Abstract
Stargardt disease is an ABCA4-associated retinopathy, which generally follows an autosomal recessive inheritance pattern and is a frequent cause of macular degeneration in childhood. ABCA4 displays significant allelic heterogeneity whereby different mutations can cause retinal diseases with varying severity and age of onset. A genotype–phenotype model has been proposed linking ABCA4 mutations, purported ABCA4 functional protein activity and severity of disease, as measured by degree of visual loss and the age of onset. It has, however, been difficult to verify this model statistically in observational studies, as the number of individuals sharing any particular mutation combination is typically low. Seven founder mutations have been identified in a large number of Caucasian Afrikaner patients in South Africa, making it possible to test the genotype–phenotype model. A generalised linear model was developed to predict and assess the relative pathogenic contribution of the seven mutations to the age of onset of Stargardt disease. It is shown that the pathogenicity of an individual mutation can differ significantly depending on the genetic context in which it occurs. The results reported here may be used to identify suitable candidates for inclusion in clinical trials, as well as guide the genetic counselling of affected individuals and families.
Keywords: ABCA4, age of onset, generalised linear model, genotype–phenotype, Stargardt disease
Introduction
Stargardt disease (STGD) (OMIM: 248200) is a juvenile onset form of macular dystrophy, characterised by central vision loss due to the degeneration of cone photoreceptor cells in the macula.1, 2 The disease can follow both autosomal recessive (ar) and autosomal dominant (ad) inheritance patterns with symptoms usually presenting within the first two decades of life.3, 4, 5, 6 The incidence of STGD is estimated to be 1 in 10 000 individuals, and it accounts for ∼ 7% of all retinal disease.7
Mutations in the ATP-binding cassette subfamily A group 4 (ABCA4) gene give rise to the recessive form of STGD, but are also associated with other retinal degenerative disorders such as cone-rod dystrophy (CRD) and autosomal recessive retinitis pigmentosa (arRP),2, 8, 9 thereby characterising these dystrophies as ABCA4-associated retinopathies (AARs).3, 10, 11, 12 It is proposed that malfunctioning ABCA4 protein prevents complete removal of retinoid products from the outer segment disc membrane of photoreceptors, resulting in the accumulation of downstream derivatives such as di-retinoid-ethanolamine in the retinal pigment epithelium (RPE). Such products can trigger RPE dysfunction through various mechanisms, consequently leading to degeneration of photoreceptors and ultimately vision loss.3, 13
A high level of allelic heterogeneity is displayed in arSTGD, as over 600 mutations in ABCA4 have been identified.14 The vast number of identified mutations in ABCA4, their respective effects on the protein and the combination of mutations within a particular individual are largely accountable for the phenotypic heterogeneity in arSTGD patients.15
The considerable amount of both allelic and phenotypic heterogeneity associated with ABCA4 mutations has led to the proposal of a genotype–phenotype model, which attempts to define the relationship between different ABCA4 mutations and severity of the AAR phenotype.16, 17 The model suggests that phenotype may be predicted by ABCA4 mutations depending on the functionality of the remaining ABCA4 protein. Thus, the lower amount of functional ABCA4 activity, the more severe the AAR. In 2006, Valverde et al18 hypothesised that both the nature and the combination of the ABCA4 mutations have a role in the age of onset (AOO) of the AAR, and the earlier the AOO, the more severe the disease. Within the context of a single AAR, a similar rationale could be applied whereby different mutations in ABCA4 could give rise to varying phenotypes in patients with STGD.
The relative pathogenic contribution of individual alleles has been difficult to assess, since STGD is a highly heterogeneous disease and therefore, few individuals have been reported to share the same biallelic combination of mutations. This study considers a South African cohort of 118 individuals who express biallelic combinations of seven founder mutations in ABCA4 (c.454C>T (p.Arg152*), c.768G>T (p.Val256Val), c.1804C>T (p.Arg602Trp), c.2588G>C (p.Gly863Ala), c.4469G>A (p.Cys1490Tyr), c.5461–10T>C, c.6079C>T (p.Leu2027Phe)), which collectively account for 36% of STGD cases studied to date in South Africa.19, 20 These data, together with clinical information for each patient, were used to test the genotype–phenotype model. A generalised linear model (GLM) was developed to predict the AOO of patients with a biallelic combination of the seven common mutations and to assess their relative pathogenic contribution.
