ascertainment. A genealogical study performed by Hayden et al.23 traced the HD mutation through 14 generations of Caucasian Afrikaans-speaking families to a common ancestor of Dutch origin. A founder effect has also been reported in the South African population linking Caucasian Afrikaans-speaking and ‘mixed' (or mixed ancestry) families.24 The Caucasian families included in this study were not exclusively Afrikaans-speaking and were thus presumed unrelated. There has as yet been no investigation into the genetic background of HD in the black subpopulation in relation to the other groups.
In this study, the black subpopulation has a significantly lower average CAG-tract size in general population alleles when compared with the Caucasian subpopulation (Figure 1). No significant differences are evident in the size distribution of expanded HD alleles across the subpopulations (Figure 1a), and the results show that HD occurs on different haplotypes (Figure 2a). Notably, the diversity encountered required the definition of novel variants that have not been published elsewhere (Figures 2 and 3.) It is thus important that these population differences be investigated in appropriate detail, because this may shed light on the origin of HD in SA, provide an explanation for differences in HD prevalence rates and add to the knowledge of non-CAG genetic factors associated with HD expansion.
The results of this study indicate similarities between the Caucasian and mixed subpopulations, whereas the black subpopulation is markedly different. Similar to the European population, expanded CAG repeats in Caucasian and mixed South Africans are most likely to occur on haplogroup A variants A1 and A2 (Figures 2 and 4).16 The presence of the ‘high-risk' variants in both groups, and the reported founder effect linking the two,24 indicates the likelihood of a similar origin for the mutation in the Caucasian and mixed subpopulations in a small number of European ancestors as previously suggested.17, 23
In contrast, CAG-expanded alleles in black South Africans are most likely to occur on variant B2 with an equal proportion on variants of haplogroup C (Figure 2a). The unique distribution of CAG-expanded alleles in black South Africans suggests multiple local origins of the HD mutation in this group. The likelihood of several discrete origins is supported by the fact that HD has been previously reported in families representative of tribal groups with geographically distinct origins.25 In addition, none of the high-risk variants A1 and A2 were present in the black cohort making it improbable that the origin of HD in this subpopulation was European as had been suggested.23 The heterogeneous genetic background of HD alleles among black South Africans, in contrast to individuals of European ancestry (B2 and C versus A1 and A2) supports the notion that HD mutations have multiple origins specific to ethnically distinct populations.
Historically, HD was believed to have a very low prevalence in African peoples,2, 26 which may have led to a bias in diagnosing HD in black South Africans and therefore a low estimated prevalence.17 Several factors have continued to contribute to under-reporting of HD in this group including a lack of access to health-care services, a shorter average life-span, cultural beliefs that lead affected individuals to consult traditional healers rather than medical practioners and traditional medical training that HD is rare in black Africans.27, 28 The black South African families in the patient cohort were all from the Division of Human Genetics, NHLS/Wits in Johannesburg. It is noteworthy that there are no black South African HD families seen at the Division of Human Genetics, NHLS/UCT (unpublished observations). A systematic study is essential to ascertain the current prevalence of HD in South Africa, particularly in the black subpopulation. Further, the absence of ‘high-risk' variants (A1 and A2) found in European populations may account for proportionally lower HD prevalence in the black subpopulation relative to Caucasian and mixed groups, as has been previously suggested for East Asian populations.15
The prevalence of HD in any population is thought to be the result of a balance between the incidence of de novo mutations and the loss of HD alleles due to negative selection of very large CAG tracts that result in juvenile-onset HD.29 The new mutation rate has been estimated at approximately 10%.30, 31 Factors influencing the expansion of the CAG tract are therefore of crucial importance for determining HD prevalence rates. The size of the CAG repeat and the sex of the transmitting parent are known to have a significant role in CAG-tract instability.4, 32 Longer repeats have been shown to exhibit more instability and therefore are more likely to expand on transmission. Spermatogenesis is also believed to be involved in the molecular mechanism of CAG repeat instability in HD, given that expansion is more likely to occur during paternal transmission.33
Other factors proposed to affect the stability of HTT alleles include environmental factors and genetic cis-acting factors within certain haplotypes.5, 16, 34, 35, 36 Determining the genetic background of the HTT gene may thus lead to the identification of genetic factors affecting stability and disease pathogenesis. The similarities established here in the molecular genetic background of the Caucasian and mixed subpopulations (variants A1 and A2) are conceivable, given the history of settlement and human interaction in South Africa. Thus, it is not unexpected that the prevalence of HD in these two populations (∼2 per 100 000),17 even though likely underestimated, is markedly similar. The low estimated prevalence for the black subpopulation corresponds with the absence of ‘high-risk' haplotypes (A1 and A2) in the cohort investigated.