Materials and methods
The cohort comprised 118 individuals from 106 Caucasian Afrikaner families, each carrying two of the common seven ABCA4 (RefSeq NG_009073.1) mutations and for whom a self-reported AOO was available. The seven mutations screened in this study were not novel, and were all listed in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) at the Institute of Medical Genetics in Cardiff (except for c.768G>T (p.Val256Val)) and the Retina International ABCA4 Mutation Database (http://www.retina-international.org/files/sci-news/abcrmut.htm). AOO was used, as this was the only common phenotypic information available for all patients within this large cohort. Informed consent was obtained from all individuals according to the tenets of the Declaration of Helsinki (2008), and this project obtained approval from the UCT Research Ethics Committee (UCT URC REC REF 186/2009).
Screening results for common ABCA4 mutations were previously established for 86 patients using the ABCR400 microarray chip (Asper Ophthalmics, Tartu, Estonia), while the results for the remaining 32 individuals were obtained by screening only for the seven common ABCA4 mutations. Reproducible, sensitive assays had been designed to detect the seven ABCA4 mutations at the genomic level, using a multiplex SNaPshot reaction, allele-specific PCR assays, and denaturing high-performance liquid chromatography, and were described previously.19 Segregation analysis was performed wherever additional family members were available to ensure alleles were in-trans and not in-cis.19
The non-parametric Kruskal–Wallis test was used initially to determine whether there were significant differences in the median AOO between biallelic mutation combinations with five or more observations. AOO was then modelled as a function of the mutations and mutation pairs using a gamma GLM with an inverse link function.21 Binary indicators for each of the seven mutations, as well as interaction terms corresponding to mutation combinations observed in at least five subjects, were included as predictors in the model. The best model was then selected using a backward stepwise procedure based on the Akaike information criterion and by performing F tests of the model deviance when additional predictors were added or removed. The deviance residuals were found to be normally distributed using the Shapiro–Wilk test (P=0.961). The robustness of the final model was assessed by repeating the model selection procedure on 5000 bootstrap replicate samples. All statistical analyses were performed in the R language and environment for statistical computing.22
Results
Table 1 shows the 23 mutation combinations observed in this cohort, out of a potential 28 different combinations of the seven mutations, with the number of patients, the average AOO, and median AOO for each group. The c.4469G>A (p.Cys1490Tyr) mutation, in combination with either itself or another mutation, was the most common, as it was detected in 64 of the 118 patient samples (54%). The median AOO differed significantly between mutation combinations (P=0.003).
Table 1. A summary of the 23 mutation combinations observed in 118 patients with STGD, showing the number of patients per combination and the average and median AOO (in years) per combination.
| Mutation 1 | Mutation 2 | Number of patients | Average AOO (years) | Median AOO (years) |
|---|---|---|---|---|
| c.5461–10T>C | c.768G>T (p.Val256Val) | 1 | 6 | 6 |
| c.5461–10T>C | c.5461–10T>C | 2 | 6.5 | 6.5 |
| c.1804C>T (p.Arg602Trp) | c.768G>T (p.Val256Val) | 3 | 6.7 | 6 |
| c.5461–10T>C | c.454C>T (p.Arg152*) | 3 | 7 | 8 |
| c.4469G>A (p.Cys1490Tyr) | c.454C>T (p.Arg152*) | 6 | 7.8 | 8 |
| c.768G>T (p.Val256Val) | c.454C>T (p.Arg152*) | 4 | 8 | 9 |
| c.768G>T (p.Val256Val) | c.768G>T (p.Val256Val) | 1 | 8 | 8 |
| c.4469G>A (p.Cys1490Tyr) | c.4469G>A (p.Cys1490Tyr) | 9 | 8.1 | 8 |
| c.6079C>T (p.Leu2027Phe) | c.454 C>T (p.Arg152*) | 7 | 8.2 | 7 |
| c.4469G>A (p.Cys1490Tyr) | c.1804C>T (p.Arg602Trp) | 13 | 8.6 | 8 |
| c.4469G>A (p.Cys1490Tyr) | c.5461–10T>C | 13 | 8.8 | 9 |
| c.4469G>A (p.Cys1490Tyr) | c.768G>T (p.Val256Val) | 10 | 10.3 | 9 |
| c.4469G>A (p.Cys1490Tyr) | c.6079C>T (p.Leu2027Phe) | 12 | 10.3 | 9.5 |
| c.6079C>T (p.Leu2027Phe) | c.5461–10T>C | 3 | 11 | 10 |
| c.1804C>T (p.Arg602Trp) | c.5461–10T>C | 1 | 11 | 11 |
| (c.1804C>T (p.Arg602Trp) | c.6079C>T (p.Leu2027Phe) | 8 | 12 | 11 |
| c.6079C>T (p.Leu2027Phe) | c.768G>T (p.Val256Val) | 6 | 12.5 | 13 |
| c.768G>T (p.Val256Val) | c.2588G>C (p.Gly863Ala) | 3 | 16.7 | 18 |
| c.1804C>T (p.Arg602Trp) | c.2588G>C (p.Gly863Ala) | 2 | 17.5 | 17.5 |
| c.4469G>A (p.Cys1490Tyr) | c.2588G>C (p.Gly863Ala) | 4 | 18.5 | 19 |
| c.5461–10T>C | c.2588G>C (p.Gly863Ala) | 1 | 20 | 20 |
| c.6079C>T (p.Leu2027Phe) | c.6079C>T (p.Leu2027Phe) | 2 | 28 | 28 |
| c.2588G>C (p.Gly863Ala) | c.454C>T (p.Arg152*) | 4 | 28 | 30 |
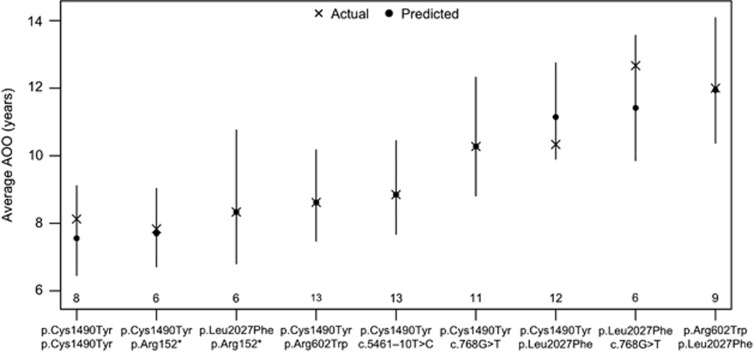
The GLM presented in Table 2 was used to examine the effects of these mutations on the average AOO. To illustrate the utility of this model, consider the population of individuals expressing the mutation pair c.4469G>A (p.Cys1490Tyr) and c.1804C>T (p.Arg602Trp). The average AOO for this group of individuals is predicted as (0.0460+0.0528+0.0468−0.0295)−1=8.61 years, where 0.0460 is the intercept term, 0.0528 is the regression coefficient for c.4469G>A (p.Cys1490Tyr), 0.0468 is the regression coefficient for c.1804C>T (p.Arg602Trp) and −0.0295 is the regression coefficient for c.4469G>A (p.Cys1490Tyr): c.1804C>T (p.Arg602Trp). The predicted averages and 95% confidence intervals for the other mutation pairs with five or more observations are illustrated in Figure 1.
Table 2. Covariates included in the generalised linear model (with inverse link function) with their respective coefficients, standard errors and P-values.