The differences in the genetic background of HD alleles from black South Africans compared with other populations (European and East Asian) suggest that different factors are likely responsible for CAG expansion in each population. Moreover, the occurrence of HD on each of the three major haplogroups among distinct ancestries argues that specific genetic motifs associated with each haplogroup lineage may not have a central role in mediating CAG repeat mutability and de novo HD mutation as has been previously suggested.15 Nevertheless, the markedly higher prevalence of HD among populations of European ancestry allows the possibility that CAG repeats on A1 and A2 haplotypes mutate at different rates due to novel cis elements associated with these two haplotype lineages. Experiments directly assessing CAG repeat instability on different haplotype backgrounds would be required to test this hypothesis.
An additional contribution to the difference in HD prevalence rates in the South African population may be the significant differences in mean CAG between the Caucasian and black subpopulations. The lower mean CAG-tract size and decreased prevalence of HD in black South Africans raises the possibility that these findings are related. The contribution of de novo mutations to HD prevalence is significant.30, 37 If de novo mutations arise mostly from alleles in the 27–35 range, the likelihood of alleles reaching that size may be greater in those close to that range. Indeed it is notable that in the general population, 21% (N=42) of Caucasian alleles have more than 20 repeats compared with only 8% (N=30) of black alleles (Figure 1b). These data are consistent with step-wise expansion of larger CAG alleles into the IA range, which over time may predispose to new mutations.16 We hypothesise that a lower new mutation rate in the black subpopulation, as exemplified by lower CAG repeat lengths, would be consistent with a lower prevalence rate of HD. Indeed, HTT allele size may provide an indication of mutation rate and prevalence rates of HD in other populations under investigation.
In conclusion, the data presented show novel and distinct haplotypes associated with HD in the different subpopulations in South Africa. The study further supports the need to identify population-specific therapies, when envisaging gene-silencing methodology targeting SNPs present on the expanded HTT allele.38, 39, 40 The subpopulations in this study emphasise the diversity that is present on the African continent. It is thus important to recognise that one population group cannot be used as a proxy for another, especially when these groups have different ethnic and geographical origins. The development of allele-specific gene silencing therapeutics will need to take into account the genetic backgrounds for HD in populations from different geographical regions around the world.
Acknowledgments
We thank especially the patients and their family members, without whom none of this work would be possible. Thanks to the clinical team at the Division of Human Genetics (UCT) and the Neurogenetics clinic, Groote Schuur Hospital. We thank the members of the Division of Human Genetics, NHLS/Wits for their support and in particular Ms T Wessels. We thank Ms D Smith and Dr L Watson for critical reading of the manuscript, along with Professor A Morris for discussion on aspects of the South African population. This work was supported by a Ripples of Hope Trainee Award (Global Health) and the University of Cape Town International Students' Scholarship to FKB; and the University of British Columbia Graduate Student International Research Mobility Award, the Princess Beatrix Fund, an Award for Excellent Students from the University of Groningen and the Huygens Scholarship for Talented Students from the Dutch government to MEK. The project was also supported in part by funds from the University Research Committee, University of Cape Town.
The authors declare no conflict of interest.
References
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Harper PS. The epidemiology of Huntington's disease. Hum Genet. 1992;89:365–376. doi: 10.1007/BF00194305. [DOI] [PubMed] [Google Scholar]
- Kremer B, Goldberg P, Andrew SE, et al. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. New Engl J Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- Ranen NG, Stine OC, Abbott MH, et al. Anticipation and instability of IT-I5 (CAG)N repeats in parent-offspring pairs with Huntington disease. Am J Hum Genet. 1995;57:593–602. [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol EK, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Myers RH, MacDonald ME, Koroshetz WJ, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington's disease. Nat Genet. 1993;5:168–173. doi: 10.1038/ng1093-168. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Kremer B, Andrew SE, et al. Molecular analysis of new mutations for Huntington's disease: intermediate alleles and sex of origin effects. Nat Genet. 1993;5:174–179. doi: 10.1038/ng1093-174. [DOI] [PubMed] [Google Scholar]