| Covariate | Coefficient | Standard error | P-value |
|---|---|---|---|
| (Intercept) | 0.0460 | 0.0106 | 3.53e−05 |
| c.4469G>A (p.Cys1490Tyr) | 0.0528 | 0.0076 | 2.71e−10 |
| c.1804C>T (p.Arg602Trp) | 0.0468 | 0.0085 | 2.33e−07 |
| c.6079C>T (p.Leu2027Phe) | −0.0090 | 0.0089 | 0.3117 |
| c.5461–10T>C | 0.0562 | 0.0106 | 6.59e−07 |
| c.768G>T (p.Val256Val) | 0.0507 | 0.0083 | 2.06e−08 |
| c.2588G>C (p.Gly863Ala) | −0.0413 | 0.0081 | 1.58e−06 |
| c.454C>T (p.Arg152*) | 0.0311 | 0.0080 | 0.0002 |
| c.4469G>A (p.Cys1490Tyr) (homozygous) | 0.0336 | 0.0144 | 0.0214 |
| c.4469G>A (p.Cys1490Tyr): c.5461–10T>C | −0.0419 | 0.0146 | 0.0049 |
| c.4469G>A (p.Cys1490Tyr): c.1804C>T (p.Arg602Trp) | −0.0295 | 0.0145 | 0.0442 |
| c.4469G>A (p.Cys1490Tyr): c.768G>T (p.Val256Val) | −0.0520 | 0.0134 | 0.0002 |
| c.6079C>T (p.Leu2027Phe): c.454C>T (p.Arg152*) | 0.0519 | 0.0158 | 0.0013 |
Figure 1.
Graph depicting the actual and predicted average AOO with 95% confidence bands for each mutation combination observed in five or more patients. Statistically significant differences in the AOO between mutation combinations are identified by non-overlapping confidence intervals. The numbers of subjects with each mutation combination are indicated above the horizontal axis.
In some cases, the average AOO of a given mutation differed significantly depending on the mutational context in which it occurred. Statistically significant differences in the AOO between mutation combinations are identified by non-overlapping confidence intervals. For example, the average AOO is significantly lower among individuals who are homozygous for the Cys1490Tyr mutation compared to individuals who express a single Cys1490Tyr mutation and the Leu2027Phe mutation.
Discussion
Significant allelic and phenotypic heterogeneity associated with the ABCA4 gene has led to the proposal of a genotype–phenotype model. Numerous studies have attempted to define the relationship between different ABCA4 mutations and the severity of the AAR. In 1998, Van Driel et al stated that ABCA4 mutations could be classified according to severity, and phenotype would vary depending on the total remaining ABCA4 protein activity/functionality.17 Alleles which result in a non-functional protein have been associated with a more severe phenotype such as arRP, whereas alleles resulting in partial ABCA4 protein activity have been associated with milder phenotypes such as CRD and STGD.
The size of the current cohort of fairly well-phenotyped STGD patients in whom two mutations were identified in the ABCA4 gene permitted statistical analysis to further investigate the proposed genotype–phenotype model within the context of this disorder. As most patients in this study are of Caucasian Afrikaner ancestry and have a biallelic combination of founder mutations, they are likely to share similar genetic backgrounds. Therefore, any phenotypic changes or trends observed are most likely attributable to the mutations of interest and not to other variations in the genome.
An aim of this study was to assess the relative pathogenicity of individual disease-causing alleles associated with STGD. This issue was recently explored by Schindler et al,23 who ranked alleles based on the coefficients of a linear regression model for visual field and visual acuity. In doing so, the authors implicitly assumed that the pathogenicity of a biallelic mutation combination is the sum of the pathogenicities associated with each of the individual mutations.23 This need not be the case; the transmembrane domains of the ABCA4 protein form a channel, which regulate passing of retinoid substrates and ligands. This channel is formed by a heterodimer whereby the components of the dimer are different from each other.24, 25 This implies that a particular mutation could have different effects depending on the other allele and mutation pathogenicity will vary subject to the domain, the dimer component and the interaction effects of the two mutations.
Indeed, our findings indicate that mutations do interact to alter the overall pathogenicity of the mutation combination. This is supported by the inclusion of interaction terms in the GLM that significantly improve the fit of the model to the data (P<0.0001). For example, consider again the population of individuals expressing the mutation combination c.4469G>A (p.Cys1490Tyr) and c.1804C>T (p.Arg602Trp). The average AOO for this group of individuals is predicted to be 8.61 years, as shown above, and is the same value as the actual average AOO depicted in Table 1. However, if the regression coefficient for c.4469G>A (p.Cys1490Tyr):c.1804C>T (p.Arg602Trp) (−0.0295) was not included, ie, (0.0460+0.0528+0.0468)−1=6.868, the predicted average AOO would be an underestimation of the true value.
The mutation combinations not detected in the cohort were c.454C>T (p.Arg152*) (homozygous), c.1804C>T (p.Arg602Trp) (homozygous), c.2588G>C (p.Gly863Ala) (homozygous), c.454C>T (p.Arg152*):c.1804C>T (p.Arg602Trp) and c.2588G>C (p.Gly863Ala): c.6079C>T (p.Leu2027Phe). There are three possible reasons why these mutation combinations were not identified. Firstly, the sample size could be too small to detect all 28 possible mutation combinations of the seven mutations and by chance, the above five combinations were absent from the patient cohort. Secondly, the above mutation combinations may not be sufficient to cause STGD as adequate functional protein may be produced even in the presence of the mutations. The c.2588G>C (p.Gly863Ala): c.6079C>T (p.Leu2027Phe) combination may be an example of this, as this combination has not yet been reported in the literature, to the best of our knowledge, and c.2588G>C (p.Gly863Ala) was previously proposed as a mild mutation.16 Lastly, and least likely, the mutation combination may be so severe that it leads to panretinal atrophy and therefore would not be identified in this STGD patient cohort. However, should these mutation combinations be identified in STGD patients in the future, this data could be used to update the GLM.
The four mutation combinations that did not produce significant interaction effects were removed from the GLM. For these cases, the effect on AOO from having two mutations in combination with each other did not differ significantly from the sum of the individual effects. The five mutation combinations which remained in the model, did significantly influence the AOO over and above the contribution of each of the single mutations on their own. However, only mutation combinations which had five or more observations were initially included. Data from additional patients with these mutation pairs should be included into the model as and when this information becomes available. It should also be noted that, since AOO is self-reported and the sample size within each mutation combination is small, the average AOO for any mutation combination would be fairly sensitive to misreported AOO values.
The GLM described here could be utilised to predict the expected AOO for a patient in whom a biallelic combination of these seven mutations is reported. This will be particularly useful in selecting candidates for inclusion in clinical trials for STGD, as monitoring could begin immediately prior to the expected AOO. Furthermore, this model could be employed to forecast the point at which gene therapy should be administered, as in many cases the disease has already progressed considerably when patients begin to experience symptoms.26 The model could aid in the testing of other ABCA4 mutations where phenotypic data exist. Finally, it has significant practical clinical utility in both selecting suitable candidates for inclusion in clinical trials as well as the counselling of individuals and families affected with this disorder.
Acknowledgments
This work was supported by Retina South Africa, the Medical Research Council of South Africa and the University of Cape Town. We are indebted to patients with STGD and family members who participated in our investigations.
The authors declare no conflict of interest.
References
- Stargardt K. Über familiäre, progressive degeneration in der maculagegend des auges. Albrecht von Graefes Arch Klin Exp Ophthalmol. 1909;71:534–550. [Google Scholar]
- Lewis RA, Shroyer NF, Singh N, et al. Genotype/phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet. 1999;64:422–434. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;17:122. doi: 10.1038/ng0997-122a. [DOI] [PubMed] [Google Scholar]
- Stone EM, Nichols BE, Kimura AE, Weingeist TA, Drack A, Sheffield VC. Clinical features of a Stargardt-like dominant progressive macular dystrophy with genetic linkage to chromosome 6q. Arch Ophthalmol. 1994;112:765–772. doi: 10.1001/archopht.1994.01090180063036. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kniazeva M, Han M, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Donoso L, Ritter R. A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest Ophthalmol Vis Sci. 2001;42:2652–2663. [PubMed] [Google Scholar]