- Semaka A, Creighton S, Warby S, Hayden MR. Predictive testing for Huntington disease: interpretation and significance of intermediate alleles. Clin Genet. 2006;70:283–294. doi: 10.1111/j.1399-0004.2006.00668.x. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Wexler NS, Lorimer J, Porter J, et al. The US–Venezuela Collaborative Research Project and Wexler NS: Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Hayden MR, Almqvist EW, et al. A genome scan for modifiers of age at onset in Huntington disease: The HD MAPS study. Am J Hum Genet. 2003;73:682–687. doi: 10.1086/378133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hayden MR, Warby SC, et al. Genome-wide significance for a modifier of age at neurological onset in Huntington's disease at 6q23-24: the HD MAPS study. BMC Med Genet. 2006;7:71. doi: 10.1186/1471-2350-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayán J, Brocklebank D, Andresen JM, et al. Genomewide linkage scan reveals novel loci modifying age of onset of Huntington's disease in the Venezuelan HD kindreds. Genet Epidemiol. 2008;32:445–453. doi: 10.1002/gepi.20317. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntington's disease: the case for genetic modifiers. Genome Med. 2009;1:80. doi: 10.1186/gm80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Visser H, Collins JA, et al. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet. 2011;19:561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MR, MacGregor JM, Beighton PH. The prevalence of Huntington's chorea in South Africa. S Afr Med J. 1980;58:193–196. [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Theilmann J, Zeisier J, Hayden MR. A CCG repeat polymorphism adjacent to the CAG repeat in the Huntington disease gene: implications for diagnostic accuracy and predictive testing. Hum Mol Genet. 1994;3:65–67. doi: 10.1093/hmg/3.1.65. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins T. Medical genetics in South Africa. J Med Genet. 1990;27:760–779. doi: 10.1136/jmg.27.12.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MR. Huntington's chorea. New York: Springer; 1981. [Google Scholar]
- Greenberg LJ, Martell RW, Theilmann J, Hayden MR, Joubert J. Genetic linkage between Huntington disease and the D4S10 locus in South African families: further evidence against non-allelic heterogeneity. Hum Genet. 1991;87:701–708. doi: 10.1007/BF00201729. [DOI] [PubMed] [Google Scholar]
- Hayden MR, Hopkins HC, Macrae M, Beighton PH. The origin of Huntington's chorea in the Afrikaner population of South Africa. S Afr Med J. 1980;58:197–200. [PubMed] [Google Scholar]
- Scholefield J, Greenberg J. A common SNP haplotype provides molecular proof of a founder effect of Huntington disease linking two South African populations. Eur J Hum Genet. 2007;15:590–595. doi: 10.1038/sj.ejhg.5201796. [DOI] [PubMed] [Google Scholar]
- Silber E, Kromberg J, Temlett JA, Krause A, Saffer D. Huntington's disease confirmed by genetic testing in five African families. Mov Disord. 1998;13:726–730. doi: 10.1002/mds.870130420. [DOI] [PubMed] [Google Scholar]
- Folstein SE, Chase GA, Wahl WE, McDonnell AM, Folstein MF. Huntington disease in Maryland: clinical aspects of racial variation. Am J Hum Genet. 1987;41:168–179. [PMC free article] [PubMed] [Google Scholar]
- Futter MJ, Heckmann JM, Greenberg LJ. Predictive testing for Huntington disease in a developing country. Clin Genet. 2009;75:92–97. doi: 10.1111/j.1399-0004.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- Sizer EB, Haw T, Wessels T, Kromberg JGR, Krause A. The utilization and outcome of diagnostic, predictive, and prenatal genetic testing for Huntington disease in Johannesburg, South Africa. Genet Test Mol Biomarkers. 2011;16:1–6. doi: 10.1089/gtmb.2011.0007. [DOI] [PubMed] [Google Scholar]
- Rubinzstein DC, Amos W, Leggo J, et al. Mutational bias provides a model for the evolution of Huntington's disease and predicts a general increase in disease prevalence. Nat Genet. 1994;7:525–530. doi: 10.1038/ng0894-525. [DOI] [PubMed] [Google Scholar]
- Falush D, Almqvist EW, Brinkmann RR, Iwasa Y, Hayden MR. Measurement of mutational flow implies both a high new-mutation rate for Huntington disease and substantial underascertainment of late-onset cases. Am J Hum Genet. 2000;63:373–385. doi: 10.1086/318193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaka A, Collins JA, Hayden MR. Unstable familial transmissions of Huntington disease alleles with 27–35 CAG repeats (intermediate alleles) Am J Med Genet. 2010;153B:314–320. doi: 10.1002/ajmg.b.30970. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Sturrock A, Leavitt BR. The clinical and genetic features of Huntington disease. J Geriatr Psychiatry Neurol. 2010;23:243–259. doi: 10.1177/0891988710383573. [DOI] [PubMed] [Google Scholar]
- Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR. on behalf of an International Huntington's disease collaborative group: A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- Wheeler VC, Persichetti F, McNeil SM, et al. Factors associated with HD CAG repeat instability in Huntington disease. J Med Genet. 2007;44:695–701. doi: 10.1136/jmg.2007.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D. Haplotype background, repeat length evolution and Huntington disease. Am J Hum Genet. 2009;85:939–942. doi: 10.1016/j.ajhg.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almqvist EW, Elterman DS, MacLeod PM, Hayden MR. High incidence rate and absent family histories in one quarter of patients newly diagnosed with Huntington disease in British Columbia. Clin Genet. 2001;60:198–205. doi: 10.1034/j.1399-0004.2001.600305.x. [DOI] [PubMed] [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C, et al. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, et al. Five siRNAs targeting three SNPs in Huntingtin may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JB, Warby SC, Southwell AL, et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]