- Blacharski P.Fundus flavimaculatusin Newsome DA (eds): Retinal Dystrophies and Degenerations New York: Raven Press; 1988135–159. [Google Scholar]
- Cremers FPM, Van de Pol DJR, Van Driel M, et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet. 1998;7:355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- Rozet JM, Gerber S, Ghazi I, et al. Mutations of the retinal specific ATP binding transporter gene (ABCR) in a single family segregating both autosomal recessive retinitis pigmentosa RP19 and Stargardt disease: evidence of clinical heterogeneity at this locus. J Med Genet. 1999;36:447–451. [PMC free article] [PubMed] [Google Scholar]
- Rivera A, White K, Stöhr H, et al. A comprehensive survey of sequence variation in the ABCA4 (ABCR) gene in Stargardt disease and age-related macular degeneration. Am J Hum Genet. 2000;67:800–813. doi: 10.1086/303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozet JM, Gerber S, Souied E, et al. Spectrum of ABCR gene mutations in autosomal recessive macular dystrophies. Eur J Hum Genet. 1998;6:291–295. doi: 10.1038/sj.ejhg.5200221. [DOI] [PubMed] [Google Scholar]
- Nasonkin I, Illing M, Koehler MR, Schmid M, Molday RS, Weber BHF. Mapping of the rod photoreceptor ABC transporter (ABCR) to 1p21—p22.1 and identification of novel mutations in Stargardt's disease. Nat Genet. 1998;236:21–26. doi: 10.1007/s004390050649. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Aleman TS, Swider M, et al. Mutations in ABCA4 result in accumulation of lipofuscin before slowing of the retinoid cycle: a reappraisal of the human disease sequence. Hum Mol Genet. 2004;13:525–534. doi: 10.1093/hmg/ddh048. [DOI] [PubMed] [Google Scholar]
- Zernant J, Schubert C, Im KM, et al. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci. 2011;52:8479–8487. doi: 10.1167/iovs.11-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevering BJ, Deutman AF, Maugeri A, Cremers FPM, Hoyng CB. The spectrum of retinal phenotypes caused by mutations in the ABCA4 gene. Graefes Arch Clin Exp Ophthalmol. 2005;243:90–100. doi: 10.1007/s00417-004-1079-4. [DOI] [PubMed] [Google Scholar]
- Maugeri A, van Driel MA, van de Pol DJR, et al. The 2588G—>C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet. 1999;64:1024–1035. doi: 10.1086/302323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driel MA, Maugeri A, Klevering BJ, Hoyng CB, Cremers FPM. ABCR unites what ophthalmologists divide(s) Ophthalmic Genet. 1998;19:117–122. doi: 10.1076/opge.19.3.117.2187. [DOI] [PubMed] [Google Scholar]
- Valverde D, Riveiro-Alvarez R, Bernal S, et al. Microarray-based mutation analysis of the ABCA4 gene in Spanish patients with Stargardt disease: evidence of a prevalent mutated allele. Mol Vis. 2006;12:902–908. [PubMed] [Google Scholar]
- Roberts LJ, Nossek CA, Greenberg LJ, Ramesar RS. Stargardt macular dystrophy: common ABCA4 mutations in South Africa-establishment of a rapid genetic test and relating risk to patients. Mol Vis. 2012;18:280–289. [PMC free article] [PubMed] [Google Scholar]
- September AV, Vorster AA, Ramesar RS, Greenberg LJ. Mutation spectrum and founder chromosomes for the ABCA4 gene in South African patients with Stargardt disease. Invest Ophthalmol Vis Sci. 2004;45:1705–1711. doi: 10.1167/iovs.03-1167. [DOI] [PubMed] [Google Scholar]
- Dobson A.An Introduction to Generalized Linear Models2nd edn.Boca Raton, FL, USA: Chapman & Hall/CRC; 2001 [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Schindler EI, Nylen EL, Ko AC, et al. Deducing the pathogenic contribution of recessive ABCA4 alleles in an outbred population. Hum Mol Genet. 2010;19:3693–3701. doi: 10.1093/hmg/ddq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meer G, Halter D, Sprong H, Somerharju P, Egmond MR. ABC lipid transporters: extruders, flippases, or flopless activators. FEBS Lett. 2006;580:1171–1177. doi: 10.1016/j.febslet.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Sun H, Nathans J. ABCR: rod photoreceptor-specific ABC transporter responsible for Stargardt disease. Methods Enzymol. 2000;315:879–897. doi: 10.1016/s0076-6879(00)15888-4. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV, Swider M, Aleman TS, et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum Mol Genet. 2009;18:931–941. doi: 10.1093/hmg/ddn421. [DOI] [PMC free article] [PubMed] [Google Scholar]